Mapping Quantitative Trait Loci for Anxiety in Chromosome Substitution Strains of Mice (original) (raw)
Abstract
Anxious behavior in the mouse is a complex quantitative phenotype that varies widely among inbred mouse strains. We examined a panel of chromosome substitution strains bearing individual A/J chromosomes in an otherwise C57BL/6J background in open-field and light-dark transition tests. Our results confirmed previous reports of quantitative trait loci (QTL) on chromosomes 1, 4, and 15 and identified novel loci on chromosomes 6 and 17. The studies were replicated in two separate laboratories. Systematic differences in the overall activity level were found between the two facilities, but the presence of the QTL was confirmed in both laboratories. We also identified specific effects on open-field defecation and center avoidance and distinguished them from overall open-field activity.
THE anxiety-related behavioral phenotype termed “emotionality” has been a primary focus for behavioral research in rodents for decades (Crawley 2000). Typically, emotionality is measured by placing a mouse in an unfamiliar, stressful environment and evaluating its movement. A variety of tests that yield correlated results have been used to assay this phenotype, including the open-field activity test, the light-dark (LD) exploration test, elevated-plus, elevated-zero, and y mazes, and the Vogel conflict test (Crawley 2000; Flint 2003b). Performance in these studies can be modulated with anxiolytic drugs, supporting the assertion that they measure a factor analogous to human anxiety (Pellow et al. 1985; Pellow and File 1986; File and Pellow 1987; Mathis et al. 1994, 1995).
It is clear that there is a large genetic component to emotionality in rodents because different inbred mouse strains, raised under identical laboratory conditions, perform very differently in these assays (Festing 1979; Crawley et al. 1997). In various tests of activity, susceptible (anxious) strains such as A/J and BALB/c are typically reluctant to explore the environment, remain close to walls or away from exposed heights, move with a flattened posture, and frequently defecate during the test. Less anxious strains, such as C57BL/6J, readily explore the test area, including open or exposed areas, walk with a normal posture, and rarely defecate (Crawley et al. 1997).
No single locus accounts for differences in emotionality phenotypes among inbred strains. Instead, these behaviors appear to be complex, involving relatively small individual contributions from a variety of genetic loci (Flint et al. 1995; Flint 2003b) as well as environmental influences. Historically, studies of emotionality were among the first to use new genetic tools for identifying quantitative trait loci (QTL). These include intercross analysis between inbred strains, breeding of artificially selected strains, intercross and breakpoint analysis among selected strains, and high-resolution mapping in heterogeneous stocks (Flint et al. 1995; Gershenfeld et al. 1997; Gershenfeld and Paul 1997; Talbot et al. 1999; Turri et al. 1999, 2001a,b; Mott et al. 2000).
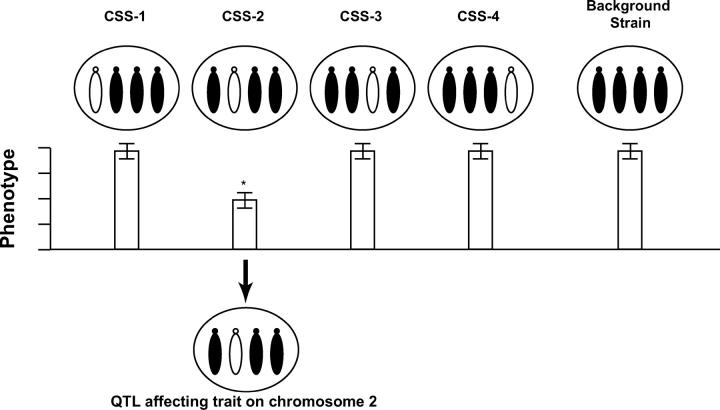
Chromosome substitution strain (CSS) analysis is a novel method of QTL identification that we proposed (Nadeau et al. 2000) and that we and others have recently demonstrated (Matin et al. 1999; Cowley et al. 2001; Koumproglou et al. 2002; Liang et al. 2002; Youngren et al. 2003; Singer et al. 2004). It involves selectively breeding inbred strains in which a single chromosome from one parental strain has been introgressed into a background derived entirely from a second strain. In its most powerful form, a complete panel of CSSs in which all chromosomes have been substituted is available. The phenotype of interest is measured in each CSS as well as in the host (background) strain; divergence between the host strain and a given CSS indicates a locus or loci on the substituted chromosome affecting that phenotype. This method offers significant improvements in speed over recombination-based methods as intercross progeny do not need to be bred and genotyped. It also provides increased sensitivity in QTL detection (Nadeau et al. 2000; Belknap 2003) because each chromosome is tested in a controlled background, free from the confounding effects of other randomly segregating loci affecting the same trait (Figure 1). CSSs also greatly accelerate any subsequent fine mapping of discovered loci, because the defined and controlled background eliminates or greatly reduces the need to spend years constructing congenic strains.
