Repeat-Induced Point Mutation and the Population Structure of Transposable Elements in Microbotryum violaceum (original) (raw)
Abstract
Repeat-induced point mutation (RIP) is a genome defense in fungi that hypermutates repetitive DNA and is suggested to limit the accumulation of transposable elements. The genome of Microbotryum violaceum has a high density of transposable elements compared to other fungi, but there is also evidence of RIP activity. This is the first report of RIP in a basidiomycete and was obtained by sequencing multiple copies of the integrase gene of a _copia_-type transposable element and the helicase gene of a _Helitron_-type element. In M. violaceum, the targets for RIP mutations are the cytosine residues of TCG trinucleotide combinations. Although RIP is a linkage-dependent process that tends to increase the variation among repetitive sequences, a chromosome-specific substructuring was observed in the transposable element population. The observed chromosome-specific patterns are not consistent with RIP, but rather suggest an effect of gene conversion, which is also a linkage-dependent process but results in a homogenization of repeated sequences. Particular sequences were found more widely distributed within the genome than expected by chance and may reflect the recently active variants. Therefore, sequence variation of transposable elements in M. violaceum appears to be driven by selection for transposition ability in combination with the context-specific forces of the RIP and gene conversion.
TRANSPOSABLE elements make up a diverse group of molecular parasites found in nearly all organisms. Their activity can be harmful to the host organism because insertion of new copies often disrupts gene expression (Wright et al. 2003) and because the similarity between copies at different loci promotes ectopic recombination and destabilizes chromosomes (Charlesworth et al. 1992; Petrov et al. 2003). Therefore, it is not surprising that parallels are drawn between transpositional activity and the virulence of conventional parasites (Jordan and McDonald 1999). Moreover, the term “arms race” has been used when there is evidence that the host genome has evolved mechanisms to defend against transposable elements (Jordan et al. 1999; Lovsin et al. 2001).
A genome defense called repeat-induced point mutation (RIP) is known to occur in ascomycete fungi (reviewed by Selker 2002). RIP is a homology-dependent gene-silencing mechanism that directly mutates multicopy DNA sequences. RIP has been credited with preventing transposable element accumulation in fungi, which generally have relatively small genomes that are said to be “streamlined” because they contain little repetitive DNA (Kidwell 2002; Selker 2002; Wöstemeyer and Kreibich 2002; Brookfield 2003; Galagan and Selker 2004).
The mechanistic details and specific protein-DNA interactions involved in target recognition and site-specific mutation are not completely resolved, but the general properties of the RIP process have been well characterized in several ascomycete species (for reviews see Selker 2002; Galagan and Selker 2004). RIP induces C-to-T mutations in duplicated DNA sequences. Cytosine residues of particular nucleotide combinations are targeted; for example, in Neurospora crassa CA dinucleotides are the targets of RIP activity. RIP is known to occur within haploid nuclei following mating but prior to meiosis (i.e., in the dikaryotic stage that precedes karyogamy). Cytosine methylation is frequently associated with RIP-mutated sequences; however, it remains undetermined whether this is a required step in a deamination process to yield C-to-T mutations. RIP acts in a pairwise manner on duplicated DNA sequences, such that they not only are altered but also become dissimilar because not all the same cytosine residues are changed in both copies. Finally, RIP occurs at a much higher rate between a pair of duplicated sequences when they are located on the same chromosome, and more so when they are in close linkage as compared to unlinked duplications. In addition to inactivating transposable elements by point mutations, RIP is thought to discourage ectopic recombination by decreasing the sequence similarity between copies at different loci (Cambareri et al. 1991).
RIP has been studied mostly from a mechanistic perspective and as a gene-silencing tool. However, there is little understanding of how RIP affects the intragenomic “population” of transposable elements or their evolutionary history. The homology-dependent and linkage-dependent nature of RIP may cause the genetic changes in transposable elements to be largely influenced by the particular genomic or chromosomal contexts in which they reside. This presents a strong contrast to our paradigm of sequence evolution based on random mutation and may seriously complicate the interpretation of transposable element evolution based on their sequence variation.
