Replication and Expansion of Trinucleotide Repeats in Yeast (original) (raw)
Abstract
The mechanisms of trinucleotide repeat expansions, underlying more than a dozen hereditary neurological disorders, are yet to be understood. Here we looked at the replication of (CGG)n · (CCG)n and (CAG)n · (CTG)n repeats and their propensity to expand in Saccharomyces cerevisiae. Using electrophoretic analysis of replication intermediates, we found that (CGG)n · (CCG)n repeats significantly attenuate replication fork progression. Replication inhibition for this sequence becomes evident at as few as ∼10 repeats and reaches a maximal level at 30 to 40 repeats. This is the first direct demonstration of replication attenuation by a triplet repeat in a eukaryotic system in vivo. For (CAG)n · (CTG)n repeats, on the contrary, there is only a marginal replication inhibition even at 80 repeats. The propensity of trinucleotide repeats to expand was evaluated in a parallel genetic study. In wild-type cells, expansions of (CGG)25 · (CCG)25 and (CAG)25 · (CTG)25 repeat tracts occurred with similar low rates. A mutation in the large subunit of the replicative replication factor C complex (rfc1-1) increased the expansion rate for the (CGG)25 repeat ∼50-fold but had a much smaller effect on the expansion of the (CTG)25 repeat. These data show dramatic sequence-specific expansion effects due to a mutation in the lagging strand DNA synthesis machinery. Together, the results of this study suggest that expansions are likely to result when the replication fork attempts to escape from the stall site.
Trinucleotide repeats, specifically (CGG)n · (CCG)n, (CAG)n · (CTG)n, and (GAA)n · (TTC)n, have attracted wide attention since their expansion leads to numerous hereditary neurological disorders in humans, including fragile X syndrome, Huntington's disease, myotonic dystrophy, Friedreich's ataxia, etc. (reviewed in reference 49). The inheritance of these diseases is characterized by the so-called anticipation, i.e., an increase in the probability, onset, and the severity of a disease as it passes through generations. The molecular basis for anticipation is that trinucleotide repeats are stably inherited and cause no harm unless the number of repeats exceeds a threshold of roughly 25, upon which an intergenerational transmission of expanded versions of these repeats becomes progressively more common (reviewed in reference 1).
The mechanisms responsible for trinucleotide repeats expansion remain unclear. The largest volume of data supports an idea that abnormal replication of repeated stretches is responsible for their expansion. First, it is generally believed that the length dependence of expansion is linked to the ability of repeated DNAs to form unusual secondary structures, since the stability of such structures increases with repeats' lengths (reviewed in reference 27). Formation of these unusual DNA structures by trinucleotide repeats significantly compromises DNA polymerization in vitro (11, 21, 47). This polymerization blockage facilitates occasional misalignment between the newly synthesized and the template DNA strand, leading to repeat expansions in vitro (31).
Second, (CNG)n stretches, when cloned into bacterial plasmids, cause a length- and orientation-dependent replication blockage in vivo, and the lagging strand in the vicinity of the repeats is under-replicated (39).
Third, stabilities of trinucleotide repeats in bacterial, yeast, and cultured mammalian cells drastically depend on their orientation with regard to replication origins (2, 6, 9, 20, 28, 33, 43). Long (CTG)n or (CGG)n stretches situated in the lagging strand template preferably delete, while the same stretches in the leading strand template can expand, though with low probability. It was suggested, therefore, that formation of unusual secondary structures by trinucleotide repeats in either the lagging strand template or the newly synthesized lagging strand attenuates DNA replication, producing deletions or expansions, respectively.
Fourth, mutations in the replication apparatus of bacteria and yeast increase the instability of trinucleotide repeats and sometimes their expansion rates. These include deletions of the yeast FLAP endonuclease (rad27), missense mutations in the yeast DNA polymerase δ and PCNA, and proofreading mutants of Escherichia coli DNA polymerase III (8, 15, 16, 41, 42, 46, 50).
Finally, analyses of repeats expansion in families with fragile X syndrome indirectly support a replication model of expansion. In normal individuals, trinucleotide repeats are interrupted by several unrelated triplets. Expansions in carriers and affected individuals appear to occur at the 3′ rather than at the 5′ flank of CGG repeated stretch, and the expanded part lacks interrupting triplets (25). In yeast (37), interruptions were also shown to stabilize trinucleotide repeats, especially when the interruptions were located in the 5′ region of the (CTG)n · (CAG)n tracts. The rare expansions that occur also show additions 3′ of the interruption, although in yeast these expansions occur with retention of the interruption. These observations could be explained by erroneous replication of repeats from an upstream replication origin leading to expansions during the lagging strand synthesis.
While consistent with the replication hypothesis, none of these data decisively prove that expansions occur during DNA replication. Moreover, some cases of repeats expansion could not be explained by replication errors. The strongest argument comes from studies of (CAG)n repeats in transgenic mice showing that expansions occur in developing sperm that do not replicate (23). Alternative mechanisms could involve DNA recombination or gap repair. (CAG)n repeats were shown to stimulate homologous recombination in bacteria (17, 18) and gene conversion induced by double-stranded breaks in DNA in yeast (35) undergoing expansion during these events. Further, mutations affecting DNA repair increase the probability of repeats expansion (19, 32, 34, 40, 48). Note, however, that the majority of alternative models still includes some DNA synthesis through repeated stretches to account for the large-scale nature of expansions.
Thus, further studies are warranted to elucidate the link between DNA replication and trinucleotide repeats expansion. Here, we analyzed the effects of trinucleotide repeats on plasmid replication in yeast. Impediment of replication fork progression through (CGG)n · (CCG)n stretches was found to increase with longer repeat lengths. Replication stalling becomes detectable around 10 repeats, while maximum impediment is reached at about 30 to 40 repeats. Quantitative analysis of these data shows that long repetitive stretches slow replication down approximately twofold. For (CTG)n · (CAG)n repeats, on the contrary, we detected only marginal replication blockage for relatively long repeats (n, ≈80). We looked at the rate of the (CNG)25 repeats expansion in the same strain where the replication studies were carried out. The rates of expansion were ∼10−5 per cell/generation for both (CGG)25 and (CTG)25 repeats. Mutation in the subunit of the replication factor C (RFC) complex, which is vital for the lagging strand DNA synthesis, increases the expansion rate for the (CGG)25 repeat by almost 2 orders of magnitude but has a much smaller effect on the (CTG)25 repeat expansion. These data further emphasize the link between replication and expansion of (CGG)n · (CCG)n repeats: expansions are likely to result from errors of the lagging strand DNA synthesis when the replication fork attempts to escape from the stall site.
