Intron Size and Exon Evolution in Drosophila (original) (raw)
Abstract
We have found a negative correlation between evolutionary rate at the protein level (as measured by _d_N) and intron size in Drosophila. Although such a relation is expected if introns reduce Hill-Robertson interference within genes, it seems more likely to be explained by the higher abundance of _cis_-regulatory elements in introns (especially first introns) in genes under strong selective constraints.
NONCODING DNA is a major component of eukaryotic genomes but we know little about the forces that affect its evolution. In particular, intron size varies within the genome and among genomes, but the reasons for this are unclear. Intron size is influenced by various factors (Comeron 2001; Duret 2001): the insertion of transposable elements (Bartolomé et al. 2002; G. Marais, unpublished data), the presence of regulatory elements controlling gene expression (Bergman and Kreitman 2001), the presence of RNA genes (Maxwell and Fournier 1995) or RNA involved in gene regulation (e.g., miRNA) (Mattick 2001), the frequency and size of deletion events (Petrov et al. 2000; Petrov 2002), selection for reducing the energetic cost of transcription (Carvalho and Clark 1999; Castillo-Davis et al. 2002), selection for keeping active chromosomal domains relatively small (Prachumwat et al. 2004), and reduction in Hill-Robertson interference between exons (Comeron and Kreitman 2000).
Hill-Robertson interference occurs when several genetically linked sites are undergoing selection at the same time (Hill and Robertson 1966; Gordo and Charlesworth 2001). When advantageous alleles initially arise in the population, they will usually not be associated with each other, because mutations appear at random in separate individuals. In the absence of recombination, one advantageous mutation will therefore tend to displace all the others (Fisher 1930; Muller 1932). In the presence of recombination, advantageous alleles can be combined together to generate the optimal genotype. A similar argument can be made for the effects of recurrent deleterious mutations on the spread of advantageous alleles (Fisher 1930; Charlesworth 1994; Peck 1994; Orr 2000). Selection is thus expected to be more efficient in the presence of recombination than in its absence. Hence, selective events occurring in one region of the genome would be facilitated if there were an enhancer of recombination nearby. Introns could act as such enhancers, because they increase the chance that a crossover occurs between sites in two different exons by spacing them apart, thereby allowing more efficient selection on variants in different coding regions of the same gene (Comeron and Kreitman 2000).
If such interference is important, we would expect the efficacy of selection to be greater in genes with larger introns, all else being equal. Comeron and Kreitman designed a test based on the effect of intron size on selection on codon usage in Drosophila melanogaster (Comeron and Kreitman 2002). This type of selection is believed to be very weak (N_e_s ∼ 1, where _N_e is the effective population size and s the selection coefficient against a mutation to a nonoptimal codon), and is thus particularly prone to generating interference effects, because the chance that several synonymous sites are segregating in the same gene at the same time is high (Gordo and Charlesworth 2001). They found that the average level of codon bias among genes drawn from the D. melanogaster genome sequence was not affected by the presence/absence of introns. They then examined codons located in the middle of the gene (called “central” codons), which are more subject to interference because they have more neighboring codons. Comeron and Kreitman (2002) found that the level of codon bias for these central codons was slightly but significantly increased in genes with central introns, compared with genes lacking such introns, in agreement with the interference hypothesis.
Introns could also reduce interference between weakly selected mutations at amino acid sites within the same gene. This implies that the rate of nonsynonymous substitutions per site (_d_N) would be influenced by intron size. However, the extent to which _d_N is influenced by purifying selection vs. positive selection is unclear (Akashi 1999; Hurst 2002). If protein sequence evolution is caused mainly by the fixation of advantageous, weakly selected mutations (positive selection), the correlation between _d_N and intron size should be positive. In contrast, if it is driven by the fixation by drift of weakly selected, deleterious mutations (purifying selection), the correlation should be negative (Hurst 2002). To distinguish between these two alternatives and test for an effect of intron size on _d_N, we used 630 orthologous gene pairs from D. melanogaster and D. yakuba (from Domazet-Loso and Tautz 2003) to estimate _d_N using PAML with default parameters (Goldman and Yang 1994). Further details are given in Marais et al. (2004) (the data set can be downloaded at http://biomserv.univ-lyon1.fr/~marais/dataIntronSize/). We examined the correlation between _d_N values and intron size in D. melanogaster. We used only the 570 genes that are likely to be located in regions of high recombination in this species (Marais et al. 2004), since genes in regions of low recombination (near the centromeres or telomeres and on chromosome 4) are known to accumulate transposable elements in their noncoding regions, and their intron sizes may have unusual evolutionary dynamics (Bartolomé et al. 2002; Rizzon et al. 2002). The results did not differ significantly when gene pairs from regions of low recombination were included (data not shown).