In this study, we determined the sequence variation for transposable elements in the fungus Microbotryum violaceum, which is unusual among fungi because this species contains a high density and diversity of transposable elements (Hood et al. 2004; Hood 2005). We provide evidence of RIP activity in M. violaceum by identifying the target site for C-to-T mutations, the first such reported for a basidiomycete. We also compare the sequence variation of element copies within and between the different chromosomes and provide evidence that gene-conversion acts in addition to RIP to determine the population structure of transposable elements in M. violaceum.
MATERIALS AND METHODS
M. violaceum (Basidiomycota; formerly Ustilago violacea) is a pathogenic fungus of plants in the Caryophyllaceae. It is commonly studied as a model for disease ecology and fungal genetics (Antonovics et al. 2002; Garber and Ruddat 2002, and references therein). Collections of the fungus from different host plants probably represent cryptic species, but the taxonomy of these lineages is not resolved. The study reported here is based on a collection from the host Silene latifolia from Italy (Lamole in Chianti, collection no. IT00-15.1). This collection has been used in previous studies (Hood et al. 2004; Hood 2005).
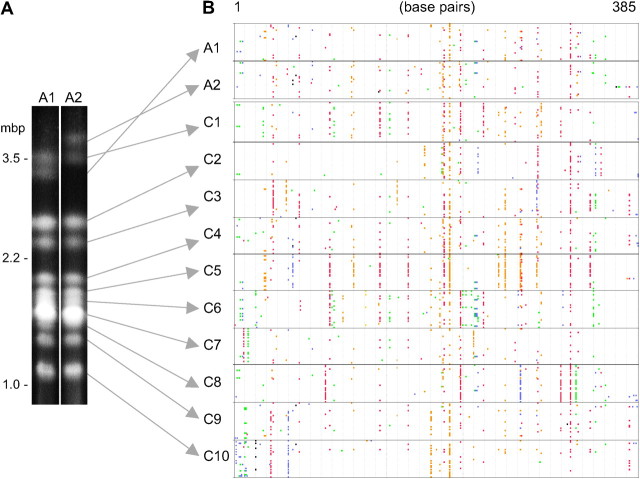
To isolate chromosome-specific DNA, karyotypes of the haploid meiotic products were separated by pulsed-field gel electrophoresis following the isolation of linear tetrads by micromanipulation (Figure 1A), as described in Hood (2002) and Hood et al. (2004). Electrophoresis conditions using a CHEF-DRII pulsed-field system (Bio-Rad, Hercules, CA) were 14 C, 96 hr, 2.8 V/cm, and 0.8% agarose, 200 sec initial and 1100 sec final switch times. DNA was extracted from each of the autosome bands and the A2 sex chromosome band using a culture of the A2 mating type. DNA from the A1 sex chromosome band was obtained from a culture from the same meiotic tetrad as the A2 culture. To extract DNA, an agarose plug was removed from the gel at each chromosome band using a glass Pasteur pipette, and DNA was isolated from the plugs by the phenol-based methods of Favre (1992). Most of the sampled locations in the gel corresponded to distinct single-chromosome bands, but one sample (plug 7 of Figure 1A) was in a region where bands are not as well resolved; this extract may therefore have included DNA from multiple chromosomes.
Figure 1.—
Genome sampling of _copia_-type retrotransposon sequences from M. violaceum. (A) Electrophoretic karyotypes of M. violaceum (collection no. IT00-15.1). Lanes represent haploid cultures of opposite mating types (A1 and A2) isolated from the same meiotic tetrad. In these cultures, the autosomes are homozygous for size, but the sex chromosomes are dimorphic. Chromosome bands were sampled and used to isolate transposable element sequences as indicated by arrows. (B) Alignment of coding strand sequences of integrase gene fragment of _copia_-type retrotransposon, ∼30 clones per chromosome. Deviations from consensus shown in color: red, C-to-T mutations; orange, G-to-A mutations; green, A-to-G or T-to-C mutations (other transitions); blue, transversion mutations; black, deletion mutations.
PCR primers were designed for a retrotransposon fragment found previously by sequencing random AFLP products from M. violaceum (Hood 2002; Hood and Antonovics 2004): internal primers for the retrotransposon sequence were ECA/MGA700BR.2 forward 5′ TGGAACCTGTACGTTGATGG and reverse 5′ ATTTTCTGACCCGTTTGACG. The sequence is in the integrase region of a Ty1/_copia_-type element, the most common type of transposable element in M. violaceum. The resulting 367-bp sequence begins approximately with the first histidine of the HHCC motif that characterizes the N terminus of the Psuedoviridae integrase gene (as defined in Peterson-Burch and Voytas 2002).