Figure 1.—
Mapping quantitative trait loci with chromosome substitution strains. A phenotype was tested in the various CSSs as well as in the C57BL/6J background strain. If a significant difference was found between a particular CSS (here, CSS-2) and the background strain, the substituted chromosome carried at least one locus affecting that phenotype. For simplicity, single chromosomes are shown; the actual CSSs are homozygous for both the substituted and the background chromosomes.
We recently described (Singer et al. 2004) the first complete CSS panel, in which chromosomes from the A/J strain were introgressed into the C57BL/6J host strain (henceforth abbreviated as B6). The strong existing body of work on QTL affecting emotionality and the pronounced phenotypic differences between the A/J and B6 strains make this CSS panel a promising tool for the study of genetic variation in emotionality using a pair of behavioral tests: open-field activity and light-dark transition. In the open-field test, a mouse is released in an enclosed, brightly lit area ruled with a grid. During a 4-min trial, the number of squares traversed (open-field activity, or OFA), the number of center squares traversed and the number of fecal pellets left behind (open-field defecation, or OFD) are recorded. The LD transition test uses a box divided into a large, brightly illuminated chamber and a smaller dark chamber. A mouse is released in the dark chamber and, in a 5-min trial, the time until the mouse first emerges into the lighted chamber, the total time spent in the lighted chamber, and the number of transitions between the chambers are recorded. In both tests, we found multiple loci affecting emotionality and validated and mapped them in independent studies.
MATERIALS AND METHODS
Behavioral testing:
Male mice were weaned at 4 weeks after birth, housed in same-sex groups of three to four, and tested at 8 weeks after birth. In the initial screening of the CSS panel, 10–12 mice/CSS were typically tested. Open-field testing was performed in a 60 × 60-cm box made of white Formica, ruled with a 4 × 4 grid, enclosed with 15-cm-high walls of clear acrylic, and illuminated by a 1280-lm lamp. A mouse was released into a center-side square of the grid and, during a 4-min trial, the number of boxes and center boxes traversed were observed and recorded. A compound emotionality phenotype, termed EMO (Talbot et al. 1999), was generated by standardizing OFA and OFD scores for a given sample set to a mean of zero and a variance of 1, reversing the sign of the normalized OFA result, and taking the mean of the two scores for each individual.
Light-dark exploration was performed in a 19 cm high × 19 cm deep Plexiglas chamber, divided into a 14-cm-wide dark chamber and a 28-cm-wide clear chamber separated by a 10 × 10-cm opening and illuminated by a 1280-lm lamp. A mouse was released into the dark chamber and its transitions between the chambers were recorded during a 5-min trial. A transition was scored when the entire body of the mouse passed into the new chamber. Times were reported in the format mm:ss±mm:ss.
To minimize variability, all behavioral testing was performed between 3 and 5 pm (while mice were kept on a 7 am–7 pm light/dark cycle). Before each day's testing, several mice were placed in the apparatus for 10 min to avoid testing the first mice in a fresh environment. Mice from different CSSs were rotated randomly through the testing to minimize the effect of any transient environmental factor on a particular CSS. The chambers were cleaned and sterilized at the end of each day of testing.
Statistical analysis:
Significance of results in the initial CSS panel screen was determined in a _t_-test between each CSS result and the B6 control. Significance levels were subjected to a Bonferroni correction to account for multiple hypothesis testing with 22 CSS strains. Results were considered significant if the corrected significance level was <0.05. This implies an expected rate of only 0.05 false positives/trait across the entire panel.
Intercross analysis:
Genotyping was performed with PCR amplification of simple sequence length polymorphism (microsatellite) markers at a spacing of 15–20 cM. Chromosomal localizations are according to the Mouse Genome Database 2001 Chromosome Committee Reports (http://www.informatics.jax.org). QTL analysis was performed with Map Manager (Manly and Olson 1999), using various models as described below. All QTL reported as exceeding Lander-Kruglyak significance also exceeded the significance threshold estimated with Map Manager's permutation function.
RESULTS
Screening in the CSS panel:
The first stage of analysis was performed in the Animal Resource Center at Case Western Reserve University (CWRU). Eight-week-old male mice from each strain of the B6-ChrA CSS panel (with the exception of CSS-5 and the mitochondrial CSS, which were not yet completed) were examined in both tests, as were mice from the A/J and B6 parental strains. As demonstrated previously (reviewed by Crawley et al. 1997), the results for A/J and B6 were extremely divergent, and several of the CSSs also differed significantly from the B6 host strain.
