Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex (original) (raw)
. Author manuscript; available in PMC: 2006 Mar 29.
Published in final edited form as: Nature. 2005 Jun 2;435(7042):687–692. doi: 10.1038/nature03588
Abstract
The Small Ubiquitin-related Modifier SUMO-1 belongs to the ubiquitin (Ub) and ubiquitin-like (Ubl) protein family. SUMO conjugation occurs on specific lysine residues within protein targets, regulating pathways involved in differentiation, apoptosis, the cell cycle, and responses to stress by altering protein function through changes in activity, cellular localization, or by protecting substrates from ubiquitination1,2. Ub/Ubl conjugation occurs in sequential steps and requires the concerted action of E2 conjugating proteins and E3 ligases1,2. In addition to being a SUMO E3, the nucleoporin Nup358/RanBP2 localizes SUMO conjugated RanGAP1 to the cytoplasmic face of the nuclear pore complex (NPC) via interactions in a complex that also includes Ubc9, the SUMO E2 conjugating protein3–6. We present the 3.0 Å crystal structure of a four protein complex between Ubc9, a Nup358/RanBP2 E3 ligase domain (IR1-M), and SUMO-1 conjugated to the C-terminal domain of RanGAP1. Structural insights, combined with biochemical and kinetic data using additional substrates, support a model whereby Nup358/RanBP2 acts as an E3 by binding both SUMO and Ubc9 to position the SUMO-E2-thioester in an optimal orientation to enhance conjugation.
Ub/Ubls are activated by E1 and transferred to E2 to form E2-Ub/Ubl-thioesters. While competent for Ub/Ubl ligation to lysine ɛ-amino groups, E2s generally require E3 ligases to specifically recognize substrate lysine residues. Most E3s belong to either RING or HECT families and while RING E3s recruit substrate and bind the E2-Ub/Ubl via a zinc domain to promote conjugation to lysine residues7, HECT E3s recruit E2-Ub/Ubl to generate E3-Ub/Ubl-thioesters for conjugation8. The SUMO E2 can directly bind the consensus sequence Ψ-K-x-D/E where Ψ is hydrophobic, K is the substrate lysine, x is any amino acid, and D or E is acidic9, although several SUMO E3s facilitate conjugation in vivo and in vitro and include RING-type E3s10,11, Nup358/RanBP212, and Pc213. Nup358/RanBP2 and Pc2 appear unrelated to either RING or HECT E3 ligases.
One of the first discovered functions for SUMO-1 was its role in nucleocytoplasmic trafficking3–5. SUMO conjugation localizes RanGAP1 to the NPC in a complex that includes the SUMO E2 Ubc9 and Nup358/RanBP2, a multi-domain 3,224 amino acid (aa) nucleoporin that also interacts with Ran and other nuclear transport factors3–6,14,15. The SUMO-RanGAP1-Ubc9-Nup358/RanBP2 complex does not dissociate upon entry into mitosis and NPC disassembly, but redistributes to kinetochores and contributes to the stability of kinetochore-microtubule interactions16. A ~30kDa Nup358/RanBP2 fragment named IR1-M-IR2 binds Ubc9 and promotes SUMO E3 activity in vitro and in vivo12,17–19, although domains flanking IR1-M-IR2 also contribute to SUMO-RanGAP1 interactions6. IR1-M-IR2 was parsed into three elements: IR1 (aa 2633–2685), M (aa 2686–2710), and IR2 (aa 2711–2761). IR1 and IR2 were named for two internal sequence repeats. IR1-M-IR2, IR1-M, and M-IR2 all promote SUMO-1 conjugation in vitro18,19.
To determine the molecular details of this system, SUMO-1 was conjugated to RanGAP1, combined with Ubc9 and Nup358/RanBP2, purified by gel filtration, and crystallized. The model containing SUMO-1 (aa 20–97), RanGAP1 (aa 432–587), Ubc9 (aa 2–157), and Nup358/RanBP2 (aa 2631–2693) was refined to 3.0Å (R=24.7, Rfree=29.0; Table 1, Supplemental Table I and Methods). The structure reveals Ubc9 as the central component of the quaternary complex, contacting SUMO, RanGAP1, and Nup358/RanBP2 (Figure 1). SUMO contacts Ubc9 and Nup358/RanBP2, but only contacts RanGAP1 via the covalent bond between SUMO Gly97 and Lys524. Nup358/RanBP2 contacts SUMO and Ubc9, but does not contact RanGAP1.