A separate PCR reaction was conducted with each chromosome-specific sample of DNA that was isolated from the karyotype gel. The resulting PCR products were then cloned using the TA cloning kit from Invitrogen and ∼30 clones per chromosome were sequenced using dye termination techniques with an ABI 377 automated DNA sequencer. Sequence alignment was carried out manually in Sequencher (Gene Codes Corporation) and is available under accession nos. AY729078, AY729484 in the PopSet section of GenBank (http://www.ncbi.nlm.nih.gov). All deviations from the consensus sequence were confirmed to represent unambiguous base pair signals in the electropherograms.
As a test to determine whether evidence of RIP activity is also present in the sequence variation of the other types of transposable elements in M. violaceum, PCR primers were designed for a _Helitron_-type sequence (accession no. BZ782234) identified in a previous study (Hood 2005). Helitrons are recently discovered DNA-based transposable elements that replicated by a rolling circle mechanism and are found in a broad range of organisms (Kapitonov and Jurka 2001; Poulter et al. 2003). Internal primers for a 301-bp region of BZ782234 were 44-forward 5′ CACGGTGAGTAGCCATTCC and 374-reverse 5′ CCGTTTACTGCGTGATCTCC. PCR was conducted using whole genomic DNA, 12 products of which were then cloned and sequenced as described above, and sequences are available under accession nos. AY939912, AY939913, AY939914, AY939915, AY939916, AY939917, AY939918, AY939919, AY939920, AY939921, AY939922, AY939923 in GenBank.
As a control to confirm that DNA isolated from karyotype gels was chromosome specific, the methods described above were replicated with a separate culture of the same fungal strain. DNA was isolated as before from chromosome bands corresponding to plugs 2 and 3 in Figure 1A and were similarly used for PCR amplification and sequencing of 30 clones per chromosome. In this case, sequencing was carried out by Genoscope-CNS (France). Also considering that one PCR reaction may produce amplification products from the same locus, the results were analyzed using the entire data set as well as only the nonredundant sequences from within each chromosome (indicative of separate loci). In determining sequence redundancies, base pair ambiguities or gaps in the alignments were conservatively considered to be equivalent to base pairs in otherwise identical sequences.
RESULTS
A total of 359 cloned sequences were obtained for the 367-bp integrase region of the Ty1/_copia_-type retrotransposon. There were ∼30 sequences from each of 12 isolated chromosome bands of the electrophoretic karyotype (Figure 1): 32 sequences for chromosome band C1; 29 sequences for C5, C6, and C7; and 30 sequences for the other bands. Variations from the consensus sequence are shown in the alignment of Figure 1B. The consensus sequence was strongly supported. Specifically, there was complete identity across all 359 sequences at 62% of the nucleotide positions (sequence length, 367 bp) and there were very few (<1%) nucleotide positions with <80% identity across all the sequences.
Pattern of repeat-induced point mutations:
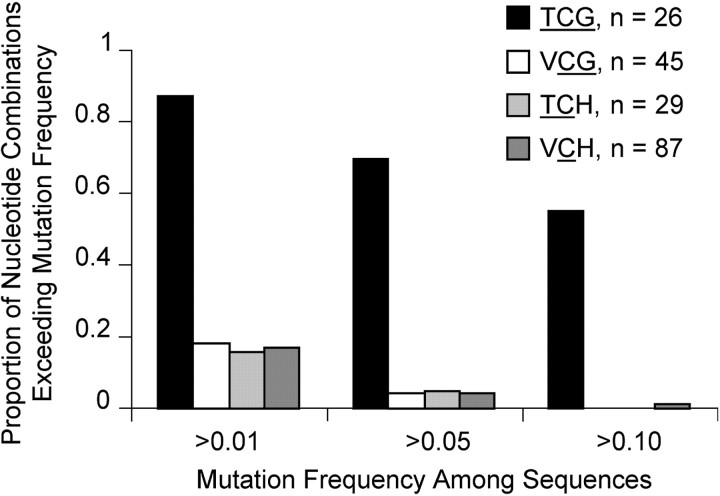
The majority of deviations from the consensus _copia_-type retrotransposon sequence were C-to-T or G-to-A changes, consistent with RIP activity (the latter are assumed to represent C-to-T mutations on the noncoding strand). In contrast to analyses of RIP in other fungi (Selker 2002; Clutterbuck 2004), the “target site” for hypermutation of cytosine residues was strictly the trinucleotide TCG rather than a dinucleotide. A partial match to this target site that was different from TCG immediately 3′ or 5′ to the cytosine residue did not show elevated C-to-T changes above the baseline for other cytosine residues (Figure 2). There were 14 RIP target sites in the coding strand and 12 in the noncoding strand of the consensus sequence. The RIP target site was not present in the PCR primers used in this study.