We find that _d_N is (1) almost two times lower in genes with introns than in genes without introns (a nonparametric Kolmogorov test was significant with P = 0.02, see Figure 1A) and (2) negatively correlated with total intron size (Spearman nonparametric correlation coefficient _R_s = −0.19, P < 10−4, see Figure 1B). Figure 1C shows that there are clear-cut differences in the mean _d_N values among intron size categories and that the observed correlation is not due to the effects of outliers. A similar relation was found between _d_N/_d_S and intron size (_R_s = −0.10, _P_ < 10−4), where _d_S is the rate of synonymous substitution per site. This eliminates the possibility that a correlation between point mutation rates (reflected in _d_S) and deletion rates (potentially affecting intron size) explains the results. Total intron size is influenced by both individual intron size and the number of introns. We have also defined a new index (relative distance between sites, RDS), which gives a better measure of the effect of introns on the distance between codons within a gene. It is the sum of the pairwise distances (in bases) for all codons within a gene, divided by the sum of pairwise distances for all codons within the coding sequence with the introns spliced out. A gene without introns would have RDS = 1, and a gene with introns would have RDS > 1, with a value that depends on the number, position, and size of these introns. We find a slightly stronger correlation of _d_N with RDS than with intron size (_R_s = −0.24, P < 10−4).
Figure 1.—
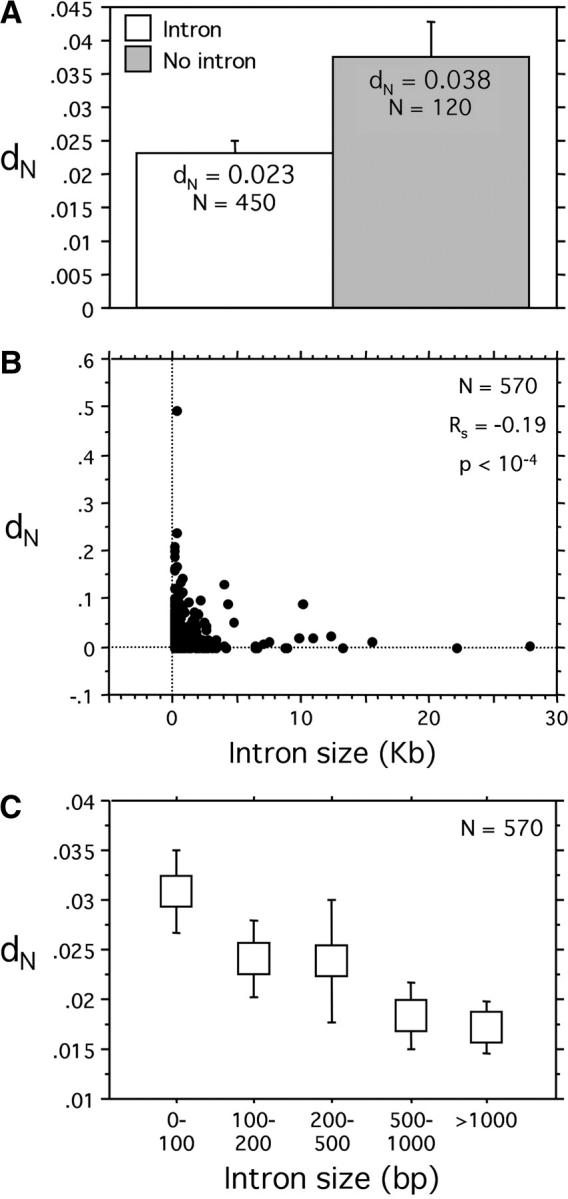
The relationship between intron size and rate of nonsynonymous substitutions per site (_d_N) in D. melanogaster. (A) _d_N is 1.6 times lower in genes with introns than in genes without introns (a nonparametric Kolmogorov, P = 0.02). As in the rest of the article, we used nonparametric statistics because most of the variables with which we are dealing do not follow the normal distribution. Error bars are 95% confidence intervals. (B) _d_N is negatively correlated with total intron size (Spearman nonparametric correlation coefficient _R_s = −0.19, P < 10−4). (C) _d_N for different intron size categories (each category contains ∼20% of the genes). Error bars are 95% confidence intervals.