MATERIALS AND METHODS
Strains.
All cloning was carried out in the E. coli XL1-Blue strain (Stratagene). The majority of yeast replication studies were performed in Saccharomyces cerevisiae strain CH1585 (MATa leu2_Δ_1 trp_Δ_63 ura3-52 his3-200) (ATCC 96098). Strain VL6-48N, lacking the 2μm plasmid (30), was kindly provided by Vladimir Larionov.
Mutant construction.
The rfc1-1 mutant was constructed as follows. Complementary primers 1 (5′-CTGGCGGATATTACCTCTTGTA) and 2 (5′-TACAAGAGGTAATATCCGCCAG) contained point substitutions corresponding to the rfc1-1 mutation (underlined). Primer 1 was used in PCR of the 5′ portion of the rfc1 gene from yeast genomic DNA together with primer 3 (5′-AAAGAAGCAGAATTGCTTGTTA). Primer 2 was used in PCR of the 3′ portion of the gene together with primer 4 (5′-GAC_CTGCAG_AACGGCTCACAAAATCAA), which contained an extra _Pst_I site at its 5′ end (italicized). The two PCR products were mixed together, denatured, and reannealed. This was followed by PCR with primers 3 and 4, resulting in a DNA fragment containing rfc1 sequence with the required point substitution. It was then digested by _Hin_dIII (which cleaves internally to primer 3) and _Pst_I and cloned together with the ura3 cassette (an _Eco_RI-_Nsi_I fragment from the YEp24 plasmid) between the _Hin_dIII and _Eco_RI sites of the pUC19 polylinker. The resultant plasmid was linearized at the _Bsh_13651 site inside the rfc1-1 open reading frame (ORF), followed by pop-in into the CH1585 strain using URA+ selection. The pop-out was achieved by selecting for 5-fluoroorotic acid (5FOA) resistance. Mutant pop-outs were distinguished from the wild-type ones at the DNA level, since the rfc1-1 mutation destroys the _Eco_RV site within the gene, and by their cold-sensitive phenotype.
A similar PCR-based strategy was used to construct the pol32_Δ mutant. DNA sequence upstream of the pol32 initiation codon was amplified by PCR of yeast genomic DNA using primers 5 (5′-GTA_GAGCTCATTAATGGAAGTACCTTA) and 6 (5′-GATGATTCAGCTGGTTGTTA GTTAATG) (sequences homologous to yeast DNA are bolded). Primer 5 also contains a Sac_I restriction site (italicized). The 3′ portion of the pol32 gene was amplified by PCR using primers 7 (5′-TAACAACCAGCTGAATCATCGAAAATTA) and 8 (5′- GCC_TTTAAAAAAAAGAAGATTATTTTG) (sequences homologous to yeast DNA are in bold). Primer 8 also contains a _Dra_I site (italicized). Complementary 5′ portions of primers 6 and 7 (underlined) allowed us to reanneal the two PCR products. Subsequent PCR with primers 5 and 8 generated a deletion derivative of the pol32 gene lacking the entire 5′ portion of the ORF. Upon digestion with _Sac_I and _Dra_I, this fragment, together with the ura3 cassette (an _Eco_RI-_Sma_I fragment from the YEp24 plasmid), was cloned between the _Sac_I and _Eco_RI sites of the pBluescript SK II(−) polylinker. The resultant plasmid was linearized at the _Mlu_I site upstream of the pol32 ORF, followed by pop-in into the CH1585 strain via URA+ selection. The pop-out was selected for 5FOA resistance. The _pol32_Δ mutant contained a designed deletion, as verified by PCR, and its cold-sensitive phenotype.
Plasmids.
YEp24 plasmids with trinucleotide repeats of various lengths and sequences at their _Sma_I sites were obtained by cloning blunt-ended _Eco_RI-_Hin_dIII fragments from previously described pBluescript derivatives that carry trinucleotide repeats (39). The blunt-ended _Eco_RV-_Nsi_I fragment from the above YEp derivatives containing the (CGG)40 · (CCG)40 repeat was then recloned in two orientations into the _Ase_I site of YEp24, positioning the repeat into the nontranscribed area.
Isolation of replication intermediates from yeast cells.
Cells were grown in 400 ml of yeast extract-peptone-dextrose medium until reaching an optical density at 600 nm of 2.0. The growth was stopped by adding 4 ml of 10% NaN3 and incubating for 2 min. Eighty milliliters of frozen 0.2 M EDTA was added, cultures were pelleted, and pellets were washed with 50 ml of ice-cold water and resuspended in 4 ml of the NIB buffer (17% glycerol, 50 mM morpholinepropanesulfonic acid, 150 mM NaOAc, 2 mM MgCl2, 0.5 mM spermidine, 0.15 mM spermine; pH 7.2). An equal volume of glass beads was added, and cells were disrupted by vortexing for 7 min and chilling on ice for 30 s after every 30 s of vortexing. Beads were allowed to settle, and supernatant was pooled and cleared by centrifugation at 13,000 × g for 25 min. The pellet was resuspended in 1.5 ml of YSTE buffer (100 mM NaCl, 50 mM Tris-HCl [pH 8.0], 20 mM EDTA) and deproteinized for 1 h at 37°C by adding 0.225 ml of 10% sarcosyl and 30 μl of a 20-mg/ml proteinase K solution. Reaction mixtures were cleared by centrifugation at 12,000 × g for 5 min. DNA was further purified by overnight equilibrium centrifugation in CsCl gradient with 87 μg of Hoechst 33258 trihydrochloride/ml at 400,000 × g. The low band, corresponding to replicative intermediates, was collected, purified from the dye by butanol extraction, precipitated with ethanol, and dissolved in 30 μl of standard Tris-EDTA buffer.
Two-dimensional gel electrophoresis.