Figure 2.—
Hypermutations at RIP target site in the integrase gene fragment of _copia_-type retrotransposon of M. violaceum. Mutation rates for cytosine residues in various 3′ to 5′ nucleotide combinations are shown. Coding and noncoding strands are combined. Standard IUB codes are used for incompletely specified nucleotides: V, not T; H, not G. Numbers of nucleotide combinations are determined using the consensus sequence.
For the region of the _Helitron_-type element, the 12 cloned sequences showed a bias in mutations to the same target site trinucleotide as shown in Figure 2. Specifically, 37% of TCG sites across the consensus sequence (including CGA sites for the complimentary strand) contained at least one sequence with transition mutations (n = 19), whereas the value for VCG was 11% (n = 37), for TCH was 3% (n = 34), and for VCH was 6% (n = 77) (using standard International Union of Biochemistry, IUB, codes for incompletely specified nucleotides).
RIP most often affects just one strand of the DNA duplex in a single event and thus yields C-to-T mutation in only the coding strand or only the noncoding strand, the latter of which appear as G-to-A mutations in the coding strand (Cambareri et al. 1991; but see Watters et al. 1999). However, nearly half (47%) of cloned _copia_-type retrotransposon sequences contained mutations in RIP target sites on both the coding and noncoding strands. This pattern results from multiple rounds of RIP that alternately affect the coding and noncoding strands. Nevertheless, sequences with RIP mutation only in the coding or only in the noncoding strands were more common than expected when they had <∼20% of RIP target sites mutated. Specifically, 46 of 79 sequences with two RIP target sites mutated had the changes only in the coding or only in the noncoding strands (binomial distribution probabilities, P = 0.110), as did 19 of 30 sequences with three RIP mutations (P = 0.035), and 23 of 44 sequences with four RIP mutations (P < 0.001). Sequences with higher numbers of mutated RIP sites did not differ from chance expectation as to whether all the changes were only on one strand. These results may indicate that a small percentage (i.e., 10–20%) of TCG sites are mutated in a single round of RIP, but that many element copies have undergone multiple rounds of RIP activity.
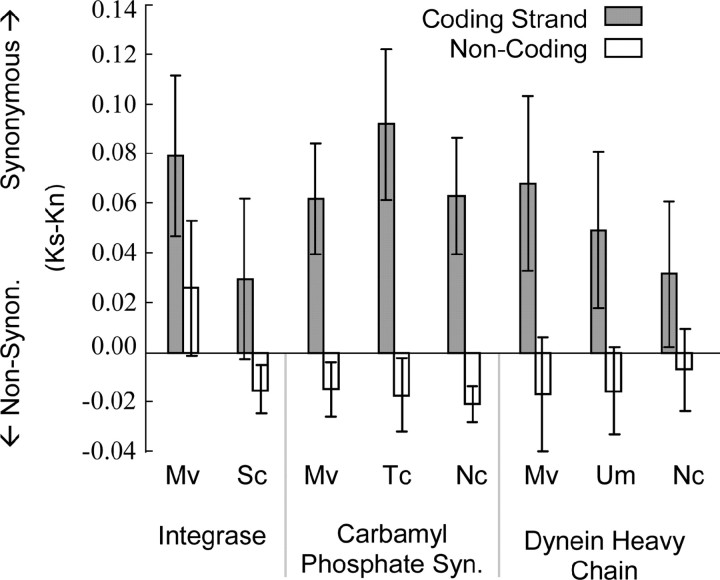
RIP target sites (TCG) in the _copia_-type retrotransposon were distributed such that the induced changes would cause synonymous substitutions more often than expected by chance alone. A comparison of the consensus sequence with a copy mutated at all RIP sites (i.e., all instances of TCG substituted with TTG) resulted in a highly significant excess of synonymous over nonsynonymous substitutions (Mega 2.1 software, _K_s > _K_a _P_-value < 0.001). The tendency toward inducing synonymous substitutions is also true for RIP mutations in the model system of N. crassa (at CA target sites in that species), but only for C-to-T mutations in the coding strand (Selker 2002). However, in the transposable element of M. violaceum, the distribution of RIP target sites in both the coding strand and the noncoding strand tended to produce synonymous substitutions (Figure 3). This result was in contrast to a sample of housekeeping genes from M. violaceum that are not expected to be as greatly exposed to RIP; simulated RIP mutations induced more synonymous substitutions in the coding strand but not in the noncoding strand. The more synonymous changes at TCG sites only in the coding strand of housekeeping genes were not due to a codon bias particular to M. violaceum, but instead were general to other fungal species. Moreover, a comparison of the integrase gene of Ty1 from yeast, where RIP has not been found (Selker 2002), also fit the pattern of housekeeping genes and not the pattern of the M. violaceum transposable element (Figure 3).