Table 1.
X-ray Statistics
| Refinement | |
|---|---|
| Resolution (Å) | 30-3.0 |
| _R_work/ _R_free | 0.247/0.290 |
| No. atoms | |
| Protein | 3564 |
| Water | 28 |
| B-factors | |
| Protein | 90 |
| Water | 51 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (º) | 1.2 |
Figure 1. Structure of SUMO-RanGAP1-Ubc9-Nup358/RanBP2 complex.
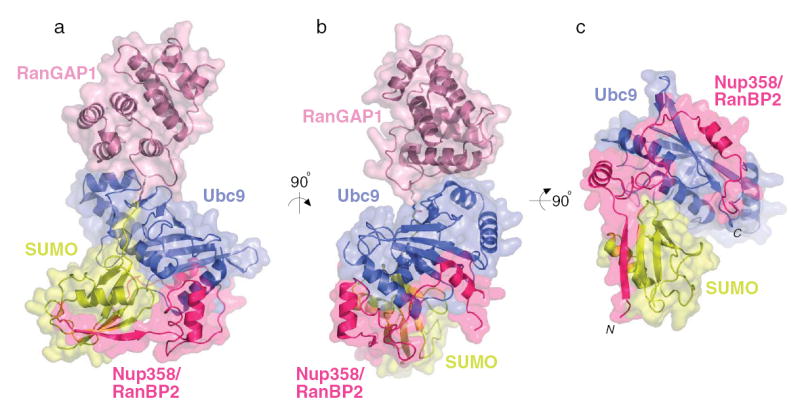
A) Ribbon and transparent surface representation for the complex between SUMO-1 (yellow), Ubc9 (blue), RanGAP1 (pink), and Nup358/RanBP2 (magenta). Each protein is labeled. The SUMO C-terminal glycine (Gly97) and RanGAP1 Lys524 are represented by solid bonds located near the image center. B) Orthogonal view of the complex. C) Orthogonal view of the complex to highlight the extended Nup358/RanBP2 structure. N- and C- termini of Nup358/RanBP2 are denoted in italics. Structural graphics generated with PYMOL30.
The human SUMO-1-RanGAP1-Ubc9 complex is similar to our previously described complex between mouse RanGAP1 and human Ubc9 in that most interactions to the RanGAP1 Ψ-K-x-D/E SUMO motif (L-K-S-E) are preserved9 (Figure 2). Despite covalent attachment to SUMO Gly97 via an isopeptide bond, Lys524 remains within a groove created by Ubc9 Asp127, Pro128, Ala129, and Tyr87. SUMO-1 Gln29 and Arg63 and the C-terminal tail (Gln92, Gln94, Thr95, Gly96, and Gly97) contact Ubc9 helix B and Ubc9 active site residues, respectively (Figure 2), burying only 650 Å2 of total surface area20. For comparison, Sae1/Sae2-SUMO-1 and Senp2-SUMO-1 buries 1650 Å2 and 1800 Å2, respectively21. Residues in the interface between SUMO-1 and Ubc9 are conserved among SUMO and Ubc9 family members. The small interface between SUMO and Ubc9 is consistent with the complex being primed to dissociate after conjugation. It is also consistent with Ubc9-SUMO specificity being achieved by the SUMO E1 activating enzyme which brings SUMO and Ubc9 together to form the thioester adduct21.
Figure 2. E2 active site in complex with RanGAP1-SUMO-1.
