Lineage-Specific Loss of Function of Bitter Taste Receptor Genes in Humans and Nonhuman Primates (original) (raw)
Abstract
Since the process of becoming dead genes or pseudogenes (pseudogenization) is irreversible and can occur rather rapidly under certain environmental circumstances, it is one plausible determinant for characterizing species specificity. To test this evolutionary hypothesis, we analyzed the tempo and mode of duplication and pseudogenization of bitter taste receptor (T2R) genes in humans as well as in 12 nonhuman primates. The results show that primates have accumulated more pseudogenes than mice after their separation from the common ancestor and that lineage-specific pseudogenization becomes more conspicuous in humans than in nonhuman primates. Although positive selection has operated on some amino acids in extracellular domains, functional constraints against T2R genes are more relaxed in primates than in mice and this trend has culminated in the rapid deterioration of the bitter-tasting capability in humans. Since T2R molecules play an important role in avoiding generally bitter toxic and harmful substances, substantial modification of the T2R gene repertoire is likely to reflect different responses to changes in the environment and to result from species-specific food preference during primate evolution.
NO doubt, organisms have increasingly acquired new genes by tandem duplication and/or polyploidization (Ohno 1970). However, as evolution has proceeded, loss of genes has also occurred whenever they became dispensable under certain circumstances of interplay between genes and their environments. In animals, nutrition that is ingested daily from surroundings must have been one of the most important environmental factors and must have permitted genes involved in vitamin C biosynthesis (Nishikimi et al. 1994) and essential amino acid biosynthesis (Lehninger 1996) to be nonfunctional. A similar phenomenon may be found in genes for the sense of taste since tasting plays a crucial role in providing animals with information about proper assessments of foods and appropriate behaviors. Mammals can basically sense tastes of sweet, sour, bitterness, salt, and umami (the taste of monosodium glutamate). Of these five modalities, sweet, bitter, and umami substances are perceived by G-protein-coupled receptor (GPCR) signaling pathways (Wong et al. 1996). Two GPCR families are involved in these pathways: One is T1R, which is associated with sweet and umami substances (Nelson et al. 2001; Li et al. 2002), and the other is T2R, which is associated with bitter substances (Adler et al. 2000; Chandrashekar et al. 2000; Matsunami et al. 2000). Unfortunately, however, the correspondence between bitter substances (ligands) and receptors is poorly known. One exception is one of the human T2R genes, which is proven to be responsible for perception of phenylthiocarbamide (Kim et al. 2003).
Both T1R and T2R have seven transmembrane domains as a common character of GPCRs. Despite this structural similarity, these two molecules exhibit no obvious amino acid sequence similarities. Rather, T1R and T2R are closely related to the pheromone receptors V2R and V1R, respectively (Matsunami and Amrein 2003). T2R genes form a larger multigene family than T1R genes. In contrast to the presence of only 3 T1R genes in the mammalian genome (Nelson et al. 2001; Li et al. 2002), >30 T2R genes exist (Conte et al. 2003; Shi et al. 2003). The recent chicken genome project (Hillier et al. 2004) ascertained that the T2R gene repertoire had expanded in the ancestral mammalian lineage after its divergence from reptiles/birds and that the capacity for sensing bitter substances has broadened in mammals relative to chickens, which possess only 3 T2R genes. Between mammalian orders such as humans and mice, the orthologous relationships of T2R genes are found to be either one to one or one to multiple (Conte et al. 2003; Shi et al. 2003). Since the one-to-multiple orthology has resulted from lineage-specific gene duplication, the T2R gene family has continuously undergone gene duplication even after the mammalian radiation. It is also known that the human and mouse T2R gene families contain eight and three pseudogenes, respectively (Conte et al. 2003; Shi et al. 2003). The absence of orthologous pseudogenes between humans and mice immediately indicates that pseudogenization [the process of becoming pseudogenes by disrupting intact open reading frames (ORFs)] of T2R genes occurred independently in mammalian lineages. Since pseudogenization can occur rather rapidly under certain circumstances, we can regard it as one possible determinant for characterizing species specificity. This view was actually exemplified by the olfactory receptor (OR) gene (Gilad et al. 2003, 2004) and pheromone receptor (VR) gene families (Zhang and Webb 2003). In these families, more pseudogenes have accumulated in hominoids and Old World monkeys (OWMs) than in other mammals and it has been argued that the transition from smell or pheromone dependency to full trichromatic color vision dependency in their life might have relaxed functional constraints against OR and VR genes (Gilad et al. 2003, 2004; Zhang and Webb 2003).
Because of important roles of T2R in avoiding generally bitter, toxic, and harmful substances, changes in the T2R gene repertoire could in turn affect the behavior of animals, especially the feeding behavior. Here, to gain insights into evolutionary changes in the T2R gene repertoire, we have conducted a comparative analysis of T2R genes among humans, nonhuman primates, tupais, and mice. We show that lineage-specific gene duplication and pseudogenization played important roles in altering the T2R gene repertoire in individual primates.
MATERIALS AND METHODS
Sources of DNA samples:
Genomic DNAs used here were for the chimpanzee (Pan troglodytes; Patr), the gorilla (Gorilla gorilla; Gogo), the orangutan (Pongo pygmaeus; Popy), the agile gibbon (Hylobates agilis; Hyag), the rhesus monkey (Macaca mulatta; Mamu), the silvered leaf monkey (Trachypithecus cristatus; Trcr), the white-tufted-ear marmoset (Callithrix jacchus; Caja), the brown capuchin (Cebus apella; Ceap), the thick-tailed bush baby (Otolemur crassicaudatus; Otcr), the Senegal galago (Galago senegalensis; Gase), the slow loris (Nycticebus coucang; Nyco), the ring-tailed lemur (Lemur catta; Leca), and the common tree shrew (Tupaia glis; Tugl). These DNAs were generously provided by Primate Research Institute, Kyoto University. Genomic DNAs for human (Homo sapiens; Hosa) populations were purchased from Coriell (Camden, NJ).
PCR amplification and sequencing:
Genomic PCR was performed with the polymerase KOD-Plus under conditions recommended by the manufacturer (TOYOBO). Because all T2R genes are intronless and ∼900 bp in size, the nucleotide sequences of the entire coding region were obtained by single PCR. The first primer sets for PCR were designed for each T2R coding region to amplify ORFs as long as possible. For some closely related T2R genes, degenerated primers were designed. In case of failure of amplification by the first primer sets, other primer sets were designed for conserved regions between human and mouse T2R homologous genes. The primer sequences are available in supplemental Table 1 at http://www.genetics.org/supplemental/. Amplifications were carried out under the following standard conditions: denaturation at 94° for 3 min; 35 cycles of 94° for 20 sec, 45°–52° for 30 sec, and 68° for 30 sec with an additional extension at 68° for 5 min. PCR products were electrophoresed using 1.5% agarose gel and purified by the QIAquick gel extraction kit (QIAGEN, Chatsworth, CA). PCR products were directly sequenced or sequenced after cloning (Zero Blunt TOPO PCR cloning kit, QIAGEN).
Data analysis:
Sequences were aligned by CLUSTAL_X (Thompson et al. 1997) and the resulting alignment was manually adjusted. Codon positions specified in this study correspond to those in the aligned T2R sequences, in which the numbering starts at the first codon of Hosa-T2R39. Phylogenetic trees were reconstructed by the neighbor-joining (NJ) (Saitou and Nei 1987) and/or maximum parsimony (Fitch 1971; Hartigan 1973) methods implemented in MEGA2 (Kumar et al. 2001). In the NJ trees, the p distances for nucleotide or amino acid sequences were used to infer the topology (Saitou and Nei 1987). To examine whether natural selection has been exerted on individual amino acids, the computer program of DataMonkey (http://www.datamonkey.org/) was used (Kosakovsky Pond and Frost 2005). This program involves five steps. First, to obtain a reliable estimate of the number of substitutions occurred at a single amino acid site, a nucleotide substitution model fitted to the data was selected by the likelihood ratio test and/or Akaike information criteria score. Second, to obtain ancestral sequences, a codon-based substitution model fitted to the data was selected using the maximum-likelihood method with the MG94 model (Muse and Gaut 1994). Third, the average numbers of synonymous (_E_S) and nonsynonymous (_E_N) sites in a particular phylogenic tree for each codon were computed, as in Suzuki and Gojobori (1999), by considering the nucleotide substitution biases estimated from the data. Fourth, the total numbers of synonymous (_N_S) and nonsynonymous (_N_N) substitutions throughout the phylogenetic tree were counted for each codon site. Finally, we tested whether the number of nonsynonymous substitutions per nonsynonymous site (_d_N; _N_N/_E_N) is significantly different from the number of synonymous substitutions per synonymous site (_d_S; _N_S/_E_S) using the extended binominal distribution. In addition, to estimate the numbers of per-site synonymous substitutions (_b_S) and per-site nonsynonymous substitutions (_b_N) for each branch of a given tree, the computer program Bn-Bs (Zhang et al. 1998) was used.