Figure 3.—
Effects of simulating RIP-induced mutations on amino acid sequences. All TCG sites were substituted with TTC in coding and noncoding strands. The difference between the substitutions per synonymous site (_K_s) and per nonsynonymous site (_K_a) is shown (analyses conducted in Mega 2.1 software). The analyzed integrase sequences were the consensus sequence _copia_-type retrotransposon fragment from M. violaceum (Mv) and an aligned sequence of Ty1 from Saccharomyces cerevisiae (Sc, NCBI accession no. M18706). Housekeeping genes were analyzed over aligned coding regions: carbamyl phosphate synthase from M. violaceum (Mv, BZ782437), Trichosporon cutaneum (Tc, L08965), Neurospora crassa (Nc, XM_331875), and dynein heavy chain from M. violaceum (Mv, BZ782578), Ustilago maydis (Um, AF403740), and N. crassa (Nc, XM_327261). Bars, standard error.
Chromosome-specific sequence variation:
The _copia_-type retrotransposon copies showed chromosome-specific patterns in their deviations from the consensus sequence. Such patterns were apparent for mutations at both RIP-target sites and other sites and included transition and transversion mutations (Figure 1B). Consequently, by including all sequences, the chromosome-of-origin explained a significant amount of the sequence variation (AMOVA, Table 1; Arlequin 2.0 software; Schneider et al. 2000). Using the conservative approach of removing any identical (i.e., redundant) sequences from within the 30 sequences per chromosome, a smaller but still significant substructuring could be attributed to sequences being more similar within a chromosome than expected by chance alone (Table 2). The mean number of unique sequences per chromosome was 18 (SE = 1.11), with no chromosome differing significantly from the mean. Chromosome-specific patterns were confirmed by replicating the sampling methods with two chromosomes, which most closely resembled the same chromosomes from the first run of the experiment using a distance based on _F_st statistics of sequence variation (Figure 4). The depth of the branches containing the replicated chromosomes reflects a measure of error in the sampling method and furthermore suggests a relationship between the transposable element variation on other chromosomes, specifically chromosomes 9 and 10.
TABLE 1 .
AMOVA using all cloned sequences, with chromosome-of-origin as population substructure
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among chromosomes | 11 | 288.49 | 0.76 | 16.82* |
| Within chromosomes | 345 | 1289.47 | 3.74 | 83.18 |
TABLE 2 .