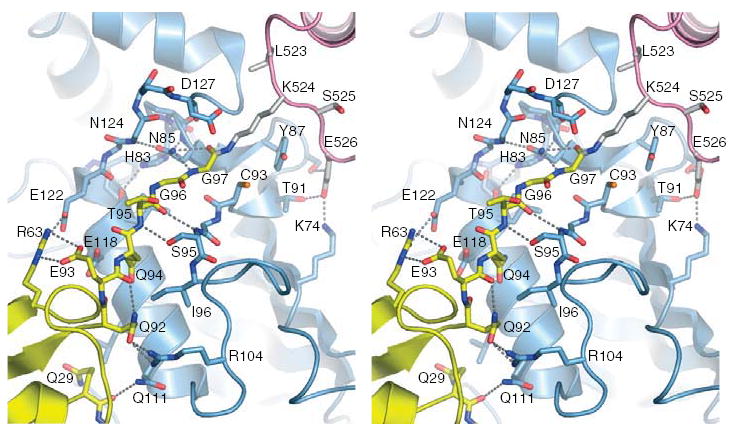
Stereo view of the E2 active site in complex with SUMO-1-RanGAP1 in ribbon and solid bond representation. Residues are labeled and hydrogen bonding interactions indicated by dashed lines. SUMO-1, RanGAP1 and Ubc9 colored yellow, pink, and blue as in Figure 1.
Several observations suggest that the Ub/Ubl-E2-thioester adopts a similar configuration to that observed in our complex prior to conjugation. First, NMR chemical shift perturbations observed for a steady state Ub-Ubc1-thioester were consistent with contacts between Ub, E2 helix B, and an E2 channel that coordinates the Ub Gly-Gly22. Second, Ub/Ubls and E2s are structurally conserved. Third, a rotation about Ubc9 Cys93 Chi1 brings the Cys sulfur atom to within 2 Å of the SUMO Gly97 carbonyl carbon (Figure 2). We previously suggested that E2s coordinate the lysine near the labile E2-Ub/Ubl-thioester to promote chemistry9. The absence of other E2 catalytic residues led to recent studies that implicated the conserved E2 His-Pro-Asn amino acid motif (HPN) in catalysis, namely that Asn Nδ contributes to formation of an oxyanion hole that stabilizes the transition state23. Our structure is consistent with this proposal insomuch as Asn85 Nδ moves away from the Ubc9 main chain and approaches the C-terminal Gly97 carbonyl oxygen, although definitive evidence for Asn85’s catalytic role is precluded by the resolution limits of our structure (3.0 Å), and that our complex includes a conjugated product rather than a E2-SUMO-thioester substrate complex.
Nup358/RanBP2 IR1-M adopts a non-globular and extended structure in the complex (Figure 3). IR1-M elements were divided into motifs I–V based on interactions in the complex (Figure 3a). Motif I forms an anti-parallel β-strand with SUMO β-strand 2. In addition to main chain contacts, motif I includes two acidic and five hydrophobic residues that contribute ionic and VDW contacts to SUMO-1 residues, respectively (Figure 3c). The C-terminal end of IR1 α-helix A contacts the Ubc9 N-terminal helix in addition to Pro69, Pro105, and Ala106. Ubc9 Arg8 bridges four main chain carbonyl oxygen atoms, two from Ubc9 and two from Nup358/RanBP2 (Figure 3d). Motif II forms a coil that packs hydrophobic residues into the interface formed by the Ubc9 N-terminal helix and residues between β1 and β2 of SUMO-1 (Figure 3d). Motif III acidic residues bridge the Ubc9 N-terminal helix and contact Ubc9 Arg13, Arg17, and Lys30 (Figure 3e). IR1 then forms α-helix B, packing motif IV hydrophobic residues onto Ubc9 β1-β3 (Figure 3f) before contacting Ubc9 β2, β3, and Phe22 via Leu2688-Tyr2689-Leu2690 in motif V (Figure 3g). The extended structure buries 1460 Å2 in the SUMO interface and 830 Å2 in the Ubc9 interface. Based on our structure, new sequence alignments between IR1 and IR2 were used to generate IR1*, IR2*, and IR1-M-IR2* (Figure 3a).
Figure 3. Nup358/RanBP2 sequence alignment and E3-E2-SUMO-1 structure.