Replication intermediates of YEp derivatives with (CNG)n repeats in the _Sma_I site were digested with _Cla_I and _Pvu_II restriction enzymes, while those with repeats in the _Ase_I site were cleaved with _Ssp_I and _Pvu_II. This was followed by phenol-chloroform extraction and ethanol precipitation. The pellets were dissolved in 15 μl of loading buffer, loaded on the 0.4% agarose gel, and electrophoresed in the first dimension. The lanes were then cut out of the 0.4% gel and embedded into a 1.5% agarose gel with 0.3 μg of EtBr/ml for the electrophoresis in the second dimension (39). The gel was vacuum transferred onto a Nytran N membrane (Schleicher & Schuell) and hybridized with a 32P-labeled probe obtained with the Random Primers DNA labeling system (Invitrogen). For intermediates with repeats in the _Sma_I site, a 1.5-kbp _Sph_I-_Pvu_II fragment of YEp24 was used as a probe. The probe for intermediates with repeats in the _Ase_I site was a 631-bp _Ssp_I-_Ase_I fragment of YEp24. Quantitative analysis was performed on a Storm 860 PhosphorImager, using ImageQuant software (Molecular Dynamics).
Measurements of expansion rates and sizes.
Yeast strains were the parental CH1585 and its isogenic derivatives carrying either the rfc1-1 or _pol32_Δ mutations. Trinucleotide repeat-containing reporter plasmids (28) were integrated into each strain at either the URA3 locus on chromosome V or at the LYS2 locus on chromosome II. Southern blotting confirmed the placement of a single copy of the reporter at the desired location in every case. Expansions were identified by acquisition of resistance to 5FOA, as described in more detail elsewhere (28). Rates of instability at 30°C were measured by the method of the median (26). The fraction of bona fide expansions was determined by PCR amplification (for CTG repeat tracts), as judged by increased size of the products compared to the starting strain (46). Southern hybridization was used to score expanded CGG tracts. Briefly, 5FOAr colonies were grown in liquid cultures and their genomic DNA was prepared and digested with _Sph_I. The resulting products were run on a 6% denaturing polyacrylamide gel and transferred to a nylon membrane. The blot was hybridized to a probe containing (CGG)n · (CCG)n repeats. The size of the expansion was inferred by comparison to a 10-bp DNA ladder (Gibco BRL).
RESULTS
Effects of trinucleotide repeats on replication fork progression in yeast.
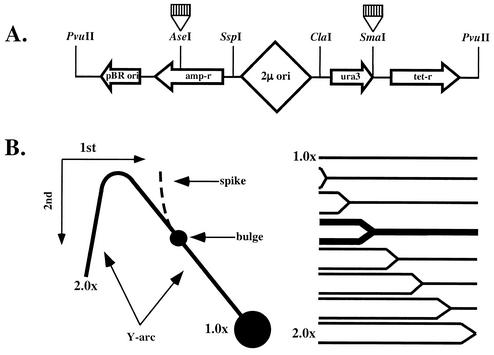
Using two-dimensional electrophoresis of replication intermediates, we have previously shown that (CNG)n repeats block replication fork progression in bacterial cells in a length- and orientation-dependent manner (39). Here we applied the same method for studying the effects of trinucleotide repeats on plasmid replication in yeast. (CNG)n repeats of various lengths were first cloned into the _Sma_I site of the yeast YEp24 plasmid (Fig. 1A), located in the 3′ untranslated region of the _ura_3 gene, 2.5 kbp downstream from the bidirectional replication ori. Due to the bidirectional character of the replication, this plasmid is separated into two domains replicated by different forks, where our inserts are in left-to-right replication fork, as depicted in Fig. 1A. Depending on a repeat's orientation, the location of each repeat can be assigned to the template for leading or lagging DNA strand synthesis for this fork. The names of our plasmids correspond to the repetitive sequence in the lagging strand template.
FIG. 1.
Electrophoretic analysis of replication intermediates in yeast. (A) Structure of the _Pvu_II-linearized YEp24 derivatives containing trinucleotide repeats. In this linear depiction, the 2μm replication origin is roughly in the middle of the plasmid. Trinucleotide repeats (striped boxes) were cloned into either _Sma_I or _Ase_I sites, being replicated by either left-to-right or right-to-left replication forks, respectively. (B) Schematic representation for the two-dimensional N/N electrophoretic analysis. Cleavage of replication intermediates with restriction enzymes _Cla_I and _Pvu_II generates Y-shaped DNA molecules, and the size of a Y increases with the replication progression. Replication blockage at a repeat leads to the accumulation of replication intermediates of a given size and shape (bold in the right panel). Upon separation by two-dimensional agarose electrophoresis, one can see a Y-arc. Partial replication blockage by a repeat should result in the appearance of a bulge on the replication arc. Complete replication blockage should lead to the appearance of a spike.
Upon isolating the replicating DNA and cleaving it with restriction enzymes _Cla_I and _Pvu_II, one might expect the appearance of Y-like structures (Fig. 1B). Y-structures differ from nonreplicated DNA in both their size and shape, which allows one to detect them by gel electrophoresis as a characteristic Y-arc (Fig. 1B). If replication is attenuated by a repeat, one should expect the appearance of a bulge on the otherwise smooth Y-arc due to the preferential accumulation of replication intermediates of a specific size and shape. A triplet repeat at the _Sma_I site is situated at approximately one-third of the distance from the end of a _Cla_I-_Pvu_II fragment relative to the replication fork entrance (Fig. 1A). One would expect, therefore, that the bulge would be located within the Y-arc's long shoulder (Fig. 1B). In the case of complete blockage, a second replication fork would approach a repeat from the opposite side, resulting in the appearance of double-Y intermediates, forming a spike on the Y-arc (reviewed in reference 10).
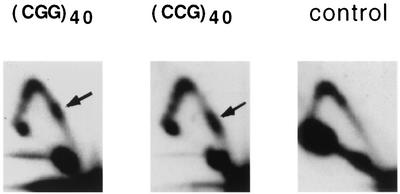
Our experimental data on the replication of plasmids with (CGG)n · (CCG)n inserts of various lengths are shown in Fig. 2A. The control plasmid, YEp24, has a relatively smooth Y-arc (Fig. 2A), except for a knob at its peak. Similar knobs are also evident in all other plasmids studied, while their intensity appeared to depend on variables (such as cooling time) during isolation of the replication intermediates (see also Fig. 3, below). Y-arcs of plasmids carrying (CGG)n · (CCG)n repeats, on the contrary, contain evident bulges in their long shoulders. These bulges were located roughly one-third of the distance from the end Y-arcs, i.e., they corresponded to the position of repeats within the analyzed restriction fragment. We do not see accumulation of spikes around these bulges, indicating that replication inhibition is not complete, even for the longest repeats studied. Note that for both control and repeat-containing plasmids, some spikes originate from the peak of the Y-arc. Those spikes, likely reflecting plasmid recombination products, were not particularly reproducible, varying in intensity between different experiments for the same plasmid.