All the above results are consistent with the interference hypothesis. At first sight, they suggest that (1) purifying selection is the main determinant of _d_N and (2) purifying selection is stronger in the presence of introns. In other words, weakly deleterious mutations at amino acid sites in the same gene seem to be more effectively eliminated when the gene possesses introns. But we need to consider alternative hypotheses. The hypotheses of selection against the energetic cost of introns (Carvalho and Clark 1999; Castillo-Davis et al. 2002) and of selection against large introns in active chromosomal domains (Prachumwat et al. 2004) both predict a negative genome-wide correlation between intron size and expression level, which has indeed been observed in Caenorhabditis elegans and humans (Castillo-Davis et al. 2002). Using a previously published data set on intron size and level of gene expression [estimated from expressed sequence tag (EST) data], compiled for the complete genome of D. melanogaster (Marais and Piganeau 2002), we find that intron size is negatively correlated with expression level in this species as well, although this correlation is very weak (_R_s = −0.01, P = 0.01). On the other hand, it is well known that protein evolution is related to gene expression: highly expressed genes tend to evolve more slowly in mammals (Duret and Mouchiroud 2000) and Drosophila (Marais et al. 2004). However, if the correlation between _d_N and intron size that we have detected is a by-product of gene expression, we should observe a positive correlation between _d_N and intron size, given the correlations between these parameters and expression level. Selection for reduced intron size (because of energetic costs or chromosomal domain size) thus does not seem to explain our results.
An alternative explanation involves the presence in introns of regulatory elements controlling gene expression. In particular, if the most conserved genes have more such elements (since the levels of expression of such genes may need to be more precisely controlled), we would expect a negative relationship between _d_N and intron size. Regulatory elements are more frequent in the first introns than in other introns in mammals (Majewski and Ott 2002; Keightley and Gaffney 2003; Chamary and Hurst 2004) and possibly also in Drosophila (Duret 2001). In agreement with this, first introns are almost two times larger than other introns in vertebrates and Drosophila (Duret 2001), which is also true for our data set (first introns, mean of 518 bp; others, mean of 294 bp; P < 10−4 on a Kolmogorov test). Second, we found that their size is significantly positively correlated with expression level (_R_s = 0.22, P < 10−4), whereas other introns do not show a significant correlation (_R_s = 0.10, NS). This is confirmed by an analysis of the whole genome (Figure 2). Third, by analyzing a previously published alignment of 163 introns from D. melanogaster and D. simulans (Halligan et al. 2004), we found that intron divergence is significantly negatively correlated with size for first introns (_R_s = −0.29, P = 0.03); although there is a negative correlation for the other introns, it is not significant (_R_s = −0.14, NS).
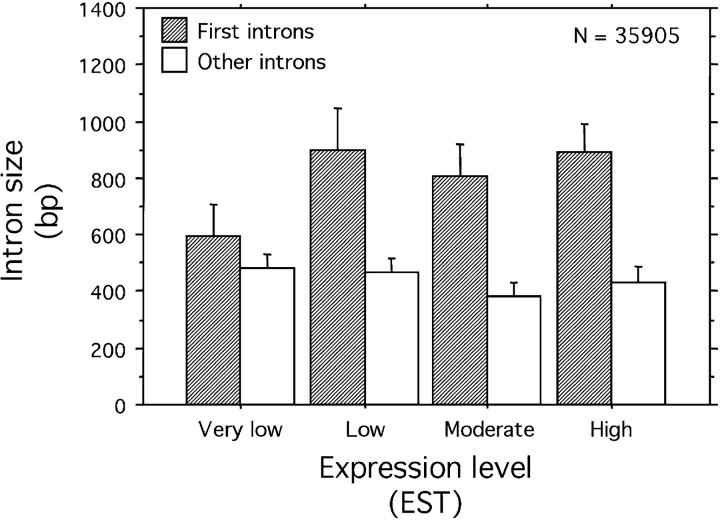
Figure 2.—
The relationship between intron size and expression level (estimated by EST counting) for the complete genome of D. melanogaster (data set from Marais and Piganeau 2002). For first introns, _R_s = 0.06, P < 10−4. For other introns, _R_s = −0.03, NS. Only introns located in regions of high recombination were included (the total number of these introns is shown). Error bars are 95% confidence intervals.