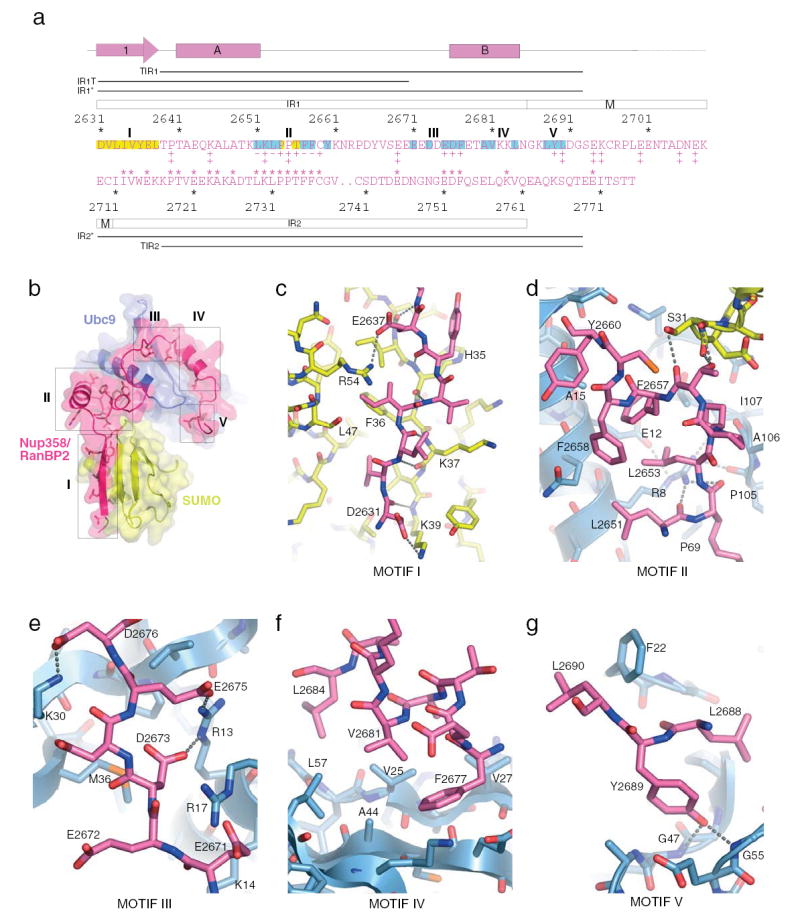
A) IR1-M-IR2 elements. Secondary structure shown above the alignment. Single aa code colored for SUMO (yellow) and Ubc9 (blue) contacts. Nup358/RanBP2 IR constructs indicated by bars. Mutational analysis18 shown below IR1-M (‡ no defect; + impaired activity; − no activity). * above IR2 indicate identical amino acid positions between IR1 and IR2. B) Nup358/RanBP2 motifs I–V (magenta), SUMO-1 (yellow), and Ubc9 (blue). C) Nup358/RanBP2 motif I. D) Nup358/RanBP2 motif II. E) Nup358/RanBP2 motif II. F) Nup358/RanBP2 motif IV. G) Nup358/RanBP2 motif V. Residues labeled and hydrogen bonding interactions indicated by dashed lines.
RanGAP1 is easily conjugated and interacts strongly with Ubc9 through surfaces adjacent to the conjugation site9. As such, it remains unclear if Nup358/RanBP2 E3 activity is required for RanGAP1 SUMO conjugation, or if it is just required to maintain a stable NPC complex. The stable interactions observed between RanGAP1 and E2; E3, SUMO and E2; and the covalent interaction between RanGAP1-SUMO suggested that our structure represented a trapped product complex. To test this, conjugation assays using RanGAP1 under multiple turnover conditions with and without Nup358/RanBP2 IR1* showed, in contrast to other substrates12,18,19, that IR1* inhibited SUMO conjugation. Importantly, SUMO-RanGAP1 accumulated to near stochiometric ratios of product, E2, and E3 (Figure 4a). Addition of SUMO-RanGAP1 also inhibited SUMO conjugation to p53 in an IR1*-dependent manner (Figure 4b). Again, stochiometric ratios of SUMO-1-RanGAP1:E2 appeared sufficient to sequester E2 and prevent further p53 conjugation. Importantly, partial disruption of the stable interface between RanGAP1 and Ubc9 alleviated IR1* dependent inhibition in multiple turnover assays and rendered RanGAP1 as a substrate for Nup358/RanBP2 E3 activity under single turnover conditions (Supplementary Figure 1).
Figure 4. Nup358/RanBP2 activities.