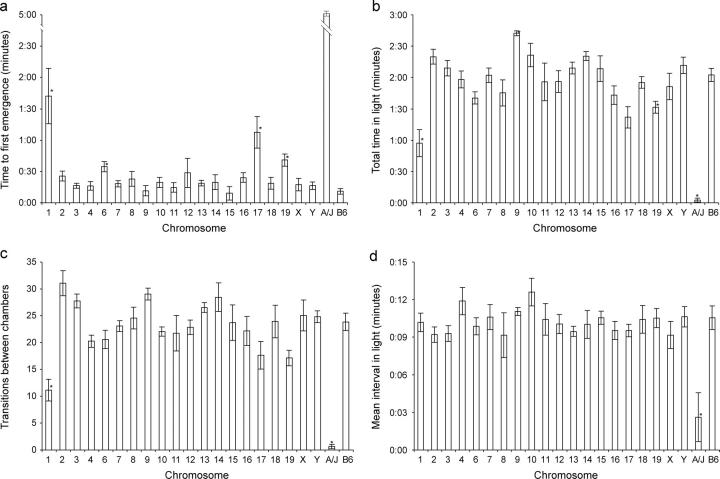
In the open-field test, B6 mice traversed an average of 133.6 ± 9.9 grid squares, including 18.9 ± 2.9 center squares or 14% of the total. No fecal pellets were seen in 13 trials (Figure 2). A/J mice traversed an average of 1.1 ± 0.3 grid squares, leaving 5.3 ± 0.5 fecal pellets. None crossed into a center square. CSSs for chromosomes 1, 4, 6, and 15 (henceforth abbreviated CSS-1, CSS-4, and so on) traversed the fewest squares: 75.2 ± 7.2, 91.3 ± 3.3, 82.5 ± 5.3, and 86.0 ± 5.3, respectively. These results agreed with previous reports of loci on chromosomes 1, 4, and 15 affecting OFA (Flint et al. 1995; Gershenfeld et al. 1997; Gershenfeld and Paul 1997; Talbot et al. 1999; Turri et al. 1999, 2001a,b; Mott et al. 2000; Zhang and Gershenfeld 2003).
Figure 2.—
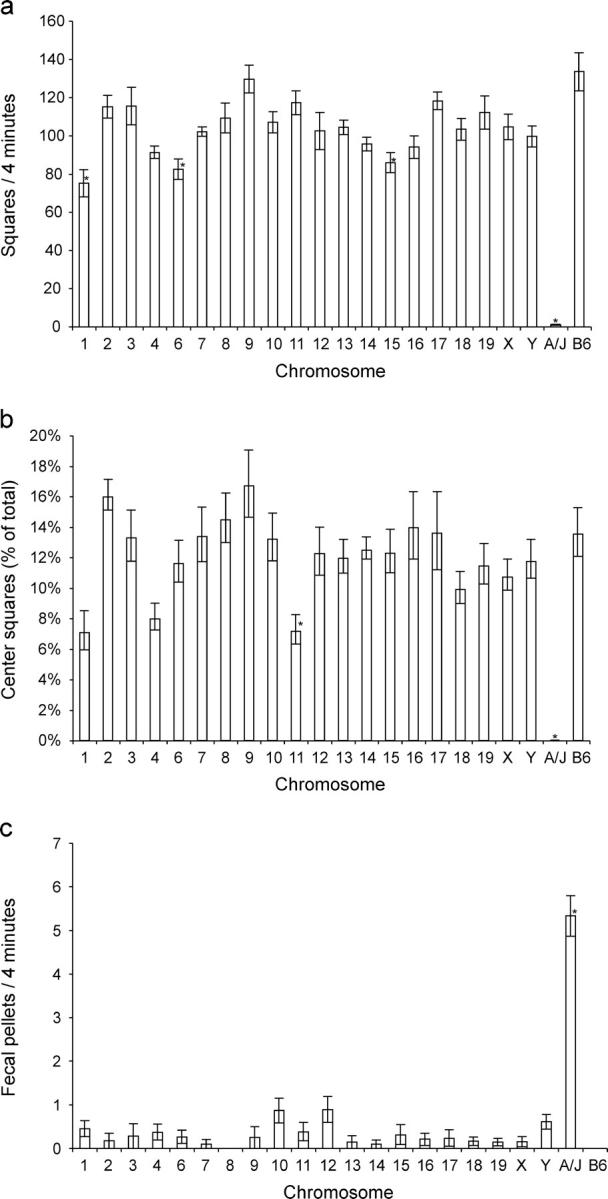
Open-field results for 20 CSSs, the B6 background strain, and A/J. Significant deviations, after correction for multiple hypothesis testing, are indicated by an asterisk. (a) OFA in number of squares traversed in a 4-min trial. (b) Center avoidance in center squares entered as percentage of total activity. (c) OFD in average number of pellets left.
The CSS for chromosome 1 entered the lowest percentage of center squares; however, a surprising finding was that while CSS-11 crossed 117.5 ± 6.2 total squares, the third most among the panel, only 7.1% were center squares. The CSS-11 mice rapidly circled the area, but remained close to the wall most of the time. All strains but one (CSS-8) left fecal pellets, a behavior that was not seen in any of the 30 B6 mice observed in this study and in an earlier pilot study of the parental strains.