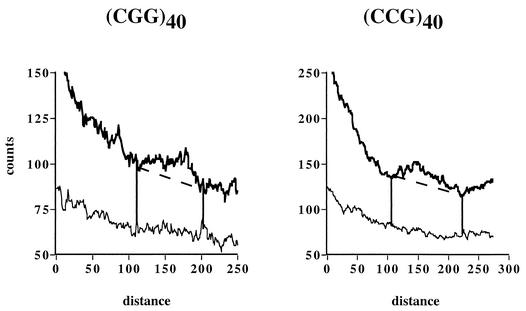
FIG. 2.
Replication of YEp24 derivatives carrying various (CGG)n · (CCG)n repeats at the _Sma_I site. (A) Electrophoretic data for plasmids (CGG)n and (CCG)n carrying corresponding repeated sequences in the lagging strand template. Arrows show replication stall sites. (B) Densitogram showing a peak on a portion of the Y-arc for the (CGG)40 plasmid. Thick line, Y-arc; thin line, background noise. The strength of replication stalling was determined as the ratio of radioactivity in the bulge area (areas 1 and 2) to that in the corresponding area of a smooth replication arc (area 2). (C) Quantitative analysis of the replication blockage for (CGG)n · (CCG)n repeats of various lengths and orientations from several experiments. Squares, (CGG)n plasmids; diamonds, (CCG)n plasmids.
FIG. 3.
Electrophoretic analysis of YEp24 derivatives carrying the (CGG)40 · (CCG)40 repeat in two orientations at the _Ase_I site. Arrows show replication stall sites.
To measure the extent of replication blockage caused by trinucleotide repeats, Y-arcs were analyzed using phosphorimaging. A typical densitogram, showing a peak on a portion of the arc, is presented in Fig. 2B. The ratio of radioactivity in the peak area (areas 1 and 2) to that in the corresponding area of a smooth replication arc (area 2) represents the extent of replication slowing. Quantitative data for (CGG)n · (CCG)n repeats of different lengths and orientations obtained from three to five independent experiments for each plasmid are presented in Fig. 2C. Replication slowdown is barely detectable at n = 8 (∼1.1-fold), sharply increases for 8 ≤ n ≤ 18, and reaches the saturation of ∼1.6-fold at about 40 repeats. This maximal replication inhibition could a priori be explained in two ways: (i) the yeast replication fork comes to a halt at a substantial fraction of plasmid molecules but manages to proceed through it smoothly in the remaining plasmid population, or (ii) the yeast replication fork slows down 1.6-fold at all repeats. The first scenario would suggest replication of a substantial fraction of repeats by an alternative replication fork, giving rise to spikes on Y-arcs. Since we do not observe those spikes, we believe that the replication fork uniformly slows down at (CGG)n · (CCG)n runs.
Our yeast results are similar to those previously obtained in bacterial systems, in that (CGG)n · (CCG)n repeats stall replication fork progression in vivo. There are, however, two notable differences between the two experimental systems: (i) much shorter repeats are sufficient to cause equivalent replication stalling in yeast compared to E. coli, and (ii) contrary to bacteria, the efficiency of replication blockage in yeast does not depend on the repeat's orientation relative to the replication ori (Fig. 2A, compare autoradiograms for plasmids with CGG or CCG repeats as lagging strand template).
The trinucleotide repeats described above were situated in the 3′ untranslated region of the ura3 gene, i.e., in the transcribed area. Our laboratories have previously shown that RNA polymerase can stall at some repetitive runs, which, in turn, attenuates replication (24). To rule out this possibility, (CGG)n · (CCG)n repeats were cloned into the _Ase_I site of the YEp24 plasmid situated in the bacterial bla gene, which is not transcribed in yeast (Fig. 1A). Triplet repeats here were situated in the right-to-left replication fork as shown in Fig. 1A. Replication intermediates were digested with the _Ssp_I and _Pvu_II restriction enzymes prior to two-dimensional electrophoresis. The location of the _Ase_I site within the _Ssp_I-_Pvu_II fragment relative to the replication fork entrance was similar to that of the _Sma_I site within the _Cla_I-_Pvu_II fragment described above. Thus, repeat-caused replication blockage should result in the appearance of a bulge in the long shoulder of the Y-arc as shown in Fig. 1B. Figure 3 shows that the (CGG)40 · (CCG)40 repeat at the _Ase_I site caused equally profound replication stops in both orientations. We conclude, therefore, that replication blockage at (CGG)n · (CCG)n repeats does not depend on their transcriptional status. We note that the position of the bulge differed slightly for the two orientations of the (CGG)40 · (CCG)40 repeat. This is an expected result, because the two orientations have slightly asymmetric flanking sequences that are acquired during recloning of the repeats (see Materials and Methods). From the replication fork direction, it was at either one-third or one-fourth of the restriction fragment for CGG or CCG orientations, respectively. The bulge is closer to 1.0×-size intermediates in (CCG)40 orientation than it is in (CGG)40 orientation, accordingly. These data, once again, emphasize the unambiguous association of triplet repeats and replication bulges.
The lack of orientation dependence in (CGG)_n_-caused attenuation of DNA replication clearly distinguishes our yeast data from previously described bacterial data. One possible explanation could lie in the peculiarities of the YEp24 plasmid maintenance in yeast cells. Stable maintenance of this plasmid depends on its integration into the resident 2μm plasmid carried out by the Flp recombinase (38). Depending on the directionality of this integration, a repeat from the same recombinant plasmid might get positioned in various orientations relative to the replication ori. This could obviously conceal orientation dependence in repeat-caused replication blockage. To address this possibility, we looked at the replication of (CGG)n repeats within the YEp24 plasmid in the yeast strain VL6-48N lacking the 2μm plasmid. The lack of a resident plasmid rules out YEp24 integration (30). Though YEp24-derived plasmids are highly unstable in this strain, the sensitivity of our assay appeared to be sufficient for detecting their replication intermediates. Quantitative analysis of replication intermediates of plasmids with the (CGG)40 repeat in both orientations relative to the ori clearly shows that replication slows down to a similar extent in both cases (Fig. 4). Thus, orientation-independent blockage of replication in yeast is intrinsic to (CGG)n repeats rather than due to fortuitous integration of YEp24-derived plasmids into the 2μm plasmid. Note that replication blockage here is less profound than that observed in the wild-type strain. One possible explanation could be that repeat-caused replication blockage is somewhat masked by the general inefficiency of YEp24 replication in strains lacking the 2μm plasmid. Supporting this, we observed a decrease in repeat-caused replication blockage upon growing the wild-type cells at lower temperatures where replication elongation is slow going (data not shown).