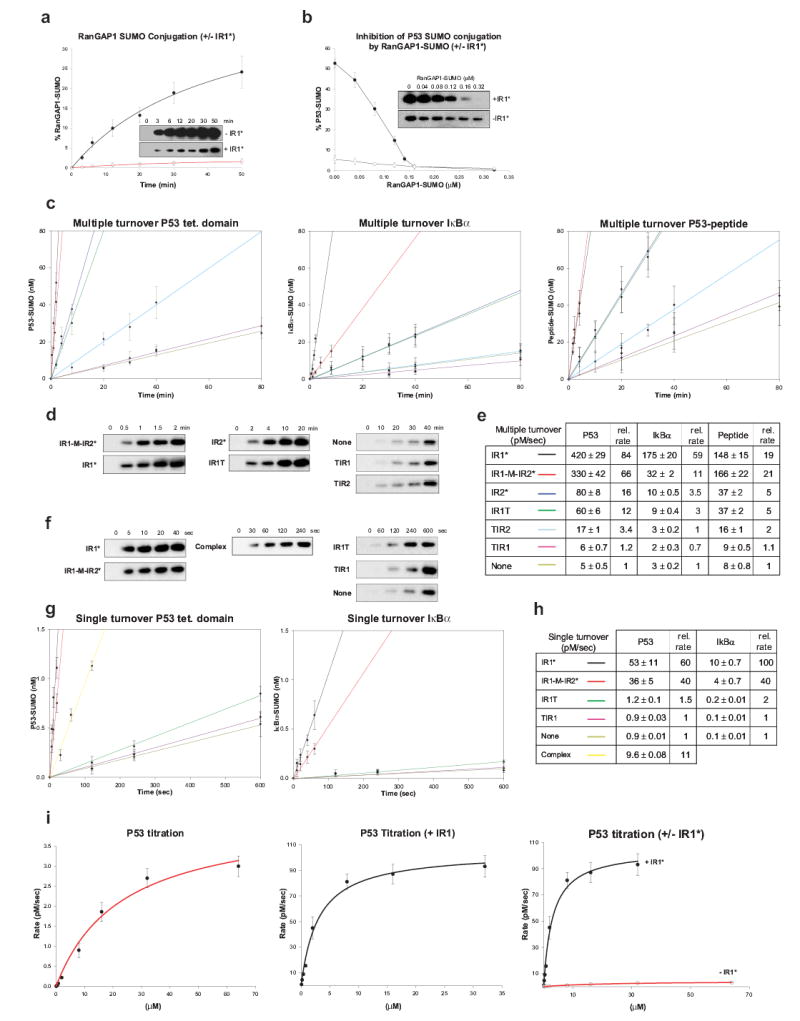
Error bars are ±1 standard deviation. A) SUMO-1 RanGAP1 conjugation under multiple turnover plus (•) or minus (○) IR1* with gel insets. B) SUMO-1 p53 conjugation plus (•) or minus (○) IR1* with SUMO-1-RanGAP1. C) SUMO-1 conjugation rates for p53, IκBα¨ peptide. Nup358/RanBP2 constructs in E. D) Gel insets for (C) using p53. E) Rates (pM/sec) and relative rates for C. F) Gel insets for (G) using p53. G) Rates for p53, IκBα using Nup358/RanBP2 constructs or IR1-M-RanGAP1-Ubc9-SUMO-Nup358/RanBP2 (yellow). H) Rates (pM/sec) for G. I) Single turnover rates for p53 plus (red) or minus (black) IR1*.
To assess the importance of contacts between Nup358/RanBP2, SUMO, and E2, assays were conducted at near-saturating substrate concentrations under multiple and single turnover conditions to quantify rate velocities using E3 constructs and substrates that included the p53 tetramerization domain, IκBα, and a peptide containing a SUMO consensus motif (Figure 3a) (See Methods). IR1* and IR1-M-IR2* enhanced conjugation rates up to 80-fold in multiple turnover assays (Figure 4c–e), while IR2* enhanced conjugation 4–16 fold. IR1T only included motifs I and II but catalyzed E3 activity at rates similar to IR2*, suggesting that motifs III–V are somewhat dispensable for activity. Deleting motif I (TIR1 and TIR2) from either IR1 or IR2 resulted in almost no E3 activity. Similar rate enhancements were observed under single turnover conditions using isolated E2-SUMO-thioester (See Methods) (Figure 4f–h).