RESULTS
Comparative analysis of T2R genes between humans and mice:
Previous studies identified 33–∼34 T2R genes in humans and 36–∼40 in mice (Conte et al. 2002, 2003; Shi et al. 2003). We reexamined T2R genes for the human (NCBI; Build 34 Version 2) and the mouse (NCBI; Build 32 Version 1) genome sequences by BLASTN and TBLASTN with all annotated human and mouse T2R genes as queries. These database searches identified 1 additional mouse T2R pseudogene (mt2r37*; an asterisk indicates a pseudogene), so that there are 41 paralogous genes in mice, including 6 pseudogenes. In humans, on the other hand, 25 T2R functional genes and 11 pseudogenes were confirmed. Among 11 pseudogenes, 6 were already identified in both Conte et al. (2002) and Shi et al. (2003) and the remaining 5 by one of these studies. The proportion of T2R pseudogenes is 31% (11/36) in humans and 15% (6/41) in mice (Table 1).
TABLE 1.
Nomenclature of the mouse t2r genes and their human orthologous genes
| Mouse t2r genes | Human T2R genes | |||||||
|---|---|---|---|---|---|---|---|---|
| This study | Chromosome | Shi et al. (2003) | Conte et al. (2003) | Category | This study | Chromosome | Shi et al. (2003) | Conte et al. (2002) |
| mt2r1 | 6 | mTAS2R33 | C | hT2R2(*) | 7 | hTAS2R2 | ||
| mt2r2 | 6 | mt2r40 | mTAS2R18 | C | hT2R16 | 7 | ht2r16 | hTAS2R16 |
| mt2r3 | 6 | mt2r41 | mTAS2R37 | C | hT2R3 | 7 | ht2r3 | hTAS2R3 |
| mt2r4 | 6 | mt2r8 | mTAS2R8 | C | hT2R4 | 7 | ht2r4 | hTAS2R4 |
| hT2R66* | 7 | hps3 | hTAS2R6 | |||||
| hT2R5 | 7 | ht2r5 | hTAS2R5 | |||||
| mt2r5 | 6 | mt2r31 | mTAS2R38 | C | hT2R38 | 7 | ht2r38 | hTAS2R61 |
| mt2r6 | 6 | mt2r34 | mTAS2R39 | C | hT2R39 | 7 | ht2r39 | hTAS2R57 |
| mt2r7 | 6 | mt2r33 | mTAS2R44 | C | hT2R40 | 7 | ht2r40 | hTAS2R58 |
| mt2r8 | 6 | mt2r36 | mTAS2R43 | C | hT2R62* | 7 | hps1 | hTAS2R62 |
| mt2r9 | 6 | mt2r38 | mTAS2R35 | C | hT2R56 | 7 | ht2r56 | hTAS2R60 |
| mt2r10 | 6 | mt2r35 | mTAS2R26 | C | hT2R41 | 7 | ht2r41 | hTAS2R59 |
| mt2r11 | 6 | mt2r42 | mTAS2R30 | C | hT2R7 | 12 | ht2r7 | hTAS2R7 |
| hT2R8 | 12 | ht2r8 | hTAS2R8 | |||||
| hT2R9 | 12 | ht2r9 | hTAS2R9 | |||||
| mt2r12 | 6 | mt2r43 | mTAS2R7 | B | ||||
| mt2r13 | 6 | mt2r44 | mTAS2R6 | B | ||||
| mt2r14 | 6 | mt2r45 | mTAS2R4 | B | hT2R10 | 12 | ht2r10 | hTAS2R10 |
| mt2r15 | 6 | mt2r5 | mTAS2R5 | B | ||||
| mt2r16 | 6 | mt2r46 | mTAS2R14 | B | ||||
| A | hT2R15* | 12 | hps8 | hTAS2R15 | ||||
| A | hT2R50 | 12 | ht2r50 | hTAS2R51 | ||||
| A | hT2R49 | 12 | ht2r49 | hTAS2R56 | ||||
| A | hT2R48 | 12 | ht2r48 | hTAS2R55 | ||||
| A | hT2R64* | 12 | hps2 | hTAS2R64 | ||||
| mt2r17 | 6 | mt2r47 | mTAS2R20 | A | hT2R63* | 12 | hps6 | hTAS2R63 |
| mt2r23 | 6 | mt2r52 | mTAS2R36 | A | hT2R68* | 12 | hps7 | hTAS2R68 |
| A | hT2R43 | 12 | ht2r43 | hTAS2R52 | ||||
| A | hT2R44 | 12 | ht2r44 | hTAS2R53 | ||||
| A | hT2R45 | 12 | ht2r45 | hTAS2R50 | ||||
| A | hT2R46 | 12 | ht2r46 | hTAS2R54 | ||||
| A | hT2R47 | 12 | ht2r47 | hTAS2R44 | ||||
| hT2R11* | 12 | hTAS2R11 | ||||||
| mt2r18 | 6 | mt2r48 | mTAS2R21 | B | ||||
| mt2r21 | 6 | mt2r50 | mTAS2R24 | B | hT2R13 | 12 | ht2r13 | hTAS2R13 |
| mt2r22 | 6 | mt2r51 | mTAS2R2 | B | ||||
| mt2r19 | 6 | mTAS2R22 | C | hT2R12* | 12 | hTAS2R12 | ||
| mt2r20 | 6 | mt2r49 | mTAS2R15 | B | ||||
| mt2r24* | 6 | mps3 | mTAS2R45 | B | ||||
| mt2r25 | 6 | mt2r54 | mTAS2R17 | B | ||||
| mt2r26 | 6 | mt2r55 | mTAS2R23 | B | ||||
| mt2r27 | 6 | mt2r56 | mTAS2R16 | B | ||||
| mt2r28 | 6 | mt2r57 | mTAS2R10 | B | ||||
| mt2r29 | 6 | mt2r58 | mTAS2R13 | B | ||||
| mt2r30* | 6 | mTAS2R42 | B | |||||
| mt2r31 | 6 | mt2r59 | mTAS2R25 | B | hT2R14 | 12 | ht2r14 | hTAS2R14 |
| mt2r32* | 6 | mps1 | mTAS2R46 | B | ||||
| mt2r33 | 6 | mt2r60 | mTAS2R29 | B | ||||
| mt2r35 | 6 | mt2r62 | mTAS2R9 | B | ||||
| mt2r36* | 6 | mps2 | mTAS2R11 | B | ||||
| mt2r37* | 6 | B | ||||||
| mt2r38 | 6 | mt2r63 | mTAS2R3 | B | ||||
| mt2r39 | 6 | mt2r64 | mTAS2R40 | B | ||||
| mt2r40* | 6 | mTAS2R41 | B | |||||
| A | hT2R65* | 12 | hps4 | |||||
| mt2r34 | 6 | mt2r61 | mTAS2R31 | A | hT2R67* | 12 | hps5 | |
| A | hT2R55 | 12 | ht2r55 | hTAS2R24 | ||||
| mt2r41 | 15 | mt2r19 | mTAS2R19 | C | hT2R1 | 5 | ht2r1 | hTAS2R1 |
The NJ tree was reconstructed on the basis of amino acid sequences of all human and mouse T2R genes (Figure 1) and the evolutionary relationships between human and mouse T2R genes can be divided into three categories with respect to their orthology (Table 1). In category A, a group of three human genes is orthologous to one mouse gene (multiple-to-one orthology). In addition, there is a group of 12 human genes with two mouse orthologs. For convenience, this group also is classified into category A. In category B, a single human gene is orthologous to a group of mouse genes (one-to-multiple orthology) and there are three such cases. The one-to-one orthologous relationships are found in 13 gene pairs and these are classified into category C (one-to-one orthology). Five uncategorized human genes (hT2R5, hT2R8, hT2R9, hT2R11*, and hT2R66*) remain due to the absence of their counterparts in mice. Among 11 human pseudogenes, 6 (hT2R15*, hT2R64*, hT2R63*, hT2R68*, hT2R65*, and hT2R67*) occur in the 15 category A genes, none in the three category B genes, 3 (hT2R2*, hT2R62*, and hT2R12*) in the 13 category C genes, and 2 (hT2R66* and hT2R11*) in the 5 uncategorized genes. On the other hand, all 6 mouse pseudogenes (mt2r24*, mt2r30*, mt2r32*, mt2r36*, mt2r37*, and mt2r40*) are found in the 25 category B genes; none exist in the 3 category A genes and 13 category C genes.