FIG. 4.
Densitograms showing replication blockage by the (CGG)40 · (CCG)40 repeat in two orientations in an S. cerevisiae strain lacking the resident 2μm plasmid. Thick line, Y-arc; thin line, background noise.
We further analyzed the effect of another expandable repeat, (CTG)n · (CAG)n, on the progression of the yeast replication fork. Only slight replication slowing for relatively long repeats, n ≈ 80, was observed (Fig. 5). Since long (CTG)n · (CAG)n repeats are known to undergo large contractions in yeast, we confirmed that, upon isolation, replication intermediates did contain the whole-size repeats in every case. Quantitative analysis of our data shows that, even for those repeats, the extent of stalling does not exceed 1.12-fold (data not shown). For (CGG)n repeats, the equivalent extent of replication inhibition was observed at n = 8. We conclude, therefore, that (CTG)n · (CAG)n repeats of premutation size have an insignificant effect on replication fork progression in yeast.
FIG. 5.
Electrophoretic analysis of YEp24-derived plasmids carrying the (CTG)80 · (CAG)80 repeat at the _Sma_I site. Names of plasmids reflect repetitive sequences in the lagging strand template.
Expansion of (CNG)n repeats in the wild type and replication mutants.
To study a relationship between replication attenuation at a repeat and its propensity to expand, we used our genetic system for monitoring repeat expansions in S. cerevisiae (28). Testing for expansions in a genetic assay in the same strain background allows direct correlations with the replication stalling results. This combinatorial approach allowed testing of the hypothesis that stalling of replication forks, or escape from stalling, promotes expansions.
Briefly, triplet repeat tracts were cloned as oligonucleotide cassettes into a reporter construct that includes the Schizosaccharomyces pombe adh1 promoter fused to a URA3 reporter gene. The reporter plasmid was then integrated into the ura3 chromosomal locus by homology-mediated recombination. The transcription initiation site in this reporter is dependent on the length of the triplet repeat. A starting tract of 25 repeats or less allows normal expression of the URA3 reporter gene with concomitant sensitivity of the cells to the cytotoxic drug 5FOA. Expansions of the tract to lengths of 30 or more repeats leads to an alternative transcription initiation upstream of the normal start site. An out-of-frame AUG codon at the very 5′ end of this alternative transcript precludes proper translation of the URA3 gene. Consequently, repeat expansions make cells resistant to 5FOA, and the expansion rate is proportional to the number of 5FOA-resistant colonies. The assay does not identify contractions or small expansions (of less than +5 repeats). Starting tracts of 25 repeats were chosen because this length falls within the region of maximal stalling (Fig. 2C) and because a large database of information is available from previous studies (36) for comparison with the present experiments.
We first looked at the rates of (CGG)n and (CTG)n repeats expansion in the yeast strain, CH1585, that we have used for our replication studies. This is a wild-type strain with regard to replication, since it does not contain any mutations in the replication apparatus. Cassettes containing either (CGG)25 or (CTG)25 repeats between the adh1 promoter and ura3 gene were integrated into the chromosomal ura3 gene. The resultant clones were URA+ and contained expected numbers of repeats, as verified by PCR analysis and/or Southern hybridization. The rate of appearance of 5FOA-resistant clones was determined using a simple fluctuation test, as described in Materials and Methods. The data in Table 1 show that, for both repeats, 5FOA-resistant clones emerge at rates of ∼10−5 per cell/generation. The genetic assay provided suitable sensitivity to detect that the CGG repeat tract is about fourfold less stable than the CTG allele.
TABLE 1.
Ratesa of trinucleotide repeat expansions in the wild-type and replication mutant strains of S. cerevisiae
| Repeat tract | Wild type rate | rfc1-1 rate (ratio to wtb) | _pol32_Δ rate (ratio to wt) |
|---|---|---|---|
| (CGG)25 | (7.2 ± 2.1) × 10−5 | (3.3 ± 1.4) × 10−3 (46) | (1.8 ± 0.4) × 10−4 (2.5) |
| (CTG)25 | (1.8 ± 0.4) × 10−5 | (4.6 ± 2.2) × 10−5 (2.6) | (2.1 ± 0.4) × 10−5 (1.2) |
To confirm that the yield of 5FOA-resistant clones reflects repeats expansion, the length of repeats in resistant clones was verified by either PCR (for CTG tracts) or Southern hybridization (for CGG tracts). Expanded CGG alleles (15 tested in all) had acquired between 4 and 18 (±3) extra repeats. Similarly, expanded CTG alleles (10 tested in all) contained between 9 and 17 (±2) added repeats.
As described in the previous section, (CGG)n repeats strongly attenuate replication fork progression, while (CTG)n repeats are far less potent. Yet the expansion rates for both repeats show a more modest difference (Table 1). This leads us to two conclusions. First, there is no obvious correlation between the strength of the replication blockage by a repeat and its propensity to expand. Second, obstruction of replication per se is not sufficient for the expansion. What remains possible is that expansions occur by accident while the replication fork attempts to escape from the stall site. Such attempts could impair the leading and lagging strand syntheses.
We, therefore, looked at the effects that replication components involved in the coordination of the leading and lagging stand synthesis have on trinucleotide repeats expansion. Using a standard pop-in, pop-out strategy, we introduced two mutations that potentially impair the leading-lagging strand equilibrium into our CH1585 strain. The cold-sensitive rfc1-1 mutation changes the ATPase activity of the large subunit of the RFC complex (4). This complex is required for loading PCNA and, subsequently, the lagging strand polymerase onto primers for Okazaki fragments (29). Since coordination of the leading and lagging strand syntheses depends on timely translocation of the lagging strand polymerase from a completed Okazaki fragment to the next Okazaki primer, this complex is vital for the coordination of DNA strand syntheses. This mutation was also shown to affect the stability of simple DNA repeats in yeast: it leads to a 10-fold-increased level of (GT)n repeat insertions (51) and substantial elongation of telomeric repeats (4).