Consistent with observations for motif I in our structure, NMR recently identified Nup358/RanBP2 IR1 amino acids (motif I) that interact with SUMO, and while E3 activity was not assessed, several mutations within motif I diminished binding to SUMO-1-RanGAP124 (Figure 3c). SUMO-1 interactions with Nup358/RanBP2 motif I might be recapitulated by other proteins that interact non-covalently with SUMO24 and Smt325. Mutational analysis revealed the importance of Nup358/RanBP2 motif II residues Leu2651, Leu2653, Phe2657, and Phe2658 in Ubc9 binding and E3 activity18, residues that contact Ubc9 in our structure (Figure 3a,d). This and another study identified Ubc9 mutations that partially diminished both E3 activity and binding to Nup358/RanBP218,19. Most detrimental Ubc9 mutations face Nup358/RanBP2 motifs IV and V in our structure. The latter study also suggested a mechanism for SUMO paralog selection by various Nup358/RanBP2 IR1-M-IR2 domains19. To confirm that SUMO-2/3-E2 is activated in an analogous manner to that proposed for SUMO-1-E2, we assayed SUMO-2 and -3 under single turnover conditions. These data revealed rate enhancements for IR1-M-IR2* and IR1*, and a strict dependence on motif I for activity (Supplemental Figure 2).
If Nup358/RanBP2 binds E2-SUMO-thioester in a manner similar to that observed in our complex, E3 activity is achieved independent of contacts to the substrate or E2 active site. So how does Nup358/RanBP2 work? Conjugation rates could be increased by an allosteric mechanism that alters the E2 active site indirectly to enhance catalysis18, possibly activating the thioester, leaving group, or E1’s ability to charge E2. However, thioester reactivity was not altered when E2-SUMO-thioester was incubated with 0, 1, or 10 mM DTT with and without E3. Differences were also not observed in E1-mediated E2-SUMO-thioester formation with and without E3, although 10:1 or 100:1 E3:E2 molar ratios inhibited E1-SUMO and E2-SUMO-thioester formation.
The importance of motif I and motif II for IR1* and IR2* E3 activity and the small interface observed between Ubc9 and SUMO suggested a model whereby Nup358/RanBP2 catalyzes E3 activity by tethering SUMO and Ubc9 together to reduce conformational flexibility, to prevent non-productive E2-SUMO conformations, and to align the complex and thioester for Ubl transfer. If true, and contrary to previous observations12,18,19 or the lack of substrate contacts, both substrate binding and catalysis should be affected. To test this, initial rate velocities were calculated over various p53 concentrations under single turnover conditions with and without IR1*. The kinetic data fit well to an equation from which Kd and K2 (Kcat) were derived (Figure 4i; See Methods). In the absence of IR1*, the Kd and K2 for p53 conjugation are 23.1±4.8 μM and 4.27±0.37 pM/s, respectively. In the presence of IR1*, the Kd and K2 are 3.04±0.47 μM and 104.7±4.3 pM/s, respectively. Thus, both Kd and K2 were affected by IR1* to increase Kd/K2 by ~185 fold.
SUMO-conjugated RanGAP1, in stochiometric quantities, sequesters Ubc9 and substantially diminishes its ability to promote conjugation of exogenous substrate in the presence of Nup358/RanBP2 (See above). Because IR1-M only contacts Ubc9 and SUMO in our complex, we predicted that an additional equivalent of Ubc9-SUMO-thioester should compete with a pre-formed E2-E3-SUMO-RanGAP1 complex for IR1-M interaction. In fact, single turnover assays indicated that IR1-M dissociated from the complex, enhancing Ubc9-SUMO-1-thioester conjugation by 11-fold (Figures 4f–h). These data suggest that while Nup358/RanBP2 may form a stable complex with SUMO-RanGAP1, it may still be available to promote E3 activity by binding additional E2-SUMO-thioesters, a scenario with important cellular implications for Nup358/RanBP2 E3 activity if E2-SUMO concentrations are sufficient to compete for Nup358/RanBP2 at the NPC.
The SUMO-RanGAP1-Ubc9-Nup358/RanBP2 structure provides data for several interactions critical for E3 assisted E2 conjugation. Perhaps most intriguing is the model that Nup358/RanBP2 enhances conjugation by coordinating the E2-SUMO-thioester optimally for substrate binding and catalysis. Comparison between Nup358/RanBP2-Ubc9-SUMO-RanGAP1 and two other E2-E3 structures26,27, E6AP-UbcH7 and c-Cbl-UbcH7, reveal distinct but overlapping E2 surfaces utilized in each complex. By aligning respective E2s, SUMO was placed into the other E2-E3 complexes. Interestingly, the RING domain and other E3 surfaces are well positioned to recognize ubiquitin within the modeled E2-Ub-thioester complexes (Supplementary Figure 3).