AMOVA using only nonredundant cloned sequences from within each chromosome, with chromosome-of-origin as population substructure
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among chromosomes | 11 | 94.32 | 0.24 | 5.27* |
| Within chromosomes | 204 | 875.41 | 4.29 | 94.73 |
Figure 4.—
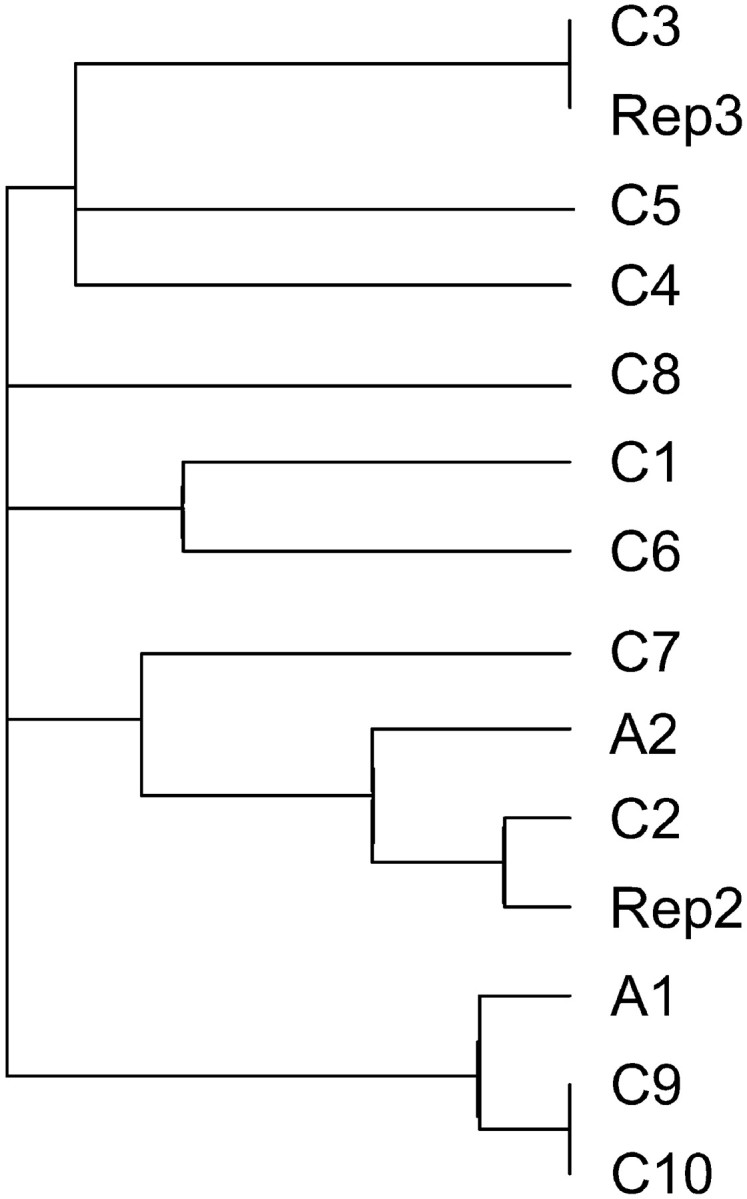
Dendrogram based upon _F_st statistics of _M. violaceum copia_-type retrotransposon sequences grouped by chromosomes-of-origin (_F_st distance analysis in DNAsp 4.0 software; tree constructed in Mega 2.1 software). Sex chromosome (A1 and A2) and autosome karyotype samples (C1–C10) are as indicated in Figure 1A. Rep2 and Rep3 correspond to methodological replication of samples C2 and C3.
A few particular _copia_-type retrotransposon sequences, however, tended to be isolated from multiple chromosomes (Figure 5). Among 139 distinct sequences found in the Microbotryum genome, 110 were isolated from only one chromosome, 12 from two chromosomes, and 3 from three chromosomes. However, assuming a Poisson distribution, sequences found on four or more chromosomes were more common than expected by chance alone at a significance level of P < 0.01. Specifically, 4 sequences were isolated from four chromosomes, 5 from five chromosomes, 3 from six chromosomes, and 2 from seven chromosomes. The consensus sequence was found on four chromosomes. Furthermore, those sequences that were isolated from larger numbers of chromosomes were also more likely to deviate from the consensus only by mutations at RIP target sites or only by silent substitutions (Figure 5) (PROC CORR procedures in SAS, P = 0.01, N = 7). One sequence containing a single-base pair deletion was isolated from five chromosomes (Figure 5).