Another cold-sensitive mutation, _pol32_Δ, deletes a small subunit of the replicative polymerase δ (12). This subunit affects polymerase processivity and is believed to play a role in the coupling of leading and lagging polymerases at the replication fork (5). At the permissive temperature, both mutations decrease the replication rate and, consequently, increase doubling time of S. cerevisiae cells in culture.
Table 1 shows the effects of these mutations on the rate of occurrence of 5FOA-resistant clones for cassettes carrying (CGG)25 and (CTG)25 repeats at the permissive temperature. The most striking results were observed for the rfc1-1 mutant. Compared to the wild type, this mutation elevates the expansion rate for the (CGG)25 tract approximately 50-fold, without changing the (CTG)25 expansion rate appreciably. The mutation _pol32_Δ, on the contrary, does not significantly affect the expansion rate for any of the trinucleotide repeats studied. In rfc1-1, PCR and Southern hybridization analysis was again used to confirm that 5FOA-resistant clones resulted from repeat expansions. Similarly to the wild-type strain, expanded CGG alleles (10 tested in all) had acquired between 5 and 17 (±3) extra repeats, while expanded CTG alleles (29 tested in all) contained between 10 and 23 (±2) extra repeats.
These data show, for the first time, that a mutation in the apparatus of the lagging strand DNA synthesis can increase the expansion rate of a trinucleotide repeat in such a dramatic sequence-specific manner. This was never previously observed for any replication mutation studied. Tellingly, the affected repeat, (CGG)n, is the one that strongly attenuates replication fork progression.
In light of these data, it seemed reasonable to analyze replication of (CGG)n · (CCG)n repeats in the rfc1-1 mutant. Even at permissive temperature, much fewer replication intermediates could be isolated from this mutant compared to the wild-type strain, making the sensitivity of our assay barely sufficient to detect replication arcs (Fig. 6). Yet one can still see bulges on Y-arcs, reflecting replication stalling caused by the (CGG)40 · (CCG)40 repeats in both orientations. Quantitative analysis of these data showed that replication blockage is only slightly (∼10%) less potent than that in the wild-type cells, but this difference is not statistically significant. Thus, the rfc1-1 mutation does not drastically change the characteristics of replication fork progression through (CGG)n repeats.
FIG. 6.
Effects of the rfc1-1 mutation on replication blockage caused by the (CGG)40 · (CCG)40 repeat. Arrows show replication stall sites.
DISCUSSION
Using electrophoretic analysis of replication intermediates, we have found that (CGG)n · (CCG)n repeats stall replication fork progression in S. cerevisiae. This is the first direct demonstration of replication attenuation by a triplet repeat in a eukaryotic system. (CTG)n · (CAG)n repeats appeared to be far less potent replication blockers, though slight inhibitory effects were still detected for relatively long repeats. This seems to be in agreement with our previous data obtained in bacterial cells, where (CGG)n · (CCG)n repeats attenuated replication much more strongly than (CTG)n · (CAG)n runs (39).
What distinguishes the yeast data from bacterial data is that the inhibitory effect of the (CGG)n · (CCG)n stretches reaches a maximal level at a much smaller number of repeats and does not depend on their orientation relative to the replication ori. While we do not know exact reasons for these differences, some hypotheses can be put forward.
The observation that shorter-length (CGG)n repeats were required to cause replication blockage in S. cerevisiae compared to E. coli could be explained by the fact that the yeast genome is much more AT rich than the bacterial one. It is possible, therefore, that yeast replication machinery is not well suited to unravel stable GC-rich DNA structures, resulting in considerable impediments from relatively short hairpins or G-quartets in the lagging strand template.
Orientation dependence in bacteria was previously explained by higher stability of unusual DNA structures formed in a (CGG)_n_-containing lagging strand template compared to the (CCG)_n_-containing template (39). The lack of such orientation dependence in yeast, at first glance, contradicts this model. This contradiction, however, might be superficial. Differential stabilities of (CGG)_n_- and (CCG)_n_-containing DNA structures are grounded in the fact that G-G pairs are quite stable at a wide range of pHs, while the stability of C-C pairs is pH dependent. Intracellular pH in E. coli is around 7.5 (45), i.e., precluding C-C pairs formation, while the more acidic (pH ∼6.0 to 6.5) environment in S. cerevisiae (13, 14) could make C-C pairs feasible.
Whatever the exact mechanism, expandable trinucleotide repeats, and particularly (CGG)n · (CCG)n, cause substantial replication stalling. Is there a link between replication blockage caused by these repeats and their expansion potential?
We believe that an argument in favor of such a link could be made for (CGG)n · (CCG)n repeats. These repeats cause potent replication inhibition in every system studied. A mutation in the lagging DNA strand synthesis apparatus, rfc1-1, drastically increases the expansion rate for these repeats, but not for (CTG)n · (CAG)n repeats that do not cause strong replication inhibition. Replication stalling occurs in nearly all replication events, but that expansion arises in only about 1 in 105. To explain these data altogether, we hypothesize that replication stalling does not automatically lead to repeat expansions; they rather result from mistakes during the lagging strand synthesis while the replication fork attempts to escape from the stall site.
Besides the lagging strand DNA synthesis, replication factor C is also involved in DNA repair and recombination (3, 22, 44). It is foreseeable, therefore, that the selective effects of the rfc1-1 mutation on CGG repeat expansions might be due to RFC involvement in repair and recombination that is required to restart stalled replication forks.
The sequence-specific enhancement of repeat expansion by the rfc1-1 mutation raises some stimulating biological questions. It was previously shown that telomeric repeats are elongated in the rfc1-1 mutant (4). Could this point to certain similarities in length maintenance of endogenous G2-3(TG)1-6 telomeric repeats and exogenous (CGG)n repeats in yeast? Another intriguing question is whether potent expansion of (CGG)n repeats in fragile X families could be accentuated by silent mutations in the replication apparatus, such as rfc1-1. Since the human homologue of the rfc1 gene has been cloned (7), it would be of interest to analyze its status in those families.