The indirect mechanism utilized by Nup358/RanBP2 to enhance SUMO conjugation might shed some light on ‘allosteric’ activation observed in other Ub/Ubl pathways such as the RING-finger and Nedd8-induced activation of the Cullin SCF complexes7, Apc11 induced activation of the APC complex27, all of which may coordinate E2-ubiquitin in a complex similar to that observed in our structure. Although less clear, mechanisms proposed here may also provide insights into activities that promote E2-Ub/Ubl-thioester activation during polyubiquitin chain formation28.
Methods
Cloning, expression, and protein purification
Preparation of human E1 (Sae1/Sae2), E2 (Ubc9), SUMO-1 (1–97), SUMO-1 (18–97), the C-terminal p53 tetramerization domain, and IκBα have been described9,21. Human RanGAP1 (aa 419–587) and Nup558/RanBP2 constructs (IR1*, IR1-M, IR1-M-IR2*, TIR1, IR1T, IR2*, TIR2; See Figure 3a) were amplified by PCR, cloned into a Smt3 vector9,21. Constructs were confirmed by DNA sequencing. E3 constructs included IR1-M-IR2* (aa 2631–2771), IR1* (aa 2631–2695), IR2* (aa 2709–2771), TIR1 (aa 2640–2695), TIR2 (aa 2718–2771), and IR1T (aa 2631–2670). The synthetic p53-derived peptide includes amino acids 380–393 (HKKLMFKTEGPDSD). Biochemical assays utilized proteins that were concentrated in buffer containing 350 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM DTT, flash frozen in liquid nitrogen, and stored at −80C. Cultures were fermented at 37°C to A600 of 3, induced with 0.75 mM IPTG for 4–6 hours at 30°C, harvested and suspended in 50 mM Tris-HCl (pH 8.0), 20% w/v sucrose, 350 mM NaCl, 20 mM imidazole, 0.1% IGEPAL, 1 mM PMSF, 1 mM β-mercaptoethanol (BME), and 10 μg/ml DNAse prior to sonication and removal of insoluble material by centrifugation. Proteins were purified by metal-affinity chromatography (Qiagen), gel filtration (Superdex75 or 200; Pharmacia), and ion exchange (MonoQ; MonoS). SUMO-RanGAP1 was prepared and combined with Ubc9 and IR1-M (aa 2631–2711), purified by gel filtration (Superdex200), and concentrated to 10 mg/ml in 50 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM DTT, frozen in liquid nitrogen and stored at −80°C.
Crystallographic analysis
Crystals were obtained at 18°C by hanging drop vapor diffusion against a well solution containing 18% PEG4000 (w/v), 0.1 M sodium citrate pH 5.0, 0.2 M ammonium acetate and transferred to crystallization solutions containing 12% ethylene glycol prior to cryoprotection. Data were processed using and the structure was solved by molecular replacement used human RanGAP1-Ubc9 coordinates to phase the complex at 4 Å. SUMO-1 was modeled based on previous structures21. Nup358/RanBP2 was modeled into electron density that became apparent after refinement of SUMO-RanGAP1-Ubc9. Nup358/RanBP2 residues 2663, 2665, 2666, 2669 and 2670 not clearly present in the electron density were modeled without side chains. Refinement and data statistics are provided in the text and Supplemental Table I.
Biochemical assays
Assays were conducted in triplicate. Samples were removed at specified times, denatured in non-reducing SDS-PAGE buffer containing 4M Urea (single turnover) or reducing SDS-PAGE buffer (multiple turnover), analyzed by SDS-PAGE and western blotting using a polyclonal rabbit antibody against SUMO-1 (Boston Biochem), and developed with ECL-Plus (Amersham). Data were imaged using a Fujifilm LAS-3000 Imager and quantified using Image Gauge v4.0 (FujiFilm).