Figure 1.—
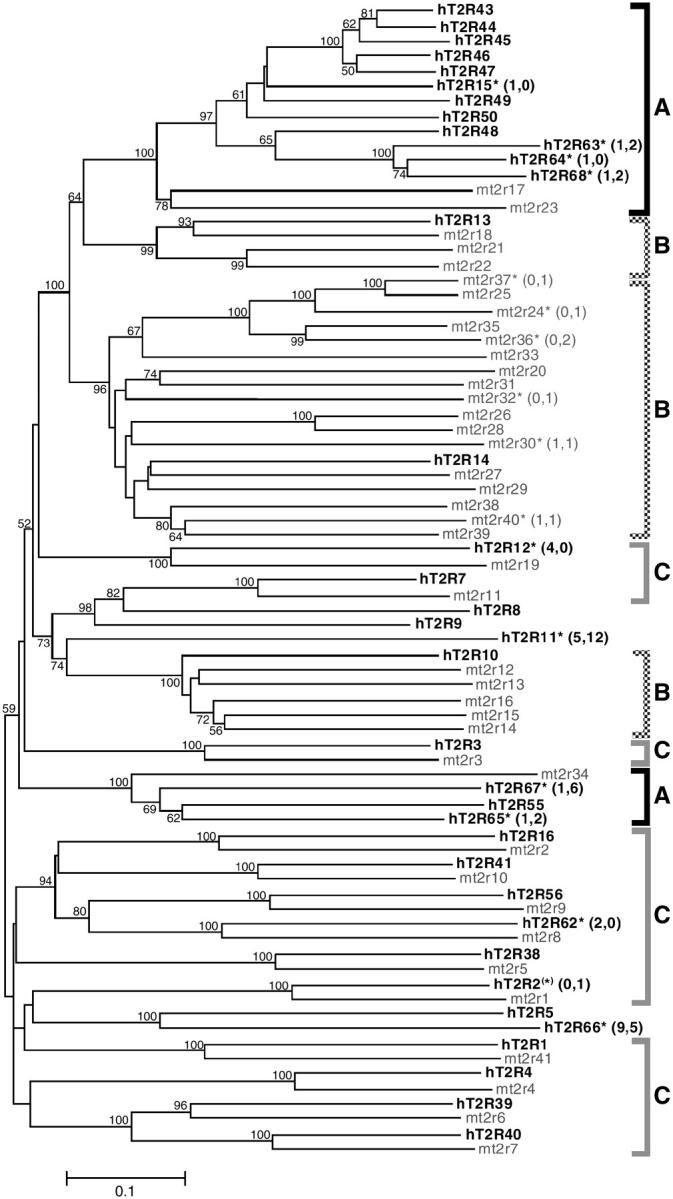
Phylogenetic tree of bitter taste receptor (T2R) genes based on amino acid sequences. Genes in humans and mice are shown as solid and shaded, respectively. The tree was reconstructed by the NJ method (Saitou and Nei 1987) on the basis of the p distances, and only >50% bootstrap values are shown at each node (1000 replications). Pseudogenes in this study are defined by the presence of ORF disruptive mutations and are indicated by asterisks (*). The codon frames in pseudogenes are inferred from those of closely related functional genes. The nomenclature of human T2R genes is the same as that in Conte et al. (2002) and Shi et al. (2003). However, because of the different designation systems of the mouse genes used by two research groups (Conte et al. 2002, 2003; Shi et al. 2003), we have renamed the mouse genes. Detailed nomenclatures in humans and mice are in Table 1. The numbers in parentheses stand for those of nonsense and indel mutations, respectively. The definitions of categories A, B, and C are given in the text.
Primate T2R genes:
To elucidate the T2R gene repertoire in primates, we amplified 348 genes for three humans [Adygei (Eastern European), Japanese (Asian), Biaka (African Pygmies)], 12 nonhuman primates, and tupais by means of the PCR technique [33 genes from Hosa (Adygei); 32, Hosa (Japanese); 33, Hosa (Biaka); 36, Patr; 29, Gogo; 29, Popy; 26, Hyag; 35, Mamu; 26, Trcr; 20, Caja; 21, Ceap; 7, Otcr; 4, Gase; 5, Nyco; 8, Leca; and 4, Tugl]. The length of these amplified sequences ranges from 707 to 960 bp with the average being ∼840 bp. As expected, the number of successfully amplified genes decreases in species distantly related to humans (such as prosimians and tupais) and this reduction is most likely due to mutations in the primer-attached sites. Among anthropoidea [hominoids, OWMs, and New World monkeys (NWMs)] in which at least 20 genes were obtained for each species, there are no significant differences in the proportion of T2R pseudogenes (Table 2, but see later results about differential rates of pseudogenization). However, the proportion differs greatly among the three categories of orthologous relationships. There are 46 pseudogenes among 115 genes in category A, none among 29 genes in category B, and 18 among 125 genes in category C. Fisher's exact test among pairs of categorized genes shows the preferential occurrence of pseudogenes in category A (P < 0.001), or pseudogenes are statistically more abundant in category A genes with extra copies than in category B or C genes without extra copies. Nonetheless, it is worth keeping in mind that in primates, pseudogenization is not a rare event in genes without extra copies (category C genes).
TABLE 2.
Primate T2R genes determined in this study
| Sample | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus_hT2R_ | Chromosomea | mt2rb | Categoryc | Hosa(Ady.) | Hosa(Japan) | Hosa(Bia.) | Patr | Popy | Hyag | Mamu | Trcr | Caja | Ceap | Otcr | Gase | Nyco | Leca | Tugl |
| 1 | 5 | 41 (51%) | C | T2R1 | T2R1 | T2R1 | T2R1 | T2R1 | T2R1 | T2R1 | T2R1 | |||||||
| 2 | 7 | 1 (64%) | C | T2R2 | 0, 1 | T2R2 | T2R2 | T2R2 | T2R2 | T2R2 | T2R2 | T2R2 | ||||||
| 16 | 7 | 2 (52%) | C | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | T2R16 | ||
| 3 | 7 | 3 (68%) | C | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | T2R3 | ||
| 4 | 7 | 4 (66%) | C | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | T2R4 | ||||
| 66 | 7 | 9, 5 | 9, 5 | 9, 5 | 8, 5 | 7, 5 | 7, 5 | 6, 6 | 6, 6 | 7, 6 | 6, 7 | 5, 9 | ||||||
| 6, 6 | 6, 5 | 6, 9 | ||||||||||||||||
| 6, 5 | 6, 7 | |||||||||||||||||
| 5 | 7 | T2R5 | T2R5 | T2R5 | T2R5 | T2R5 | T2R5 | 0, 1 | T2R5 | |||||||||
| 38 | 7 | 5 (65%) | C | T2R38 | T2R38 | T2R38 | T2R38 | T2R38 | T2R38 | T2R38 | T2R38 | T2R38 | ||||||
| 39 | 7 | 6 (54%) | C | T2R39 | T2R39 | T2R39 | T2R39 | 0, 2 | 1, 0 | 0, 2 | 0, 1 | 1, 0 | T2R39 | |||||
| 40 | 7 | 7 (66%) | C | T2R40 | T2R40 | T2R40 | T2R40 | T2R40 | T2R40 | T2R40 | T2R40 | T2R40 | ||||||
| 62 | 7 | 8 (51%) | C | 2, 0 | 2, 0 | 2, 0 | T2R62 | T2R62 | T2R62 | T2R62 | T2R62 | T2R62 | ||||||
| 56 | 7 | 9 (58%) | C | T2R56 | T2R56 | T2R56 | T2R56 | T2R56 | T2R56 | T2R56 | 1, 0 | |||||||
| 41 | 7 | 10 (68%) | C | T2R41 | T2R41 | T2R41 | T2R41 | T2R41 | ||||||||||
| 7 | 12 | 11 (67%) | C | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | T2R7 | ||
| 8 | 12 | T2R8 | T2R8 | T2R8 | T2R8 | T2R8 | T2R8 | T2R8 | T2R8 | T2R8 | 0, 5 | T2R8 | ||||||
| 9 | 12 | T2R9 | T2R9 | T2R9 | T2R9 | T2R9 | T2R9 | T2R9 | 1, 0 | T2R9 | ||||||||
| 10 | 12 | 12 (56%) | B | T2R10 | T2R10 | T2R10 | T2R10 | T2R10 | T2R10 | T2R10 | T2R10 | T2R10 | T2R10 | T2R10-1 | T2R10 | T2R10-1 | T2R10 | 0, 1 |
| T2R10-2 | T2R10-2 | |||||||||||||||||
| 0, 1 | ||||||||||||||||||
| 11 | 12 | C | 5, 12 | 5, 12 | 5, 12 | 6, 13 | 5, 13 | |||||||||||
| 12 | 12 | 19 (47%) | B | 4, 0 | 4, 0 | 4, 0 | 4, 0 | 4, 0 | 3, 3 | 3, 2 | T2R12 | |||||||
| 13 | 12 | 18 (58%) | B | T2R13 | T2R13 | T2R13 | T2R13 | T2R13 | T2R13 | T2R13 | T2R13 | T2R13 | T2R13 | |||||
| 14 | 12 | 27 (50%) | A | T2R14 | T2R14 | T2R14 | T2R14 | T2R14 | T2R14 | |||||||||
| 15 | 12 | 17 (50%) | A | 1, 0 | 1, 0 | 1, 0 | 1, 1 | T2R15 | 0, 3 | 0, 1 | T2R15 | T2R15e | ||||||
| 0, 1 | T2R15e | |||||||||||||||||
| 50 | 12 | 17 (50%) | A | T2R50 | T2R50 | T2R50 | T2R50 | T2R50 | T2R50 | T2R50 | T2R50 | 2, 3 | T2R15e | |||||
| 49 | 12 | 17 (50%) | A | T2R49 | T2R49 | T2R49 | T2R49 | T2R49 | T2R49-1 | T2R15e | ||||||||
| T2R49-2 | T2R15e | |||||||||||||||||
| T2R49-3 | T2R15e | |||||||||||||||||
| 48 | 12 | 17 (49%) | A | T2R48 | T2R48 | T2R48 | T2R48 | T2R48 | T2R15e | |||||||||
| 64 | 12 | 17 (51%) | A | 1, 0 | 1, 0 | 1, 0 | 1, 0 | T2R64 | T2R15e | |||||||||
| 63 | 12 | 17 (45%) | A | 1, 2 | 1, 2 | 1, 2 | 1, 2 | T2R15e | ||||||||||
| 68 | 12 | 17 (47%) | A | 1, 2 | 1, 2 | 1, 2 | 2, 1 | T2R15e | ||||||||||
| 1, 2 | T2R15e | |||||||||||||||||
| 43 | 12 | 17 (51%) | A | T2R43 | T2R43 | T2R43-1 | T2R43 | T2R43d | T2R15e | |||||||||
| T2R43-2 | T2R43d | T2R15e | ||||||||||||||||
| T2R43-3 | T2R43d | T2R15e | ||||||||||||||||
| 44 | 12 | 17 (52%) | A | T2R44 | T2R44 | T2R44 | T2R44 | T2R43d | T2R15e | |||||||||
| 45 | 12 | 17 (50%) | A | T2R45 | T2R45 | T2R45 | T2R45 | T2R45-1 | T2R45-1 | T2R43d | T2R15e | |||||||