To test this hypothesis further, we examined the relationship between protein evolution and intron size for first and other introns separately. For first introns, we observe a similar correlation to that for all introns (_R_s = −0.20, P < 10−4, n = 450), but there is no significant correlation for the other introns (_R_s = −0.06, NS, n = 302). Moreover, the trend is still visible when only genes with more than one intron are included (for first introns _R_s = −0.15, P < 10−4, n = 302). This result is striking, because it suggests that the presence of regulatory elements within introns is the most likely explanation for the association between protein evolution and intron size in Drosophila, since we do not expect such a result with the alternative hypotheses, including that of Hill-Robertson interference. However, first introns may have the side effect of increasing recombination within a gene. Indeed, they contribute 54% of the variability in total intron length, so that most variation in intron size is due to the first introns. To test this, we compared _d_N and intron size after removing the effects of gene expression, but did not find a significant correlation (_R_s = −0.08, NS), suggesting that the presence of regulatory elements within introns may be sufficient to explain our results.
Our observations do not appear to support the interference hypothesis, but do not allow us to rule it out. The extent of Hill-Robertson interference between amino acid sites under selection is not very well understood, either theoretically or empirically. Some recent work suggests that such interference may explain an apparent relationship between recombination rate and _d_N in a comparison of D. melanogaster and D. simulans (Betancourt and Presgraves 2002), but the meaning of this relationship has been recently challenged (Marais and Charlesworth 2003; Marais et al. 2004). If there is little or no interference between amino acid sites within the same gene, introns would have an effect only on the efficacy of selection on synonymous sites within a gene. However, this effect is very weak. Previous work shows that introns are associated with a change in mean frequency of optimal codons from 64 to 68% and only for a subset of codons (the central ones, see above). This is in agreement with the very weak correlation between intron size and recombination rates reported previously (Carvalho and Clark 1999; Comeron and Kreitman 2000) and suggests that interference explains only a very small fraction of variability in intron size in eukaryotic genomes.
We have found that intron size is globally negatively correlated with expression level in Drosophila, as reported for other eukaryotes (Castillo-Davis et al. 2002). However, when we split introns into first introns vs. the others, we found that first intron size is significantly positively correlated with expression level. This does not disagree with the hypotheses of selection for reducing the cost of transcribing introns (Carvalho and Clark 1999; Castillo-Davis et al. 2002) and selection against large introns in active chromosomal domains (Prachumwat et al. 2004), which were proposed to explain the negative relationship between intron size and expression level. It means simply that Drosophila first introns do not follow the general trend, probably because these introns are enriched in regulatory elements, which appear to be more frequent in highly expressed genes. However, this does not seem to be the case in humans, where first introns are smaller in ubiquitously expressed genes than in narrowly expressed genes, although the difference is much smaller than that for other introns (Comeron 2004). Further investigation is needed to understand this difference between Drosophila and humans.
Our results suggest that genes with more slowly evolving amino acid sequence (low _d_N) may also have more regulatory elements, particularly in their first introns, and that this generates the observed relationship between _d_N and intron size. It has already been shown that highly conserved genes have special expression patterns. Duret and Mouchiroud (2000) showed that these genes are much more broadly expressed than others in mammals. They suggested that this is because mutations in housekeeping genes affect more tissues than mutations in tissue-specific genes and will therefore have larger effects on fitness. This would cause them to be much more constrained, although other explanations are possible (Akashi 2001, 2003). More recently, Castillo-Davis et al. (2004) have shown that protein sequence divergence is correlated with that of _cis_-regulatory elements. To measure the latter, they defined a new index, _d_SM (the fraction of both noncoding sequences that does not contain a region of significant alignment), and computed it for a set of aligned genomic sequences from C. elegans and C. briggsae. They showed that (1) it correlates positively with expression differences between nematodes, (2) shared sequences correspond to experimentally known motives for gene expression, and (3) _d_SM is large in nonpromoter intergenic regions. They then observed that _d_SM and _d_N are positively correlated in nematodes, suggesting that selective pressures on gene expression and protein sequence evolution are coupled (Castillo-Davis et al. 2004). A similar conclusion has been reached on different grounds for Drosophila (Nuzhdin et al. 2004). This is entirely consistent with our observations and with the idea that selection for the presence of regulatory elements can affect the evolution of intron size.
Acknowledgments
We thank Peter Andolfatto and Laurent Duret for their comments on the manuscript. G.M. was a European Union Marie Curie Fellow, and B.C. is supported by the Royal Society.