In the light-dark transition test, B6 mice first emerged into the illuminated chamber after an average of 0:10 ± 0:02 min, spending 2:02 ± 0:06 min in the lighted area, with 23.8 ± 1.6 transitions between chambers. Most A/J mice did not emerge at all, spending an average of 0:02 ± 0:01 min in the lighted area, with 0.6 ± 0.4 transitions between chambers (Figure 3).
Figure 3.—
Light-dark transition results for 20 CSSs, the B6 background strain, and A/J. Significant deviations, after correction for multiple hypothesis testing, are indicated by an asterisk. (a) Time (in minutes) to first emergence into the lighted chamber. (b) Total time (in minutes) in lighted chamber during a 5-min trial. (c) Number of transitions between chambers. (d) Average length (in minutes) of intervals in the lighted chamber.
Again, CSS-1 diverged the most from the baseline B6 phenotype, emerging after 1:42 ± 0:26 min and spending 0:57 ± 0:12 min in the lighted chamber with 11.1 ± 2.0 transitions. CSS-6 also showed reduced activity (0:34 ± 0:04 min to emergence, 1:40 ± 0:05 min in the lighted chamber), while CSS-4 and CSS-15 were similar to B6. Surprisingly, although CSS-17 was the second most active CSS in the open-field assay, it was the second least active in the light-dark test (1:07 ± 0:15 min to emergence, 1:21 ± 0:10 min in the lighted chamber).
Intersite phenotype variation:
At this stage, testing shifted to the animal facility at the Whitehead Institute for Biomedical Research (WIBR). To confirm the results from screening the CSS panel and to map the responsible loci along the length of the chromosomes, we performed intercrosses between the implicated CSSs and the B6 parental strain. This process is analogous to traditional whole-genome intercross QTL analysis, but the testing of a single chromosome in an otherwise constant background greatly increases sensitivity while lowering the threshold for significance. LOD scores between 1.4 and 2.6 (depending on the length of the substituted chromosome and the model of dominance) are required for significance, in contrast to the 3.3–4.3 necessary in whole-genome studies (Lander and Kruglyak 1995). Because LOD scores are logarithmic, reducing the significance threshold by half greatly increases the power of a study.
Because of concerns about the reproducibility of behavioral assays between laboratories (Crabbe et al. 1999; Wahlsten et al. 2003), we began by retesting 11 of the CSSs in the new environment. The strains that displayed the most reduction in activity relative to B6 continued to do so. However, in both open-field and LD tests, most of the strains displayed less activity at WIBR than at the CWRU facility. For example, B6 traversed 116.6 ± 8.6 open-field squares at WIBR and few mice immediately ran into the lighted chamber in the LD test, as happened frequently with most strains tested at CWRU.
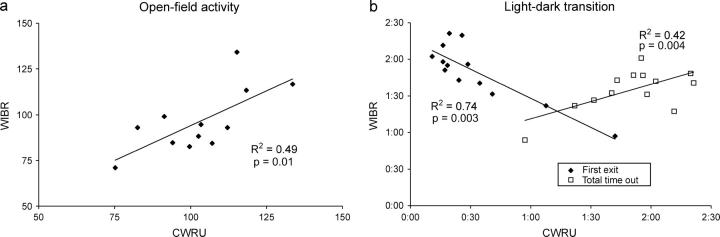
The reduction in activity was strikingly linear when results from WIBR were plotted against those from CWRU (Figure 4). OFA fit a line of y = 0.76_x_ + 17.9 squares, with an _R_2 of 0.49 (P = 0.01, from the F_-statistic calculated in an ANOVA test). Comparison of the LD results yielded fits of y = −0.79_x + 0.09 min (R_2 = 0.74, P = 0.0003) for time to first emergence and y = 0.47_x + 0.03 min (_R_2 = 0.42, P = 0.004) for total time in the lighted chamber. (Results in A/J mice were excluded from these regressions because they showed virtually no activity in either context and because including them would have led to a wildly disproportionate skewing of the line fit and inappropriately high _R_2 results.)
Figure 4.—
Comparison of performance at CWRU and WIBR for CSSs for chromosomes 1, 2, 4, 6, 10, 12, 14, 16, 17, 18, 19, and Y as well as the B6 background strain. Results for CWRU are plotted on the _x_-axis and for WIBR on the _y_-axis. (a) Open-field activity. (b) Time to first emergence (solid diamonds) and total time in the lighted chamber (open squares) in a light-dark transition test.