| T2R45-2 | T2R45-2 | T2R43d | T2R15e | |||||||||||||||
| T2R45-3 | T2R45-3 | T2R43d | T2R15e | |||||||||||||||
| T2R45-4 | T2R43d | T2R15e | ||||||||||||||||
| T2R45-5 | T2R43d | T2R15e | ||||||||||||||||
| 2, 0 | T2R43d | T2R15e | ||||||||||||||||
| 46 | 12 | 17 (51%) | A | T2R46 | T2R46 | T2R46-1 | T2R43d | T2R15e | ||||||||||
| T2R46-2 | T2R43d | T2R15e | ||||||||||||||||
| 47 | 12 | 17 (50%) | A | T2R47 | T2R47 | T2R47 | T2R47 | T2R47-1 | T2R43d | T2R15e | ||||||||
| T2R47-2 | T2R43d | T2R15e | ||||||||||||||||
| 65 | 12 | 34 (40%) | A | 1, 2 | 1, 2 | 1, 1 | 0, 1 | 0, 2 | 0, 1 | 2, 2 | ||||||||
| 2, 1 | ||||||||||||||||||
| 67 | 12 | 34 (41%) | A | 1, 6 | 1, 6 | 1, 6 | 1, 6 | 2, 6 | 2, 6 | 1, 7 | 2, 8 | 3, 6 | 1, 6 | |||||
| 55 | 12 | 34 (49%) | A | T2R55 | T2R55 | T2R55 | 0, 1 | 0, 1 | 0, 1 | T2R55 | T2R55 | T2R55 | T2R55 | 2, 4 | 0, 1 | |||
| Pseudogene (%) | 30.3 | 34.4 | 27.3 | 28.6 | 19.2 | 21.9 | 34.6 | 26.3 | 23.8 |
To infer the evolutionary history of primate T2R genes, the NJ tree was again reconstructed on the basis of amino acid sequences of all available primate and mouse sequences (Figure 2). Since none of the primates have extra copies of category C genes, duplicability (the ability of loci to be duplicated) appears to be unchanged throughout the primate and mouse lineages. By contrast, duplicability in category A and B genes differs not only between primates and mice, but also within primates. In category B, the T2R10 gene is one such example. There are two hT2R10 orthologs in slow loris (Nyco) and three in bush babies (Otcr) as well as five in mice, whereas there is only one hT2R10 ortholog in nonhuman anthropoidea (Table 2). Duplicability at this locus in prosimians appears to be as high as in mice. In category A, on the other hand, the T2R43 group (T2R15, -43–50, -63, -64, -68) experienced at least 19 duplications in anthropoidea (data not shown). This extensive duplication is in contrast to the case of mice in which only one gene duplication produced mt2r17 and mt2r23, collectively orthologous to the primate T2R43 group (Figures 1 and 2).
Figure 2.—
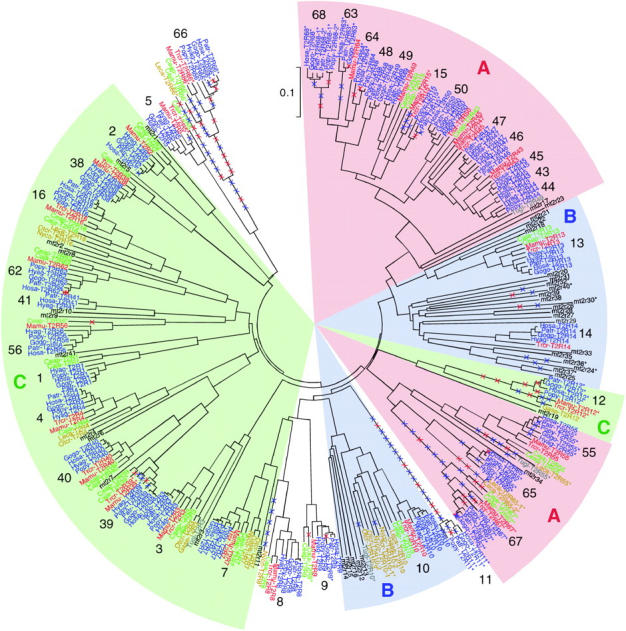
Phylogenetic tree of 305 T2R genes based on amino acid sequences in primates and mice. Genes used are from hominoids (blue), OWMs (red), NWMs (green), prosimians (yellow), tupais (gray), and mice (black). The NJ tree is reconstructed by using the p distances. The numbers in boldface type correspond to those in the locus designation system for human T2R genes. Pseudogenes are indicated by asterisks (*). Nonsense (red crosses) and indel (blue crosses) mutations responsible for pseudogenization are placed along branches. The definitions of categories A, B, and C are the same as in Figure 1.
Pseudogenization of T2R genes:
To determine a branch in the phylogenetic tree of primate T2R genes along which a particular gene was disrupted, we parsimoniously placed nonsense mutations as well as insertions and/or deletions (indels) that can cause frameshift mutations (Figure 2). Because there are no shared pseudogenes between humans and mice, no disruptive mutation was placed in the common ancestral lineage. Among primates, shared disruptive mutations result in eight pseudogenes common to more than one species (Figure 2). T2R66 is an example of the pseudogenization that predated the divergence between anthropoidea and prosimians. The remaining 23 disruptive mutations occur on terminal branches and result in lineage-specific pseudogenes: 5 in prosimians and tupais, and 18 in anthropoidea (see below).
Species-specific changes regarding pseudogenization:
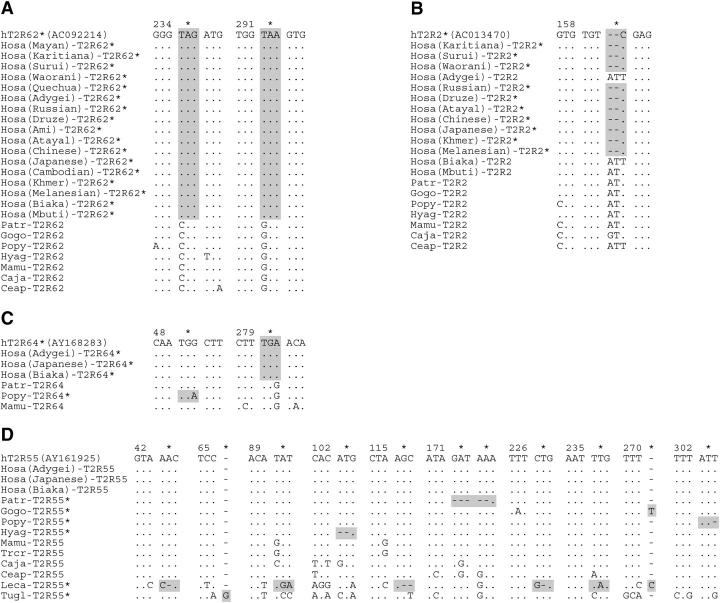
Of 23 lineage-specific pseudogenes, 3 (hT2R62*, hT2R2*, and hT2R64*) are confined in humans (Figure 3). In hT2R62*, there are two nonsense mutations (CAG → TAG at codon position 235 and GAA → TAA at 292). Since both of these mutations are fixed in the sample from 17 human populations (Figure 3A) and since the chimpanzee ortholog is functional, T2R62 in humans must have been inactivated long ago, but after the divergence between humans and chimpanzees. On the other hand, hT2R2* was produced relatively recently. It was previously annotated as a pseudogene owing to a two-base deletion at codon position 160 (Conte et al. 2002; Shi et al. 2003). However, since we found the intact form in Adygei (Eastern European), Mbuti (African Pygmies), and Biaka (African Pygmies) (Figure 3B), the hT2R2 locus is actually polymorphic in terms of the two-base deletion. Regarding hT2R64*, there is one fixed nonsense mutation of TGG → TGA at codon position 280. It is interesting to note that the orangutan ortholog (Popy-T2R64*) also suffers a nonsense mutation of TGG → TGA, but at codon position 49 (Figure 3C).
Figure 3.—
Partial alignments of the nucleotide sequences of primate T2R genes. Dots show the same nucleotides as those in the top sequence, which were retrieved from GenBank. Both asterisks and shaded boxes show the location of mutations that impair intact ORFs. (A) T2R62 is pseudogenized by two nonsense mutations (CAG → TAG at codon position 235 and GAA → TAA at 292). (B) T2R2 is polymorphic in terms of a two-base deletion at codon position 160 in human populations. Only Adygei (from Eastern Europe), Biaka, and Mbuti Pygmies (both from Africa) possess the intact genes, while the deletion is found in all sequences collected from 10 other human populations (Karitiana, Surui, Waorani Indians from South America, Russians from Eastern Europe, Druze from the Middle East, Atayal, Chinese, Japanese from Eastern Asia, and Khmers and Melanesians from Southeast Asia) and from GenBank resources. (C) T2R64 is a human-specific pseudogene (TGG → TGA at 280). This gene also became a pseudogene in the orangutan through an independent nonsense mutation (TGG → TGA at 49). (D) Independent mutations disrupt T2R55 in the four apes: chimpanzees (five-base deletion at 172–173), gorillas (one-base insertion at 271), orangutans (one-base deletion at 303), and gibbons (two-base deletion at 103); in the prosimians: ring-tailed lemurs (two one-base deletions at 43 and 227, two-base deletion at 116, one-base insertion at 271, and two nonsense mutations at 90 and 236); and in the tupai: common tree shrews (one-base insertion at 66).