References
- Akashi, H., 1999. Within- and between-species DNA sequence variation and the ‘footprint’ of natural selection. Gene 238**:** 39–51. [DOI] [PubMed] [Google Scholar]
- Akashi, H., 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11**:** 660–666. [DOI] [PubMed] [Google Scholar]
- Akashi, H., 2003. Translational selection and yeast proteome evolution. Genetics 164**:** 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé, C., X. Maside and B. Charlesworth, 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19**:** 926–937. [DOI] [PubMed] [Google Scholar]
- Bergman, C. M., and M. Kreitman, 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11**:** 1335–1345. [DOI] [PubMed] [Google Scholar]
- Betancourt, A. J., and D. C. Presgraves, 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99**:** 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, A. B., and A. G. Clark, 1999. Intron size and natural selection. Nature 401**:** 344. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C. I., S. L. Mekhedov, D. L. Hartl, E. V. Koonin and F. A. Kondrashov, 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31**:** 415–418. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C. I., D. L. Hartl and G. Achaz, 2004. _Cis_-regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 14**:** 1530–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary, J. V., and L. D. Hurst, 2004. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol. Biol. Evol. 21**:** 1014–1023. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63**:** 213–227. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., 2001. What controls the length of noncoding DNA? Curr. Opin. Genet. Dev. 11**:** 652–659. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., 2004. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics 167**:** 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., and M. Kreitman, 2000. The correlation between intron length and recombination in Drosophila: dynamic equilibrium between mutational and selective forces. Genetics 156**:** 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., and M. Kreitman, 2002. Population, evolutionary and genomic consequences of interference selection. Genetics 161**:** 389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso, T., and D. Tautz, 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13**:** 2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., 2001. Why do genes have introns? Recombination might add a new piece to the puzzle. Trends Genet. 17**:** 172–175. [DOI] [PubMed] [Google Scholar]
- Duret, L., and D. Mouchiroud, 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17**:** 68–74. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1930 The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11**:** 725–736. [DOI] [PubMed] [Google Scholar]
- Gordo, I., and B. Charlesworth, 2001. Genetic linkage and molecular evolution. Curr. Biol. 11**:** R684–R686. [DOI] [PubMed] [Google Scholar]
- Halligan, D. L., A. Eyre-Walker, P. Andolfatto and P. D. Keightley, 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14**:** 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on limits to artificial selection. Genet Res. 8**:** 269–294. [PubMed] [Google Scholar]
- Hurst, L. D., 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18**:** 486. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and D. J. Gaffney, 2003. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proc. Natl. Acad. Sci. USA 100**:** 13402–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski, J., and J. Ott, 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12**:** 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G., and B. Charlesworth, 2003. Genome evolution: recombination speeds up adaptive evolution. Curr. Biol. 13**:** R68–R70. [DOI] [PubMed] [Google Scholar]
- Marais, G., and G. Piganeau, 2002. Hill-Robertson interference is a minor determinant of variations in codon bias across Drosophila melanogaster and Caenorhabditis elegans genomes. Mol. Biol. Evol. 19**:** 1399–1406. [DOI] [PubMed] [Google Scholar]
- Marais, G., T. Domazet-Loso, D. Tautz and B. Charlesworth, 2004. Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J. Mol. Evol. 59**:** 771–779. [DOI] [PubMed] [Google Scholar]
- Mattick, J. S., 2001. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2**:** 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, E. S., and M. J. Fournier, 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64**:** 897–934. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1932. Some genetic aspects of sex. Am. Nat. 66**:** 118–138. [Google Scholar]
- Nuzhdin, S. V., M. L Wayne, K. L Harmon and L. M McIntyre, 2004. Common patterns of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21**:** 1308–1317. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2000. The rate of adaptation in asexuals. Genetics 155**:** 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, J. R., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137**:** 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. DNA loss and evolution of genome size in Drosophila. Genetica 115**:** 81–91. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., T. A. Sangster, J. S. Johnston, D. L. Hartl and K. L. Shaw, 2000. Evidence for DNA loss as a determinant of genome size. Science 287**:** 1060–1062. [DOI] [PubMed] [Google Scholar]
- Prachumwat, A., L. DeVincentis and M. F. Palopoli, 2004. Intron size correlates positively with recombination rate in Caenorhabditis elegans. Genetics 166**:** 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon, C., G. Marais, M. Gouy and C. Biémont, 2002. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12**:** 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]