Fine mapping in intercrosses:
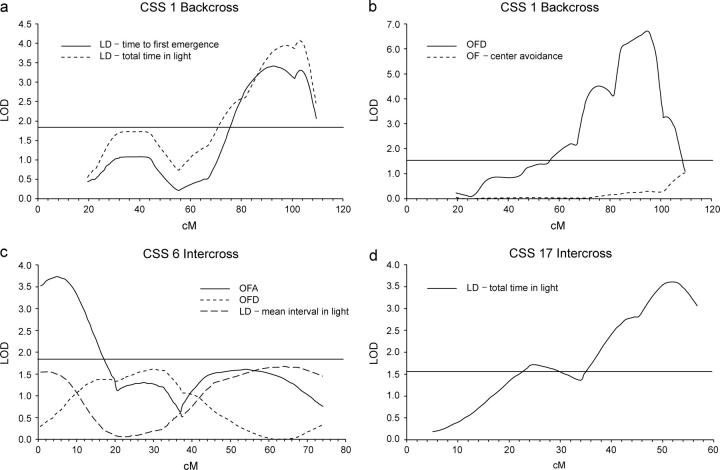
We selected the following phenotypes for fine mapping: reduced LD activity and elevated OFD in CSS-1, reduced OFA in CSS-6, and reduced LD activity in CSS-17 (Figure 5). Because analysis of heterosomic CSS-1 showed reduced activity in the LD test, and elevated OFD (not shown), we pursued a backcross of the F1 to B6. A study of 91 N2 progeny mapped a QTL between D1Mit151 and D1Mit511 at 93 cM with a LOD of 3.3 (significance threshold = 1.7) for time to first emergence in the LD test and a LOD of 4.1 in the same interval at 103 cM for total time spent in the lighted chamber. When OFD was treated as a dichotomous trait and analyzed with the penetrance scan function of Mapmaker (Gorham et al. 1996), a LOD of 6.7 at 95.5 cM was obtained with a peak between D1Mit151 and D1Mit511. These results are consistent with previous reports for a locus on the distal end of chromosome 1 (Flint et al. 1995; Gershenfeld et al. 1997; Talbot et al. 1999; Turri et al. 1999, 2001a,b; Mott et al. 2000).
Figure 5.—
QTL analysis for results in mapping crosses between implicated CSSs and B6. Significance thresholds are indicated by a horizontal line. (a) CSS-1 backcross analyzed for time to first emergence and total time in the lighted chamber in a light-dark transition test. (b) CSS-1 backcross analyzed for OFD and open-field center avoidance. (c) CSS-6 intercross analyzed for OFA, OFD, and mean interval in the lighted chamber in a light-dark transition test. (d) CSS-17 intercross analyzed for total time in the lighted chamber in a light-dark transition test.
Heterozygosity for A/J alleles at the chromosome 1 locus or loci appeared to have a weaker effect on OFA, failing to show any significant effect in this small backcross. Nonetheless, QTL analysis of OFA in the cross was consistent with the LD results, with a LOD of 0.96 for center avoidance at D1Mit511 at 113 cM. The compound EMO phenotype, a normalized measure combining OFA and OFD, indicated a significant LOD of 1.9 at D1Mit159 at 81.6 cM.
In a study of 82 F2 progeny from an intercross between CSS-6 and B6, two QTL affecting OFA were detected: one of LOD 3.7 between D6Mit138 and D6Mit274 at 4.9 cM (chromosome significance threshold, 1.6) and a second of LOD 1.6 between D6Mit36 and D6Mit339 at 52.5 cM, both with a recessive effect for the A/J allele or alleles. Analysis of OFD yielded a LOD of 1.6 between D6Mit274 and D6Mit209 at 17.0 cM while no suggestive linkages were seen for the EMO phenotype.
A suggestive peak of LOD 1.1, affecting time in the light during the LD test, was also detected between D6Mit138 and D6Mit274 at 2.1 cM, while analysis of the mean time of intervals in the lighted chamber indicated a peak of LOD 1.5, also at 2.1 cM, and another of LOD 1.7 between D6Mit36 and D6Mit339 at 63.9 cM.
A study of 70 F2 progeny from an intercross between CSS-17 and B6, analyzed for total time spent in the light during the LD test and fit to a two-parameter model because dominance of the QTL was unclear in the F1 (chromosome significance threshold, 2.3), indicated a QTL between D17Mit39 and D17Mit221 at 51.2 cM with a LOD of 3.6. No significant effect was seen on OFA or OFD.
DISCUSSION
Emotionality and CSS mapping:
The A/J and C57BL/6J strains were selected for the CSS breeding project in large part because of the many phenotypic differences known between them (Festing 1979; Mouse Phenome Database 2001). In particular, anxiety-related phenotypes were known to differ substantially and reproducibly between those strains. Therefore, this new technique for dissecting the genetic basis of complex quantitative traits offered the promise of better understanding the mechanisms underlying emotionality.