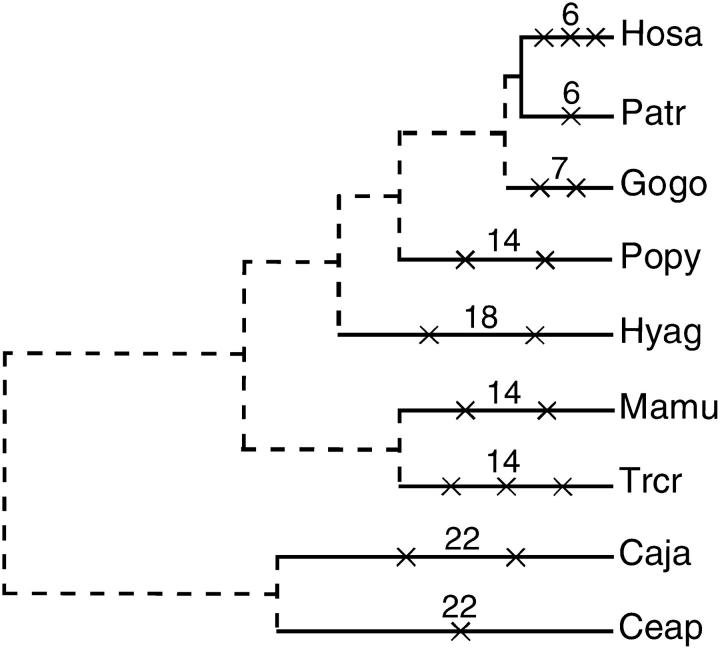
The remaining 19 lineage-specific pseudogenes are as follows: Patr (1), Gogo (2), Popy (1), Hyag (2), Mamu (2), Trcr (3), Caja (2), Ceap (1), Otcr (1), Leca (2), and Tugl (2). This observation indicates that all anthropoidea used in this study (Hosa, Patr, Gogo, Popy, Hyag, Mamu, Trcr, Caja, and Ceap) have their own pseudogenes. However, the rate of pseudogenization is high in the lineage leading to humans. Two fixed and one polymorphic pseudogene in humans arose within 6 million years (MY) after the divergence from chimpanzees, whereas 15 pseudogenes in nonhuman anthropoidea arose in a total of 117 MY (see Goodman et al. 1998 and Figure 4). The rate of pseudogenization is thus significantly different between humans and nonhuman anthropoidea (Fisher's exact test; P < 0.05, one-tailed). It is also interesting to note that disruptive mutations in a particular gene occurred several times independently in different species. Like T2R64 mentioned above, T2R55 is disrupted independently in prosimians, tupais, and four apes (chimpanzees, gorillas, orangutans, and agile gibbons), T2R39 in two apes (orangutans and agile gibbons), two OWMs (rhesus monkeys and silvered leaf monkeys), and one NWM (marmosets), T2R9 in gorillas and marmosets, and T2R10 in bush babies and tree shrews (Figures 2 and 3D). In addition, T2R15 became nonfunctional by a single nonsense mutation in the common ancestral lineage between humans and chimpanzees as well as by independent indel mutations in rhesus monkeys and silvered leaf monkeys (Figure 2).
Figure 4.—
The rate of pseudogenization in humans and nonhuman anthropoidea. Each cross on a terminal branch shows species-specific pseudogenization. The divergence times (MY) represented above each branch are from Goodman et al. (1998). While three human-specific pseudogenes have arisen within 6 MY, 15 (1 + 2 + 2 + 2 + 2 + 3 + 2 + 1) pseudogenes in nonhuman anthropoidea have accumulated within 117 MY (6 + 7 + 14 + 18 + 14 + 14 + 22 + 22). Pseudogenization that occurred along internodal branches (dashed lines) is excluded from consideration.
Selection for and against functional domains of T2R genes:
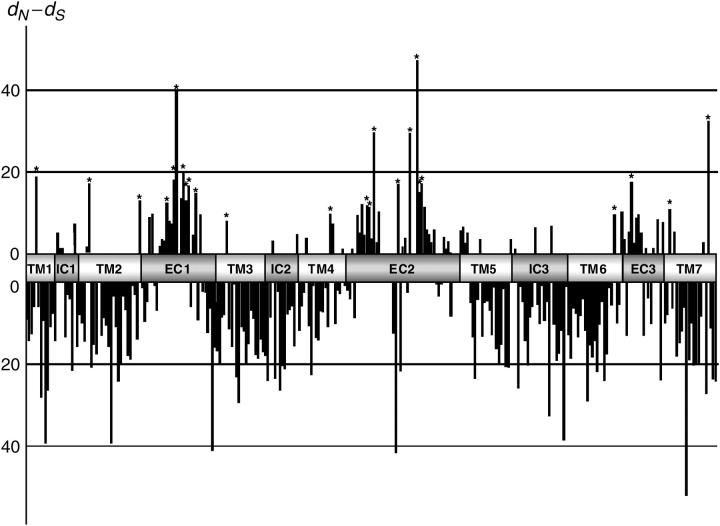
We examined whether differential selective pressure has been acting on different domains of primate functional T2R genes. T2R molecules consist of three domains: transmembrane (TMs), intracellular (ICs), and extracellular (ECs) (Adler et al. 2000). To evaluate selective pressure on each of these domains, we calculated the difference of _d_N − _d_S codon by codon (see materials and methods) for 150 undisrupted sequences from primates and mice. To reduce sampling errors in this analysis, we excluded any codon that is absent in more than half the sequences (>75). Figure 5 shows that codons with _d_N > _d_S are abundant in ECs whereas codons with _d_N < _d_S predominate in TMs + ICs. Fisher's exact test (_P_ < 0.01) also shows that positively selected amino acids (_d_N > _d_S) are much more frequently observed in ECs (15 of 84 sites) than in TMs + ICs (8 of 190) (see also Shi et al. 2003 for the comparison between human and mouse T2R genes). Thus, some amino acid changes in ECs are selected for and most amino acid changes in TMs + ICs are selected against.
Figure 5.—
The extent of positive and negative selection on individual codons. Schematic positions of three functional domains of the T2R gene (ECs, TMs, and ICs) are shown on the _x_-axis. The value above and below the _x_-axis, respectively, indicates an excess and deficiency in the number of per-site nonsynonymous substitutions (_d_N) over per-site synonymous ones (_d_S) in an individual codon. The zero value means _d_N = _d_S. Asterisks (*) show significant positive selection (P < 0.05) based on the extended binominal distribution (http://www.datamonkey.org/).
Lineage-dependent selection on T2R genes:
It is also intriguing to examine how concordantly or discordantly selection operates between the primate and mouse lineages. We used 16 T2R genes in category B and C to analyze their _b_S and _b_N values (see materials and methods) for 16 branches leading to primates (corresponding to branch numbers from 1 to 16 in Table 3) and for 1 branch leading from the common node of primates to mice (corresponding to branch number 17 in Table 3). Among 272 possible comparisons of branches and genes in primates and mice, 227 pairs of the _b_S and _b_N values were obtained because of lack of sequences for some genes in some species. These comparisons were made separately for ECs (Table 3A) and TMs + ICs (Table 3B). For the primate EC comparisons, 8 show that _b_N is significantly larger than _b_S (_b_N > _b_S), whereas for the 16 mouse comparisons, none show _b_N > _b_S but two exhibit _b_S > _b_N. The ratio of _b_N to _b_S in ECs averaged over all T2R genes is 0.94 in primates and 0.79 in mice. Although positive selection contributed to these relatively high ratios, relaxation of selective constraints in primates appears to be a more important factor. To test this conjecture, we compared the ratios in TMs + ICs between primates and mice. There is only 1 with _b_N > _b_S and 7 with _b_S > _b_N in the primate comparisons, whereas all 16 show _b_S > _b_N in the mouse comparisons. There is thus little evidence for positive selection in TMs + ICs and the average ratio of _b_N to _b_S is 0.57 in primates and 0.47 in mice. More importantly, since the increased amount of the ratio is similar in both domains, it is likely that primate T2R genes in categories B and C underwent relaxed negative selection rather than reinforced positive selection compared with the mouse orthologs.
TABLE 3.