The results with the rfc1-1 mutant provide an interesting contrast with previous results with _rad27_Δ mutants (8, 42, 46). In the rfc1-1 strain tested here, the expansion rate for (CGG)n repeats was increased ∼50-fold, whereas there was a much smaller effect on (CTG)n repeat expansion. Furthermore, the sizes of (CGG)n expansions were within twofold of the original tract size. In contrast, a previous study (46) showed that a _rad27_Δ mutant accelerates the rate of expansions for at least four triplet repeats (CTG, GAC, CAG, and CTA) by roughly the same extent, so _rad27_Δ effects are not sequence specific. Also in _rad27_Δ cells, some very large expansions, up to five times the original tract size, were observed (46). The differences between rfc1-1 and _rad27_Δ mutants suggests that expansions can occur through different mechanisms which can be distinguished by the two mutations.
The situation with the replication and expansion link for (CTG)n · (CAG)n repeats is less clear. Only relatively long repeats inhibit replication in bacteria and yeast. Even for those repeats, inhibition is weak, except for special circumstances, such as treatment of bacterial cells with chloramphenicol (39). Further, our yeast data show that certain mutations affecting the lagging strand synthesis have only a small effect on expansion rates for these repeats. At the same time, a previous study (28) concluded that replication is important for (CTG)n tracts expansions, based on experiments demonstrating an orientation effect on the repeat tract, relative to replication. It seems plausible, therefore, to speculate that mechanisms other than replication could be involved in their expansion. Supporting this, it has been demonstrated that (CTG)n · (CAG)n repeats stimulate homologous recombination in bacteria and gene conversion in yeast, frequently expanding in this process (17, 18, 35).
There are, however, some problems with this explanation as well. Expansion rates for (CGG)n · (CCG)n and (CTG)n · (CAG)n repeats are similar (Table 1). It is somewhat difficult to envision how two different processes, replication and recombination, lead to expansions with similar rates. Further, expansions of (CTG)n · (CAG)n repeats have been shown to depend on their orientation relative to the ori in bacterial, yeast, and mammalian systems (6, 9, 20, 28). Quite recently it was found that the propensity of these repeats to expand in cultured mammalian cells also depends on their distance from the ori (6). Finally, several replicative mutations were shown to affect expansion of these repeats (8, 15, 41, 42, 46). Thus, the final picture for (CTG)n · (CAG)n repeats remains more complex, and additional studies are warranted to resolve the relative contributions of replication and recombination.
Acknowledgments
We are indebted to Vladimir Larionov for his invaluable experimental advice and suggestions. We thank Cristina Bartocci for her help with replication experiments, Virginia Zakian and Robert Wells for helpful discussions, and Gerald Buldak for proofreading the manuscript.
This work was supported by grant GM60987 from NIH to S.M.M. and grant GM61961 from NIH to R.L.
Richard Pelletier and Maria M. Krasilnikova contributed equally to this publication.
REFERENCES
- 1.Ashley, C., Jr., and S. T. Warren. 1995. Trinucleotide repeat expansion and human disease. Annu. Rev. Genet. 29**:**703-728. [DOI] [PubMed] [Google Scholar]
- 2.Balakumaran, B. S., C. H. Freudenreich, and V. A. Zakian. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 9**:**93-100. [DOI] [PubMed] [Google Scholar]
- 3.Beckwith, W., and M. A. McAlear. 2000. Allele-specific interactions between the yeast RFC1 and RFC5 genes suggest a basis for RFC subunit-subunit interactions. Mol. Gen. Genet. 264**:**378-391. [DOI] [PubMed] [Google Scholar]
- 4.Beckwith, W. H., Q. Sun, R. Bosso, K. J. Gerik, P. M. J. Burgers, and M. A. McAlear. 1998. Destabilized PCNA trimers suppress defective Rfc1 proteins in vivo and in vitro. Biochemistry 37**:**3711-3722. [DOI] [PubMed] [Google Scholar]
- 5.Burgers, P. M., and K. J. Gerik. 1998. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 273**:**19756-19762. [DOI] [PubMed] [Google Scholar]
- 6.Cleary, J. D., K. Nichol, Y. H. Wang, and C. E. Pearson. 2002. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 31**:**37-46. [DOI] [PubMed] [Google Scholar]
- 7.Fotedar, R., R. Mossi, P. Fitzgerald, T. Rousselle, G. Maga, H. Brickner, H. Messier, S. Kasibhatla, U. Hubscher, and A. Fotedar. 1996. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells. EMBO J. 15**:**4423-4433. [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279**:**853-856. [DOI] [PubMed] [Google Scholar]
- 9.Freudenreich, C. H., J. B. Stavenhagen, and V. A. Zakian. 1997. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 17**:**2090-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, K. L., and B. J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262**:**613-627. [DOI] [PubMed] [Google Scholar]
- 11.Gacy, A. M., G. M. Goellner, C. Spiro, X. Chen, G. Gupta, E. M. Bradbury, R. B. Dyer, M. J. Mikesell, J. Z. Yao, A. J. Johnson, A. Richter, S. B. Melancon, and C. T. McMurray. 1998. GAA instability in Friedreich's ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell 1**:**583-593. [DOI] [PubMed] [Google Scholar]
- 12.Gerik, K. J., X. Li, A. Pautz, and P. M. Burgers. 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 273**:**19747-19755. [DOI] [PubMed] [Google Scholar]
- 13.Haworth, R. S., and L. Fliegel. 1993. Intracellular pH in Schizosaccharomyces pombe: comparison with Saccharomyces cerevisiae. Mol. Cell. Biochem. 124**:**131-140. [DOI] [PubMed] [Google Scholar]
- 14.Imai, T., and T. Ohno. 1995. Measurement of yeast intracellular pH by image processing and the change it undergoes during growth phase. J. Biotechnol. 38**:**165-172. [DOI] [PubMed] [Google Scholar]
- 15.Ireland, M. J., S. S. Reinke, and D. M. Livingston. 2000. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155**:**1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, R. R., A. Pluciennik, W. A. Rosche, R. R. Sinden, and R. D. Wells. 2000. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in Escherichia coli. J. Biol. Chem. 275**:**2174-2184. [DOI] [PubMed] [Google Scholar]
- 17.Jakupciak, J. P., and R. D. Wells. 2000. Gene conversion (recombination) mediates expansions of CTG·CAG repeats. J. Biol. Chem. 275**:**40003-40013. [DOI] [PubMed] [Google Scholar]