Results obtained from two tests of emotionality agreed well with previous studies that suggested that a small number of loci, specifically those on chromosomes 1, 4, and 15, exert a large effect on emotionality (Flint et al. 1995; Gershenfeld et al. 1997; Talbot et al. 1999; Turri et al. 1999, 2001a,b; Mott et al. 2000). Our findings agree with that assessment in both the number and the location of major players, and our mapping of a major locus on chromosome 1 to the distal end of the chromosome is consistent with all previous work. Flint (2003a) argues for two distinct QTL on chromosome 1: one polymorphic between B6 and BALB/c at ∼100 cM (Flint et al. 1995; Mott et al. 2000; Turri et al. 2001a,b) and a second polymorphic between B6 and A/J at ∼80 cM (Talbot et al. 1999). The location near 100 cM is more consistent with our findings, despite our use of B6 and A/J. The discrepancy may be due to ambiguity in our LOD peak, as the location of the putative QTL at 80 cM still exceeds significance in our studies. Alternatively, an earlier study (Gershenfeld et al. 1997) of an intercross between B6 and A/J placed a QTL at 100.5 cM, which supports our results.
We also detected additional loci with smaller effects on OFA and OFD, and on LD transition, presumably as a result of the increased sensitivity of the CSS method. The effect of loci on chromosomes 6 and 17 has not been previously reported at either a significant or a suggestive level. CSS-9 and CSS-10 also showed significant reduction in light-dark activity in the initial screening and previously unreported QTL may be present there as well.
The efficiency of CSS mapping compared favorably with that of recombination-based mapping of loci affecting the same phenotypes, detecting loci by testing 10–12 mice/strain in the initial screen and validating them in intercrosses of <100 F2 progeny. Comparable studies in intercrosses have used similar or much larger numbers of mice—from 514 to 1636 F2 progeny (Flint et al. 1995; Gershenfeld et al. 1997; Turri et al. 2001a)—in addition to the selective breeding of high- and low-activity strains that preceded two of those studies. The resolution of our F2 mapping was limited by the small sample sizes that we chose to use and the resulting paucity of informative crossovers. The resolution of those studies could, of course, be improved to any desired level by increasing the sample size. Perhaps most importantly, the whole process is greatly accelerated by eliminating the construction of congenic strains in many cases. (A more detailed treatment of the relative statistical advantages of CSS mapping is given in the supplementary material for Singer et al. 2004.)
The next steps to map these loci more closely are relatively straightforward. Several routes are available for moving to a precise localization of the QTL with relative ease (Belknap 2003; Youngren et al. 2003). The process of assigning a QTL to a specific gene remains a more difficult undertaking. However, recent insights into the haplotype block structure of laboratory mouse strains (Wade et al. 2002) suggest that focusing on regions of divergence could greatly accelerate the fine-mapping process; other options, such as the use of microarray analysis to identify candidate genes, may also offer shortcuts.
Genetic architecture of the emotionality phenotypes:
The partitioning of the total A/J-B6 genetic divergence over the 20 tested CSSs raises the question of how the sum of the phenotypic differences between the various CSSs and B6 compares to the A/J-B6 disparity for the same trait. In the case of LD transition, the aggregate reduction in activity over 20 CSSs was a 1:49 (mm:ss) increase in time to first emergence, compared to 1:59 for A/J, and a 5:23 decrease in total time in light, compared to 4:44 for A/J. Open-field activity, in contrast, had an aggregate reduction of 604.9 traversed squares, far greater than the entirety of B6 activity (133.6 squares vs. 1.1 squares for A/J). The lack of an additive effect in open-field activity may reflect gene interactions, where multiple B6 alleles are required for the B6 phenotype and substitutions of individual A/J alleles have an epistatic effect on loci from other chromosomes. Analysis of diet-induced obesity in the CSS panel (Singer et al. 2004) also indicated nonadditivity.
A novel finding was the center avoidance of CSS-11 in open-field testing. The combination of high open-field ambulation and aversion to the center of the box has not been previously described. In fact, our findings probably understate the difference between the CSS-11 and B6 phenotypes. Open-field results were scored manually, necessitating the use of a relatively coarse 4 × 4 grid with 15-cm squares that did not distinguish circular paths enclosing roughly the middle two-thirds of the box, as seen with most strains tested, from the CSS-11 behavior of rapid laps around the test area with the sides of the mice in contact with the wall much of the time. Repeating this work in an automated open-field apparatus with greater resolution is likely to indicate a more pronounced phenotypic difference than that revealed in this analysis.
Another unexpected finding was the demonstration of proportionate changes in activity levels when the same strains were tested in two different facilities. The issue of intersite variability in behavioral tests of emotionality has been addressed in several studies (Crabbe et al. 1999; Wahlsten et al. 2003) and found to be considerable (although interstrain differences still proved significant in general and for open-field activity in particular). Those studies, however, treated intersite variability primarily as an obstacle to be avoided. In this case, the testing of inbred strains with intermediate phenotypes in two locations provided a powerful confirmation of putative QTL, but also demonstrated a level of predictability in the effects of laboratory environment that has not been previously described.