Numbers of per-site synonymous substitutions ( bS) and per-site nonsynonymous substitutions ( bN) for each branch of the phylogenetic tree in category B and C genes
| Gene | Outgroup | Branchno.: | Hosa:1 | Patr:2 | 3a | Gogo:4 | 5a | Popy:6 | 7a | Hyag:8 | 9a | Mamu:10 | 11a | Trcr:12 | 13a | Caja:14 | 15a | Ceap:16 | Mumu:17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Extracellular domains | |||||||||||||||||||
| T2R1 | mt2r41 | 1.9/0.8 | 0.0/1.4 | 0.0/0.0 | 1.4/0.2 | 0.5/1.4 | 3.6/1.7 | 0.0/0.0 | 3.1/4.7 | 9.2/16.6 | 0.0/5.7# | 1.1/3.2 | 0.4/4.9# | 75.4/75.9 | |||||
| T2R2 | mt2r1 | 1.9/0.0 | 0.0/1.5 | 0.0/0.0 | 0.0/2.3# | 0.2/1.6 | 0.0/0.6 | 1.9/0.2 | 1.7/1.2 | 0.0/0.7 | 5.5/5.2 | 1.6/0.9 | 4.0/2.0 | 8.6/3.1 | 0.0/1.8 | 38.4/26.3 | |||
| T2R16 | mt2r2 | 0.0/0.6 | 0.0/0.0 | 2.2/0.1 | 0.0/1.0 | 0.0/0.6 | 1.9/1.6 | 0.1/0.6 | 0.0/1.1 | 1.4/0.6 | 0.0/3.1 | 0.0/4.9# | 1.7/0.7 | 3.1/2.6 | 4.4/3.3 | 2.7/3.1 | 1.2/3.7 | 60.4/45.0 | |
| T2R3 | mt2r3 | 1.5/0.6 | 0.0/0.0 | 0.0/0.0 | 0.0/0.0 | 0.3/0.3 | 2.8/0.7 | 0.0/0.0 | 1.4/2.2 | 0.0/2.6# | 3.3/0.4 | 0.2/1.3 | 0.0/2.2# | 0.0/3.1 | 9.1/2.4 | 8.8/6.2 | 5.2/1.0 | 32.0/20.6 | |
| T2R4 | mt2r4 | 0.0/0.0 | 1.6/0.0 | 0.2/0.0 | 0.0/0.6 | 1.5/0.9 | 2.9/1.4 | 0.0/0.0 | 0.4/2.1 | 0.0/0.5 | 3.0/0.2 | 4.2/1.0 | 0.0/0.2 | 12.4/6.9 | 4.2/1.0 | 43.7/28.8 | |||
| T2R38 | mt2r5 | 1.5/0.0 | 1.4/0.0 | 0.0/0.0 | 0.3/0.0 | 0.4/0.3 | 1.9/1.6 | 0.0/0.0 | 3.2/2.4 | 0.0/1.0 | 0.0/0.4 | 3.6/0.8 | 1.4/1.1 | 2.9/4.4 | 6.8/4.3 | 35.7/24.4 | |||
| T2R39 | mt2r6 | 0.0/0.0 | 0.0/0.0 | 0.0/0.0 | 0.0/0.0 | 0.2/0.6 | 1.4/0.0 | 0.0/0.0 | 3.4/2.9 | 0.0/1.1 | 3.3/0.2 | 2.3/2.3 | 0.0/0.8 | 0.0/2.8 | 3.0/0.6 | 6.2/2.8 | 0.2/1.0 | 46.8/55.4 | |
| T2R40 | mt2r7 | 0.0/0.6 | 1.6/0.5 | 0.0/0.5 | 1.5/1.1 | 0.5/0.0 | 0.9/1.0 | 0.5/0.7 | 1.2/0.1 | 0.0/1.0 | 2.7/2.2 | 2.2/0.1 | 2.8/0.0 | 2.9/1.0 | 3.0/2.2 | 51.1/22.2* | |||
| T2R62 | mt2r8 | 1.3/0.0 | 1.9/0.6 | 0.5/0.8 | 2.8/0.4 | 0.9/0.9 | 2.4/2.7 | 0.0/0.0 | 0.6/5.1# | 3.2/1.5 | 0.0/0.0 | 0.0/0.0 | 4.5/7.0 | 14.6/6.4 | 16.0/10.8 | 62.7/62.4 | |||
| T2R56 | mt2r9 | 0.0/1.0 | 0.0/0.6 | 0.0/0.0 | 1.7/0.8 | 0.9/1.4 | 0.7/2.8 | 0.6/0.0 | 3.6/0.5 | 4.0/0.0 | 2.1/3.0 | 0.0/3.2 | 10.3/3.9 | 45.1/27.1 | |||||
| T2R41 | mt2r10 | 0.0/0.2 | 0.0/1.4 | 0.6/1.3 | 3.7/2.8 | 60.0/28.4* | |||||||||||||
| T2R7 | mt2r11 | 0.0/0.0 | 1.4/0.0 | 0.0/0.2 | 0.0/0.4 | 2.2/0.0 | 0.1/1.5 | 0.0/0.0 | 4.8/0.5 | 3.1/2.5 | 2.5/0.9 | 0.0/1.4 | 1.4/0.0 | 0.9/0.0 | 5.2/1.7 | 2.9/2.7 | 4.3/0.8 | 22.4/19.9 | |
| T2R10 | mt2r12 | 0.0/0.5 | 0.0/0.6 | 0.3/0.2 | 1.4/0.4 | 0.0/2.0 | 2.3/1.4 | 0.0/2.3 | 2.0/2.7 | 2.7/0.0 | 5.6/2.7 | 2.8/2.0 | 3.3/1.6 | 0.0/6.3 | 3.9/3.6 | 3.8/3.8 | 0.0/4.2# | 59.3/54.8 | |
| T2R12 | mt2r19 | 0.5/0.4 | 1.2/0.2 | 0.0/0.0 | 0.0/0.7 | 0.0/0.7 | 0.4/1.6 | 0.0/4.4 | 0.0/0.8 | 6.1/0.8 | 0.4/0.3 | 54.7/58.9 | |||||||
| T2R13 | mt2r18 | 0.0/0.5 | 1.9/1.7 | 0.0/0.0 | 0.0/0.0 | 0.0/0.7 | 0.0/1.5 | 0.0/0.2 | 0.0/1.0 | 2.3/2.7 | 1.8/1.9 | 3.4/2.6 | 0.0/0.9 | 5.7/1.8 | 3.0/3.8 | 1.4/9.8 | 1.8/4.9 | 41.3/45.0 | |
| T2R14 | mt2r27 | 0.0/0.0 | 0.0/0.0 | 1.2/0.0 | 0.0/0.7 | 0.0/2.9 | 2.3/2.2 | 8.1/0.0 | 3.8/5.6 | 113.2/71.6 | |||||||||
| B. Transmembranes and intracellular domains | |||||||||||||||||||
| T2R1 | mt2r41 | 0.0/0.3 | 0.6/0.0 | 0.2/0.0 | 1.8/0.0 | 4.2/0.7 | 2.1/0.7 | 0.8/0.2 | 2.1/2.1 | 2.1/1.1 | 4.4/1.6 | 8.6/3.6 | 2.7/1.5 | 45.2/23.1** | |||||
| T2R2 | mt2r1 | 0.0/0.0 | 0.1/0.2 | 0.0/0.0 | 0.0/1.0 | 0.0/0.3 | 3.2/0.2 | 0.3/0.0 | 0.0/0.7 | 1.2/1.1 | 4.3/1.8 | 2.3/0.0 | 5.8/1.3* | 3.6/2.2 | 1.1/1.2 | 44.9/20.0** | |||
| T2R16 | mt2r2 | 0.6/0.3 | 0.0/0.0 | 1.2/0.0 | 1.5/0.0 | 0.5/0.3 | 0.7/1.3 | 0.6/0.0 | 1.4/1.1 | 1.1/1.2 | 1.9/1.0 | 1.3/0.6 | 0.5/1.6 | 1.5/3.5 | 2.5/2.7 | 3.2/2.3 | 46.2/22.6** | ||
| T2R3 | mt2r3 | 0.0/0.0 | 0.6/0.0 | 0.0/0.0 | 0.0/0.0 | 1.9/0.8 | 0.9/1.1 | 0.5/0.0 | 1.2/0.3 | 0.0/0.7 | 3.4/0.9 | 2.5/1.2 | 1.1/1.3 | 2.3/2.1 | 2.1/0.2 | 2.6/2.6 | 3.8/1.7 | 33.3/18.5* | |
| T2R4 | mt2r4 | 0.0/0.2 | 0.0/0.3 | 0.1/0.0 | 0.5/0.7 | 1.1/0.2 | 2.2/0.6 | 0.0/0.0 | 1.9/1.1 | 1.4/0.5 | 0.9/0.8 | 1.0/0.8 | 0.5/0.5 | 0.2/1.5 | 3.4/2.7 | 33.5/13.8*** | |||
| T2R38 | mt2r5 | 0.0/0.6 | 0.6/0.1 | 0.0/0.0 | 0.0/0.3 | 1.0/0.2 | 0.7/0.6 | 0.1/0.1 | 2.8/1.0 | 1.6/0.0 | 0.4/0.3 | 1.7/0.2 | 3.0/0.7 | 2.9/1.4 | 3.3/3.1 | 38.6/16.1*** | |||
| T2R39 | mt2r6 | 0.0/0.0 | 0.5/0.5 | 0.0/0.2 | 1.2/0.0 | 0.9/0.3 | 0.9/0.7 | 0.0/0.6 | 0.9/0.6 | 1.3/0.1 | 1.4/0.3 | 1.7/1.4 | 2.9/0.6 | 2.3/0.3 | 3.0/1.0 | 1.1/2.0 | 3.8/0.2* | 38.5/19.2** | |
| T2R40 | mt2r7 | 0.0/0.0 | 0.0/0.0 | 0.0/0.0 | 1.4/0.2 | 0.0/0.0 | 0.0/0.2 | 0.7/0.5 | 0.9/0.3 | 0.0/0.6 | 2.5/1.4 | 1.4/0.3 | 0.8/1.6 | 1.5/1.3 | 4.7/1.1 | 27.0/14.7* | |||
| T2R62 | mt2r8 | 1.1/0.6 | 0.7/0.3 | 0.0/0.0 | 0.9/0.2 | 1.2/0.6 | 2.8/1.4 | 0.4/0.3 | 0.9/1.1 | 3.7/2.4 | 0.0/0.0 | 0.0/0.0 | 4.6/3.4 | 10.7/3.8* | 12.4/5.1* | 43.2/25.5* | |||
| T2R56 | mt2r9 | 0.0/0.5 | 1.2/0.7 | 0.7/0.3 | 1.0/0.7 | 1.0/1.7 | 3.1/1.9 | 0.0/0.0 | 3.0/1.3 | 1.9/0.0 | 2.2/2.5 | 5.5/1.6 | 10.7/6.9 | 43.7/22.8** | |||||