Multiple turnover reaction- Reactions were performed at 37°C in using 150 nM E1, 100 nM Ubc9 (E2), 10 μM SUMO-1 (1–97), 5 mM MgCl2, 0.1 % tween-20, 20 mM Hepes pH 7.5, 50 mM NaCl, 1 mM DTT, and indicated Nup358/RanBP2 constructs (200 nM) using 8 μM p53 tetramerization domain (p53), 4 μM IkBα, or 500 μM p53-peptide as substrate. RanGAP1 conjugation was assessed in the presence or absence of IR1* (200 nM). Inhibition of p53 conjugation by RanGAP-SUMO was assessed with and without IR1* (200 nM) in reactions containing RanGAP1-SUMO-1 at 0, 0.04, 0.08, 0.12, 0.14, 0.16 or 0.32 μM. Single turnover reaction- E2-SUMO-thioester was formed at 37°C in non-reducing buffer containing 100 nM E1, 1 μM Ubc9 (E2), 5 mM MgCl2, 0.1 % tween-20, 20 mM Hepes pH 7.5, 50 mM NaCl, 1 μM mature SUMO-1 (1–97) and 1 μM ATP and stopped after 10 min by 10-fold dilution at 4°C in buffer that contained 5 mM EDTA. Diluted reactions contained 10 nM E1, 100 nM Ubc9 (E2), 100 nM mature SUMO-1, 0.5 mM MgCl2, 5 mM EDTA, 0.1 % tween-20, 20 mM Hepes pH 7.5, 50 mM NaCl, either 8 μM p53 or 4 μM IκBα, and 100 nM of the indicated Nup358/RanBP2 elements. Reactions also included p53 in the presence of 100 nM RanGAP1-Ubc9-SUMO-Nup358/RanBP2 (IR1-M). Data in Supplemental Figure 1 were generated under single turnover conditions by replacing SUMO-1 with SUMO-2 (1–93) or SUMO-3 (1–92). Rate constants were calculated for p53 by measuring rate velocities under single turnover conditions with and without IR1* (100 nM) using 0, 0.08, 0.4, 0.8, 2, 4, 16, 32 or 64μM p53. Data were fitted to a hyperbolic 2-parameter, single rectangular function to derive reaction constants (ν =k2[S]/Kd+[S]; where k2=rate, Kd=dissociation constant, ν =velocity, and S=substrate). The kinetic scheme used to evaluate Kd (k−1/k1) and k2: E + S ↔ ES → E’ + P where k−1>>k2, [S]>>[E], and the chemistry is irreversible29.
Supplementary Material
Fig S1
Fig S2
Fig S3
SI Table 1
Acknowledgments
Use of the Advanced Photon Source (APS) is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Use of the SGX Collaborative Access Team beamline facilities at Sector 31 of the APS was provided by Structural GenomiX, Inc., who constructed and operates the facility. We thank Michael J. Matunis for the original clone containing Nup358/RanBP2 (aa 2596–2836), K.R. Rajashankar and Ali Yunus for helpful discussion and reagents that contributed to this work. D.R. and C.D.L. were supported in part by National Institutes of Health grant (GM65872). C.D.L. acknowledges support from the Rita Allen Foundation.
Footnotes
Competing interests statement. The authors declare that they have no competing financial interests.
References
- 1.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 5.Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc Natl Acad Sci U S A. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO Modification Pathway Localize to Filaments of the Nuclear Pore Complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 8.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 11.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 12.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 13.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama N, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 16.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 17.Saitoh H, Pizzi MD, Wang J. Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J Biol Chem. 2002;277:4755–4763. doi: 10.1074/jbc.M104453200. [DOI] [PubMed] [Google Scholar]
- 18.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT nor RING type. Nat Struct Molec Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- 19.Tatham MH, et al. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Molec Biol. 2004;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 20.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 21.Lois LM, Lima CD. Structures of the Small Ubiquitin-like MOdifier E1 activating enzyme provide insights into SUMO activation and the mechanism for E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–890. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 23.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 26.Huang L, et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 27.Leverson JD, et al. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 29.Strickland S, Palmer G, Massey V. Determination of dissociation constants and specific rate constants of enzyme-substrate (or protein-ligand) interactions from rapid reaction kinetic data. J Biol Chem. 1975;250:4048–4052. [PubMed] [Google Scholar]
- 30.DeLano, W.L. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org (2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
SI Table 1