CWRU has a large, multi-species facility with very different levels of ambient noise, odor, and humidity compared to its WIBR counterpart, which may account for the difference. Another possible factor is that mice were housed with corncob bedding at CWRU as opposed to the pine shavings used in cages at WIBR. Mice were more difficult to catch by the tail in the coarser corncob bedding and were frequently chased around before being placed in the test apparatus, which may have led to increased liveliness during testing. Interestingly, a distinctive “face-washing” gesture seen at 2.5 min (frequently to the second) in the open-field test with most mice tested at CWRU was rarely observed at WIBR and then only at the end of a 4-min trial.
A recent report (Francis et al. 2003) demonstrated that the difference in open-field performance and other tests of emotionality between B6 and BALB is mediated partly by maternal effects and that the genotype of the dam is therefore significant. In our work, maternal effects are either inseparable from zygotic or adult effects, as in the comparison of CSSs to B6, or controlled for, as in the case of backcross or intercross progeny where all individuals in the same cross share the same parental genotype.
This work used the CSS paradigm to identify loci affecting performance in two related tests, reproducing previous results as well as identifying novel loci that had eluded earlier studies. The behavioral differences between the B6 and A/J inbred strains go well beyond emotionality (Festing 1979; Crawley et al. 1997), however, affecting learning, aggression, prepulse inhibition, consumption of ethanol and other drugs, and other phenotypes. In fact, in studies of weight gain differences between the CSSs at WIBR, we found it necessary to house CSS-4 males individually because of severe fighting, suggesting the existence of an A/J allele on that chromosome contributing to elevated aggression. The power of the CSS approach to dissect such complex traits provides an opportunity to make new findings across the spectrum of behavioral genetics.
Acknowledgments
We thank Namgyal Dolma for superb maintenance of the Whitehead Institute for Biomedical Research mouse colony, Keith Olszens for assistance in the CWRU facility, and Leeta Green and Maribel Rios for their role in designing and implementing the assays. This work was supported by National Institutes of Health National Center for Research Resources grant RR12305, National Institute of Child Health and Human Development grant HD07518, and National Institute of General Medical Sciences grant GM07250.
References
- Belknap, J. K., 2003. Chromosome substitution strains: some quantitative considerations for genome scans and fine mapping. Mamm. Genome 14**:** 723–732. [DOI] [PubMed] [Google Scholar]
- Cowley, A. W., Jr., R. J. Roman, M. L. Kaldunski, P. Dumas, J. G. Dickhout et al., 2001. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension 37**:** 456–461. [DOI] [PubMed] [Google Scholar]
- Crabbe, J. C., D. Wahlsten and B. C. Dudek, 1999. Genetics of mouse behavior: interactions with laboratory environment. Science 284**:** 1670–1672. [DOI] [PubMed] [Google Scholar]
- Crawley, J. N., 2000 What's Wrong With My Mouse? Behavioral Phenotyping of Transgenic and Knockout Mice. Wiley-Liss, New York.
- Crawley, J. N., J. K. Belknap, A. Collins, J. C. Crabbe, W. Frankel et al., 1997. Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology 132**:** 107–124. [DOI] [PubMed] [Google Scholar]
- Festing, M. F. W., 1979 Inbred Strains in Biomedical Research. Oxford University Press, Oxford.
- File, S. E., and S. Pellow, 1987. Behavioral pharmacology of minor tranquilizers. Pharmacol. Ther. 35**:** 265–290. [DOI] [PubMed] [Google Scholar]
- Flint, J., 2003. a Analysis of quantitative trait loci that influence animal behavior. J. Neurobiol. 54**:** 46–77. [DOI] [PubMed] [Google Scholar]
- Flint, J., 2003. b Animal models of anxiety and their molecular dissection. Semin. Cell Dev. Biol. 14**:** 37–42. [DOI] [PubMed] [Google Scholar]
- Flint, J., R. Corley, J. C. De Fries, D. W. Fulker, J. A. Gray et al., 1995. A simple genetic basis for a complex psychological trait in laboratory mice. Science 269**:** 1432–1435. [DOI] [PubMed] [Google Scholar]
- Francis, D. D., K. Szegda, G. Campbell, W. D. Martin and T. R. Insel, 2003. Epigenetic sources of behavioral differences in mice. Nat. Neurosci. 6**:** 445–446. [DOI] [PubMed] [Google Scholar]
- Gershenfeld, H. K., and S. M. Paul, 1997. Mapping quantitative trait loci for fear-like behaviors in mice. Genomics 46**:** 1–8. [DOI] [PubMed] [Google Scholar]