| T2R41 | mt2r10 | 0.5/0.5 | 0.6/0.0 | 1.5/0.3 | 0.8/1.6 | 43.5/17.0*** | |||||||||||||
| T2R7 | mt2r11 | 0.0/0.2 | 0.7/0.6 | 0.0/0.0 | 0.8/0.3 | 0.9/0.3 | 2.9/0.4 | 0.0/0.0 | 2.2/1.1 | 0.9/0.3 | 3.1/1.0 | 1.9/0.4 | 4.4/1.0 | 0.6/1.0 | 3.3/1.7 | 2.1/1.6 | 5.1/1.0* | 28.6/15.3* | |
| T2R10 | mt2r12 | 0.0/0.5 | 0.0/0.0 | 0.0/0.0 | 0.0/0.6 | 1.5/0.5 | 2.5/0.8 | 0.4/0.0 | 2.1/1.5 | 0.0/1.5 | 0.7/0.5 | 6.1/0.6* | 0.8/1.2 | 0.0/2.9 | 5.9/3.3 | 9.9/5.3 | 2.0/1.3 | 51.6/18.6*** | |
| T2R12 | mt2r19 | 1.7/0.0 | 1.9/0.3 | 0.0/0.3 | 0.6/0.0 | 0.3/0.3 | 1.6/0.9 | 4.6/0.3 | 0.9/2.7 | 0.0/6.1# | 0.9/2.5 | 62.2/31.2** | |||||||
| T2R13 | mt2r18 | 0.0/0.9 | 1.2/0.9 | 0.0/0.0 | 0.0/0.9 | 1.8/0.9 | 1.1/2.1 | 0.0/0.2 | 1.0/0.6 | 2.5/1.0 | 1.1/1.6 | 1.2/1.0 | 0.7/0.5 | 1.3/0.3 | 1.4/2.0 | 0.7/0.7 | 11.2/1.6** | 52.8/16.1*** | |
| T2R14 | mt2r27 | 1.4/0.4 | 0.1/0.7 | 1.4/0.6 | 1.2/0.0 | 0.5/2.1 | 1.3/1.9 | 2.8/1.5 | 4.6/4.4 | 58.4/29.0** |
Likewise, we examined the _b_S and _b_N values in primate category A genes for comparison. Figure 6 contrasts the _b_S and _b_N values between categories A and C as well as between domains EC and TM + IC. In both domains, the _b_N value tends to be higher in category A than in category C irrespective of the _b_S value. The Mann-Whitney test also revealed that the ratio of _b_N to _b_S is significantly higher in category A genes than in category C genes in both ECs (P < 0.05) and TMs + ICs (P < 0.01). Similar to the comparison between primates and mice, these results indicate that category A genes within primates underwent relaxed negative selection.
Figure 6.—
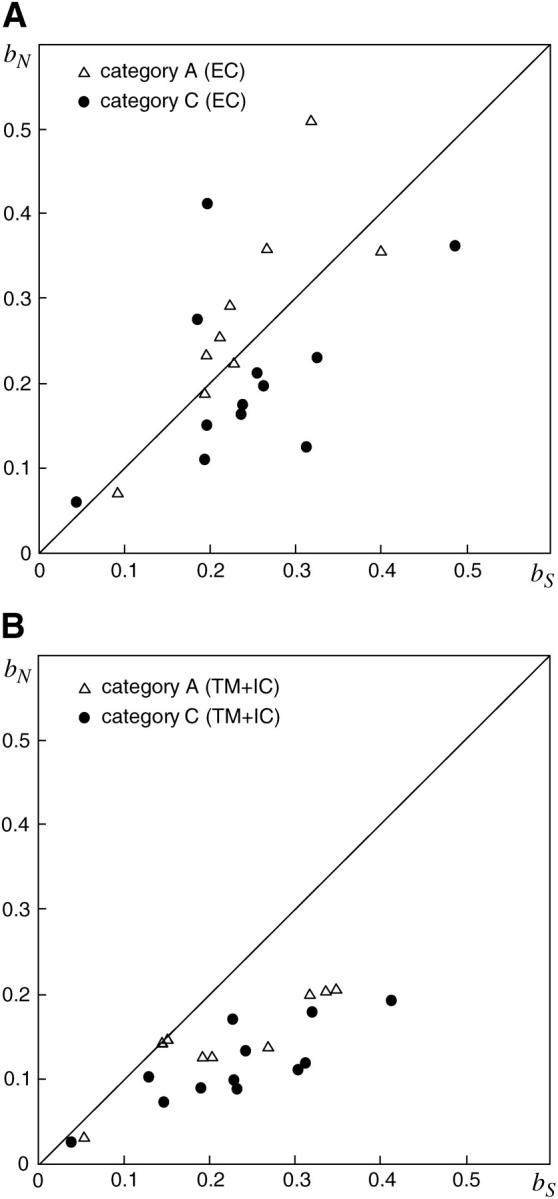
The _b_S and _b_N values of category A genes (▵) and category C genes (•) in ECs (top) and TMs + ICs (bottom). The _x_- and _y_-axis stand for _b_S and _b_N, respectively. The primate genes used for category A are T2R15, T2R43-44, T2R45, T2R46, T2R47, T2R48, T2R49, T2R50, and T2R55, and those for category C are T2R1, T2R2, T2R3, T2R4, T2R7, T2R16, T2R38, T2R39, T2R40, T2R41, T2R56, and T2R62.
DISCUSSION
In this article, we have characterized the repertoire of bitter taste receptor (T2R) genes in humans and nonhuman primates to understand the tempo and mode of the duplication and pseudogenization process. It is generally difficult to discern the complete repertoire of other chemosensory receptor genes in mammals. In the case of olfactory receptor (OR) genes, this difficulty is due mainly to the fact that they form an exceptionally large multigene family (856 genes in humans, 1404 in mice, and 971 in dogs according to the HORDE database at http://bip.weizmann.ac.il/HORDE/). On the other hand, the number of T2R genes is moderately small and suspected to be at most ∼40 in the mammalian genome. This fact has facilitated our study to discern the great majority of the T2R gene repertoire in anthropoidea. Indeed, for each nonhuman anthropoid we have identified at least 20 genes that are orthologous to 36 human T2R genes (Table 2).
In OR genes, hominoids (especially humans) and OWMs have a higher proportion of pseudogenes than do NWMs or lemurs (Gilad et al. 2003, 2004), and in pheromone receptor (VR) genes, hominoids and OWMs are thought to have completely lost the functional genes (Zhang and Webb 2003). By contrast, among anthropoidea there are no significant differences in the proportion of the T2R pseudogenes, which suggests that the tempo and mode of pseudogenization in T2R is different from that in OR and VR.
In primates, many more pseudogenes have accumulated in category A (46 pseudogenes of 115 genes) than in category B (0 of 29) and in category C (18 of 125) (Fisher's exact test; P < 0.001). In mice, all pseudogenes are restricted to category B and absent in categories A and C. This contrast strongly suggests that the more frequently duplication occurs, the more pseudogenes tend to accumulate. This trend is also evident for prosimians, in which three duplicated T2R10 genes are found and one of them is a pseudogene (Figure 2 and Table 2). It therefore appears that the evolutionary process of T2R genes exemplifies the birth-and-death model for multigene families (Nei et al. 1997), as for MHC (Hughes and Nei 1989; Nei et al. 1997), immunoglobulin (Ota and Nei 1994), histone (Piontkivska et al. 2002), and OR genes (Young et al. 2002). However, major determinants that have shaped the gene repertoire may well differ from family to family. Broadly, determinants are either internal (genomic) or external (environmental). The gene repertoire of rRNA and histone multigene families is likely to be determined by internal (physiological or genomic) requirements. On the other hand, the gene repertoire in MHC and OR genes is likely to be determined by requirements for defense against exogenous pathogens and perception of chemical substances, respectively. The situation of T2R resembles that of MHC and OR in that the gene repertoire is largely shaped by available foods.