- 18.Jakupciak, J. P., and R. D. Wells. 1999. Genetic instabilities in (CTG.CAG) repeats occur by recombination. J. Biol. Chem. 274**:**23468-23479. [DOI] [PubMed] [Google Scholar]
- 19.Jaworski, A., W. A. Rosche, R. Gellibolian, S. Kang, M. Shimizu, R. P. Bowater, R. R. Sinden, and R. D. Wells. 1995. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl. Acad. Sci. USA 92**:**11019-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, S., A. Jaworski, K. Ohshima, and R. D. Wells. 1995. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10**:**213-218. [DOI] [PubMed] [Google Scholar]
- 21.Kang, S., K. Ohshima, M. Shimizu, S. Amirhaeri, and R. D. Wells. 1995. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 270**:**27014-27021. [DOI] [PubMed] [Google Scholar]
- 22.Keller, R. C., R. Mossi, G. Maga, R. E. Wellinger, U. Hubscher, and J. M. Sogo. 1999. Electron microscopic analysis reveals that replication factor C is sequestered by single-stranded DNA. Nucleic Acids Res. 27**:**3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovtun, I. V., and C. T. McMurray. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27**:**407-411. [DOI] [PubMed] [Google Scholar]
- 24.Krasilnikova, M. M., G. M. Samadashwily, A. S. Krasilnikov, and S. M. Mirkin. 1998. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 17**:**5095-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunst, C. B., and S. T. Warren. 1994. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell 77**:**853-861. [DOI] [PubMed] [Google Scholar]
- 26.Lea, D. E., and C. A. Coulson. 1949. The distribution of the number of mutants in bacterial populations. J. Genet. 49**:**264-285. [DOI] [PubMed] [Google Scholar]
- 27.McMurray, C. T. 1999. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. USA 96**:**1823-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miret, J. J., L. Pessoa-Brandao, and R. S. Lahue. 1998. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95**:**12438-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossi, R., and V. Hubscher. 1998. Clamping down on clamps and clamp loaders—the eukaryotic replication factor C. Eur. J. Biochem. 254**:**209-216. [PubMed] [Google Scholar]
- 30.Noskov, V., N. Kouprina, S. H. Leem, M. Koriabine, J. C. Barrett, and V. Larionov. 2002. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 30**:**8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohshima, K., and R. D. Wells. 1997. Hairpin formation during DNA synthesis primer realignment in vitro in triplet repeat sequences from human hereditary disease genes. J. Biol. Chem. 272**:**16798-16806. [DOI] [PubMed] [Google Scholar]
- 32.Oussatcheva, E. A., V. I. Hashem, Y. Zou, R. R. Sinden, and V. N. Potaman. 2001. Involvement of the nucleotide excision repair protein UvrA in instability of CAG∗CTG repeat sequences in Escherichia coli. J. Biol. Chem. 276**:**30878-30884. [DOI] [PubMed] [Google Scholar]
- 33.Panigrahi, G. B., J. D. Cleary, and C. E. Pearson. 2002. In vitro (CTG)∗(CAG) expansions and deletions by human cell extracts. J. Biol. Chem. 277**:**13926-13934. [DOI] [PubMed] [Google Scholar]
- 34.Parniewski, P., A. Jaworski, R. D. Wells, and R. P. Bowater. 2000. Length of CTG.CAG repeats determines the influence of mismatch repair on genetic instability. J. Mol. Biol. 299**:**865-874. [DOI] [PubMed] [Google Scholar]
- 35.Richard, G.-F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19**:**2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolfsmeier, M. L., M. J. Dixon, L. Pessoa-Brandao, R. Pelletier, J. J. Miret, and R. S. Lahue. 2001. cis-elements governing trinucleotide repeat instability in Saccharomyces cerevisiae. Genetics 157**:**1569-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolfsmeier, M. L., and R. S. Lahue. 2000. Stabilizing effects of interruptions on trinucleotide repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 20**:**173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadowski, P. D. 1995. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 51**:**53-91. [PubMed] [Google Scholar]
- 39.Samadashwily, G. M., G. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17**:**298-304. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt, K. H., C. M. Abbott, and D. R. Leach. 2000. Two opposing effects of mismatch repair on CTG repeat instability in Escherichia coli. Mol. Microbiol. 35**:**463-471. [DOI] [PubMed] [Google Scholar]
- 41.Schweitzer, J. K., and D. M. Livingston. 1999. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics 152**:**953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweitzer, J. K., and D. M. Livingston. 1998. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7**:**69-74. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu, M., R. Gellibolian, B. A. Oostra, and R. D. Wells. 1996. Cloning, characterization and properties of plasmids containing CGG triplet repeats from the FMR-1 gene. J. Mol. Biol. 258**:**614-626. [DOI] [PubMed] [Google Scholar]
- 44.Shivji, M. K. K., V. N. Podust, U. Hubscher, and R. D. Wood. 1995. Nucleotide excision-repair DNA-synthesis by DNA polymerase- epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 34**:**5011-5017. [DOI] [PubMed] [Google Scholar]
- 45.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 78**:**6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spiro, C., R. Pelletier, M. L. Rolfsmeier, M. J. Dixon, R. S. Lahue, G. Gupta, M. S. Park, X. Chen, S. V. S. Mariappan, and C. T. McMurray. 1999. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell 4**:**1079-1085. [DOI] [PubMed] [Google Scholar]
- 47.Usdin, K., and K. J. Woodford. 1995. CGG repeats associated with DNA instability and chromosome fragility from structures that block DNA synthesis in vitro. Nucleic Acids Res. 23**:**4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Broek, W. J., M. R. Nelen, D. G. Wansink, M. M. Coerwinkel, H. te Riele, P. J. Groenen, and B. Wieringa. 2002. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11**:**191-198. [DOI] [PubMed] [Google Scholar]
- 49.Wells, R. D., and S. T. Warren (ed.). 1998. Genetic instabilities and hereditary neurological disorders. Academic Press, San Diego, Calif.
- 50.White, P. J., R. H. Borts, and M. C. Hirst. 1999. Stability of the human fragile X (CGG)n triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol. 19**:**5675-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie, Y. L., C. Counter, and E. Alani. 1999. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn_3_ insertion in RFC1, the large subunit of the yeast clamp loader. Genetics 151**:**499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]