- Gershenfeld, H. K., P. E. Neumann, C. Mathis, J. N. Crawley, X. Li et al., 1997. Mapping quantitative trait loci for open-field behavior in mice. Behav. Genet. 27**:** 201–210. [DOI] [PubMed] [Google Scholar]
- Gorham, J. D., M. L. Guler, R. G. Steen, A. J. Mackey, M. J. Daly et al., 1996. Genetic mapping of a murine locus controlling development of T helper 1/T helper 2 type responses. Proc. Natl. Acad. Sci. USA 93**:** 12467–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumproglou, R., T. M. Wilkes, P. Townson, X. Y. Wang, J. Beynon et al., 2002. STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J. 31**:** 355–364. [DOI] [PubMed] [Google Scholar]
- Lander, E., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11**:** 241–247. [DOI] [PubMed] [Google Scholar]
- Liang, M., B. Yuan, E. Rute, A. S. Greene, A. P. Zou et al., 2002. Renal medullary genes in salt-sensitive hypertension: a chromosomal substitution and cDNA microarray study. Physiol. Genomics 8**:** 139–149. [DOI] [PubMed] [Google Scholar]
- Manly, K. F., and J. M. Olson, 1999. Overview of QTL mapping software and introduction to map manager QT. Mamm. Genome 10**:** 327–334. [DOI] [PubMed] [Google Scholar]
- Mathis, C., S. M. Paul and J. N. Crawley, 1994. Characterization of benzodiazepine-sensitive behaviors in the A/J and C57BL/6J inbred strains of mice. Behav. Genet. 24**:** 171–180. [DOI] [PubMed] [Google Scholar]
- Mathis, C., P. E. Neumann, H. Gershenfeld, S. M. Paul and J. N. Crawley, 1995. Genetic analysis of anxiety-related behaviors and responses to benzodiazepine-related drugs in AXB and BXA recombinant inbred mouse strains. Behav. Genet. 25**:** 557–568. [DOI] [PubMed] [Google Scholar]
- Matin, A., G. B. Collin, Y. Asada, D. Varnum and J. H. Nadeau, 1999. Susceptibility to testicular germ-cell tumours in a 129.MOLF-Chr 19 chromosome substitution strain. Nat. Genet. 23**:** 237–240. [DOI] [PubMed] [Google Scholar]
- Mott, R., C. J. Talbot, M. G. Turri, A. C. Collins and J. Flint, 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97**:** 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Phenome Database, 2001 http://www.jax.org/phenome/.
- Nadeau, J. H., J. B. Singer, A. Matin and E. S. Lander, 2000. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 24**:** 221–225. [DOI] [PubMed] [Google Scholar]
- Pellow, S., and S. E. File, 1986. Anxiolytic and anxiogenic drug effects on exploratory activity in an elevated plus-maze: a novel test of anxiety in the rat. Pharmacol. Biochem. Behav. 24**:** 525–529. [DOI] [PubMed] [Google Scholar]
- Pellow, S., P. Chopin, S. E. File and M. Briley, 1985. Validation of open:closed arm entries in an elevated plus-maze as a measure of anxiety in the rat. J. Neurosci. Methods 14**:** 149–167. [DOI] [PubMed] [Google Scholar]
- Singer, J. B., A. E. Hill, L. C. Burrage, K. R. Olszens, J. Song et al., 2004. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304**:** 445–448. [DOI] [PubMed] [Google Scholar]
- Talbot, C. J., A. Nicod, S. S. Cherny, D. W. Fulker, A. C. Collins et al., 1999. High-resolution mapping of quantitative trait loci in outbred mice. Nat. Genet. 21**:** 305–308. [DOI] [PubMed] [Google Scholar]
- Turri, M. G., C. J. Talbot, R. A. Radcliffe, J. M. Wehner and J. Flint, 1999. High-resolution mapping of quantitative trait loci for emotionality in selected strains of mice. Mamm. Genome 10**:** 1098–1101. [DOI] [PubMed] [Google Scholar]
- Turri, M. G., S. R. Datta, J. DeFries, N. D. Henderson and J. Flint, 2001. a QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr. Biol. 11**:** 725–734. [DOI] [PubMed] [Google Scholar]
- Turri, M. G., N. D. Henderson, J. C. DeFries and J. Flint, 2001. b Quantitative trait locus mapping in laboratory mice derived from a replicated selection experiment for open-field activity. Genetics 158**:** 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, C. M., E. J. Kulbokas, III, A. W. Kirby, M. C. Zody, J. C. Mullikin et al., 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420**:** 574–578. [DOI] [PubMed] [Google Scholar]
- Wahlsten, D., P. Metten, T. J. Phillips, S. L. Boehm, II, S. Burkhart-Kasch et al., 2003. Different data from different labs: lessons from studies of gene-environment interaction. J. Neurobiol. 54**:** 283–311. [DOI] [PubMed] [Google Scholar]
- Youngren, K. K., J. H. Nadeau and A. Matin, 2003. Testicular cancer susceptibility in the 129.MOLF-Chr19 mouse strain: additive effects, gene interactions and epigenetic modifications. Hum. Mol. Genet. 12**:** 389–398. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and H. K. Gershenfeld, 2003. Genetic contributions to body weight in mice: relationship of exploratory behavior to weight. Obes. Res. 11**:** 828–838. [DOI] [PubMed] [Google Scholar]