Because T2R genes in category C are conserved to maintain the one-to-one orthologous relationships between primates and mice, we can speculate that T2R in category C can perceive bitter substances common to mammals. In this regard, it is worth noting that all three human-specific pseudogenes (hT2R2*, hT2R12*, and hT2R62*) belong to category C (Figure 2 and Table 1). Equally importantly, humans have accumulated more pseudogenes per unit of time than nonhuman anthropoidea (Figure 4). Taken together, these observations indicate accelerated deterioration of tasting particular bitter substances in humans. Interestingly, there are also lineage-specific pseudogenes in some nonhuman primates and such pseudogenization tends to repeat in different lineages. As shown in Figure 2, there are six such genes (T2R64, T2R55, T2R39, T2R9, T2R10, and T2R15) that were repeatedly dysfunctional in more than one primate species. Repeated disruptions of these genes in different primates and tupais strongly suggest that the fate of taste receptor genes is associated with environmental factors rather than with genetic ones. Nevertheless, since T2R55 and T2R39 are functional in humans, these genes are likely indispensable to tasting particular bitter substances throughout human evolution.
We have demonstrated that functional constraints are more relaxed for the primate T2R genes than for the mouse genes (Table 3). Since the number of positively selected amino acids in ECs is significantly larger than that in TMs + ICs (Figure 5), the average ratio of _b_N to _b_S in primates is greater in ECs (0.94) than in TMs + ICs (0.57). Similarly, the average ratio in mice is 0.79 in ECs and 0.47 in TMs + ICs. We have noted that increased amounts of these ratios in primates relative to mice are almost the same between ECs (0.94/0.79) and TMs + ICs (0.57/0.47), indicating that irrespective of the domains, ∼20% of the functional constraints are more relaxed in the primate lineage than in the mouse lineage. This relaxation in the primate lineage is reflected in the presence of a relatively larger number of pseudogenes in primates (20∼35%) than in mice (15%). Recently, two research groups studied the pattern of the molecular evolution of T2R genes in humans and chimpanzees (Parry et al. 2004; Wang et al. 2004). Specifically, Wang et al. (2004) examined the extent of functional constraints in 25 pairs of human and chimpanzee T2R orthologs and in 28 pairs of mouse and rat orthologs. Their conclusion is similar to ours: The T2R genes in humans and chimpanzees are under relaxed selective constraints compared with those in mice and rats. Furthermore, Fischer et al. (2005) reported the evolutionary study of T2R genes in humans, four great apes, and two OWMs and compared the ratio of per-site nonsynonymous substitutions to per-site synonymous substitutions in primates (_f_p) with that in rodents (_f_r). Since the _f_p value is significantly larger than _f_r, these authors argued that the elevated ratio in primates results from the reduced effective population size and can be attributed to fixation of slightly deleterious mutations (Ohta 1975). Theoretically speaking, if the reduced effective population size is truly the cause of the elevated ratio in primates, all T2R genes should be equally influenced by increased amounts of effectively neutral mutations in a small population. This conclusion can be tested by the f values of individual T2R genes between primates and mice. Under the slightly deleterious hypothesis, we would expect more or less the same ratio of _f_p to _f_r regardless of the genes involved. However, this is not the case. Although the _f_p/_f_r ratio averaged over 31 genes is 1.27 and consistent with the relatively small effective size in primates, the individual ratio values vary greatly from 0.58 (T2R15) to 2.21 (T2R41). This large variation in the f_p/f_r ratio among loci can be more easily accounted for by relaxed negative selection than by reduced effective size.
We have shown that functional constraints are weaker against genes in category A than in category C (Figure 6). Hence, there is a trend that newly duplicated category A genes in primates tend to have accumulated nonsynonymous substitutions and contributed to expansion of the gene repertoire, which might allow primates to perceive numerous bitter substances in a changing environment. We have also shown that primate T2R genes were subjected to extensive lineage-dependent pseudogenization of functionally diversified duplicated copies. It appears that under joint effects of gene duplication and pseudogenization as well as interplay between genes and environments, primate T2R genes have evolved toward species-specific repertoires.
Acknowledgments
This research was supported in part by the Japanese Society for Promotion of Science grant no. 12304046 (to N.T.).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AB198983, AB199308.
References
- Adler, E., M. A. Hoon, K. L. Mueller, J. Chandrashekar, N. J. Ryba et al., 2000. A novel family of mammalian taste receptors. Cell 100**:** 693–702. [DOI] [PubMed] [Google Scholar]
- Chandrashekar, J., K. L. Mueller, M. A. Hoon, E. Adler, L. Feng et al., 2000. T2Rs function as bitter taste receptors. Cell 100**:** 703–711. [DOI] [PubMed] [Google Scholar]
- Conte, C., M. Ebeling, A. Marcuz, P. Nef and P. J. Andres-Barquin, 2002. Identification and characterization of human taste receptor genes belonging to the TAS2R family. Cytogenet. Genome Res. 98**:** 45–53. [DOI] [PubMed] [Google Scholar]
- Conte, C., M. Ebeling, A. Marcuz, P. Nef and P. J. Andres-Barquin, 2003. Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol. Genomics 14**:** 73–82. [DOI] [PubMed] [Google Scholar]
- Fischer, A., Y. Gilad, O. Man and S. Pääbo, 2005. Evolution of bitter taste receptors in humans and apes. Mol. Biol. Evol. 22**:** 432–436. [DOI] [PubMed] [Google Scholar]
- Fitch, W. M., 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20**:** 406–416. [Google Scholar]
- Gilad, Y., O. Man, S. Pääbo and D. Lancet, 2003. Human specific loss of olfactory receptor genes. Proc. Natl. Acad. Sci. USA 100**:** 3324–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, Y., V. Wiebe, M. Przeworski, D. Lancet and S. Pääbo, 2004. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2**:** 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider et al., 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9**:** 585–598. [DOI] [PubMed] [Google Scholar]
- Hartigan, J. A., 1973. Minimum evolution fits to a given tree. Biometrics 29**:** 53–65. [Google Scholar]
- Hillier, L. W., W. Miller, E. Birney, W. Warren, R. C. Hardison et al., 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432**:** 695–716. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and M. Nei, 1989. Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol. Biol. Evol. 6**:** 559–579. [DOI] [PubMed] [Google Scholar]
- Kim, U. K., E. Jorgenson, H. Coon, M. Leppert, N. Risch et al., 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299**:** 1221–1225. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L., and S. D. W. Frost, 2005. Not so different after all: comparison of various methods for detecting amino-acid sites under selection. Mol. Biol. Evol. 22**:** 1208–1222. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17**:** 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lehninger, A. L., 1996 Biochemistry, Ed. 2. Kalyani Publishers, New Delhi.
- Li, X., L. Staszewski, H. Xu, K. Durick, M. Zoller et al., 2002. Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. USA 99**:** 4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami, H., and H. Amrein, 2003. Taste and pheromone perception in mammals and flies. Genome Biol. 4**:** 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami, H., J. P. Montmayeur and L. B. Buck, 2000. A family of candidate taste receptors in human and mouse. Nature 404**:** 601–614. [DOI] [PubMed] [Google Scholar]
- Muse, S. V., and B. S. Gaut, 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11**:** 715–724. [DOI] [PubMed] [Google Scholar]
- Nei, M., X. Gu and T. Sitnikova, 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94**:** 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, G., M. A. Hoon, J. Chandrashekar, Y. Zhang, N. J. Ryba et al., 2001. Mammalian sweet taste receptors. Cell 106**:** 381–390. [DOI] [PubMed] [Google Scholar]
- Nishikimi, M., R. Fukuyama, S. Minoshima, N. Shimizu and K. Yagi, 1994. Cloning and chromosomal mapping of the human nonfunctional gene for L-gulono-γ-lactone oxidase, the enzyme for L-ascorbic acid biosynthesis missing in man. J. Biol. Chem. 269**:** 13685–13688. [PubMed] [Google Scholar]
- Ohno, S., 1970 Evolution by Gene Duplication. Springer-Verlag, New York.
- Ohta, T., 1975. Statistical analyses of Drosophila and human protein polymorphisms. Proc. Natl. Acad. Sci. USA 72**:** 3194–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, T., and M. Nei, 1994. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol. Biol. Evol. 11**:** 469–482. [DOI] [PubMed] [Google Scholar]
- Parry, C. M., A. Erkner and J. Le Coutre, 2004. Divergence of T2R chemosensory receptor families in humans, bonobos, and chimpanzees. Proc. Natl. Acad. Sci. USA 101**:** 14830–14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska, H., A. P. Rooney and M. Nei, 2002. Purifying selection and birth-and-death evolution in the histone H4 gene family. Mol. Biol. Evol. 19**:** 689–697. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4**:** 406–425. [DOI] [PubMed] [Google Scholar]
- Shi, P., J. Zhang, H. Yang and Y. P. Zhang, 2003. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol. Biol. Evol. 20**:** 805–814. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., and T. Gojobori, 1999. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16**:** 1315–1328. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25**:** 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., S. D. Thomas and J. Zhang, 2004. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum. Mol. Genet. 13**:** 2671–2678. [DOI] [PubMed] [Google Scholar]
- Wong, G. T., K. S. Gannon and R. F. Margolskee, 1996. Transduction of bitter and sweet taste by gustducin. Nature 381**:** 796–800. [DOI] [PubMed] [Google Scholar]
- Young, J. M., C. Friedman, E. M. Williams, J. A. Ross, L. Tonnes-Priddy et al., 2002. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11**:** 535–546. [DOI] [PubMed] [Google Scholar]
- Zhang, J., and D. M. Webb, 2003. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc. Natl. Acad. Sci. USA 100**:** 8337–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., H. Rosenberg and M. Nei, 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95**:** 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]