Figure 5.—
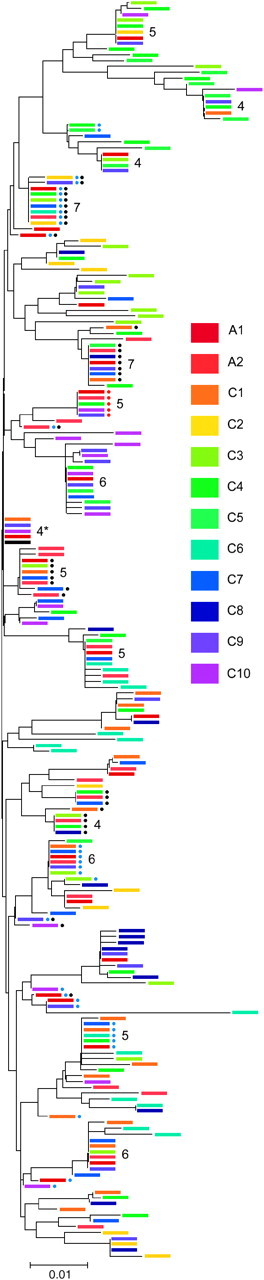
Dendrogram of _copia_-type retrotransposon sequences from M. violaceum (neighbor-joining tree constructed in Mega 2.1 software). Colors indicate chromosome-of-origin as shown in Figure 1. Only nonredundant sequences within a chromosome sample are included. Numbers reflect from how many chromosomes a sequence was isolated (four or more are shown). (*) This sequence is identical to the consensus (black). Sequences that differ from consensus only by silent mutations (black dot) or only by RIP mutations (blue dots) are marked. Sequences marked with red dots contain a single-base pair deletion.
DISCUSSION
Repeat-induced point mutations in M. violaceum:
This study has provided the first evidence of a RIP genome defense against transposable elements in a basidiomycete. The process has been sought in a small number of basidiomycetes other than M. violaceum (Selker 2002), but not necessarily by similar methods, and its distribution throughout the phylum remains to be determined. There are clear similarities between our data with M. violaceum and the RIP process in ascomycetes, specifically that there is hypermutation of cytosine residues at particular nucleotide combinations (Hamann et al. 2000; Anderson et al. 2001; Daboussi et al. 2002; Clutterbuck 2004). However, in contrast to most ascomycetes where RIP is known to occur, the RIP target site in M. violaceum is a trinucleotide (TCG) rather than a dinucleotide. Moreover, the specificity of RIP activity appears higher in M. violaceum than in the other fungi. In particular, the hypermutation was strictly limited to the TCG combination, whereas other systems show only a general preference toward cytosine residues of one or multiple dinucleotide combinations (Clutterbuck 2004). It remains to be determined whether the RIP mechanism in M. violaceum operates by the same molecular pathways as in ascomycetes. However, an important distinction must be drawn with regard to the dikaryotic stage during which RIP is active. The dikaryon is very transient in ascomycetes and occurs in a brief period between mating and karyogamy, but it is the dominant vegetative condition of basidiomycetes. This developmental difference may provide much greater opportunity for RIP activity in basidiomycetes. Transformation studies, such as those conducted in N. crassa and other ascomycetes (e.g., Bhat et al. 2004), can establish when precisely RIP mutations occur in the M. violaceum genome.
Because RIP occurs in a broader range of taxa than previously recognized, it is of greater interest to resolve its evolutionary history and adaptive significance in combating molecular parasites. For example, despite the apparent activity of RIP, the high density of transposable element in M. violaceum is inconsistent with previously studied ascomycetes (Hood et al. 2004; Hood 2005). Moreover, the mutational studies of Garber and Ruddat (1994)(1998, 2000, 2002) indicate that transposable elements continue to be active in M. violaceum. Why then does it appear that RIP is less effective at defending the M. violaceum genome than suggested for ascomycetes (Selker 2002)?
The first of four possible explanations outlined below is that active transposable elements have been repeatedly introduced into the M. violaceum genome by horizontal transfer. For example, the presence of an active Tad element in a RIP-competent strain of N. crassa was explained by a recent genome invasion (Anderson et al. 2001). Also consistent with horizontal transfer is the observation that M. violaceum is particularly rich in the diversity of transposable elements for a species that exhibits a highly selfing mating system (i.e., automixis or intratetrad mating; Hood and Antonovics 2004). There are non-LTR, gypsy, and _copia_-type retrotransposons, as well as _Helitron_-type DNA-based elements (Hood 2005). Moreover, copia elements, as investigated in this study, are the most rare type of transposable elements in fungi, especially among basidiomycetes (Kempken and Kück 1998; Goodwin and Poulter 2001; Wöstemeyer and Kreibich 2002; Díez et al. 2003). However, _copia_-type elements are the most common type in M. violaceum. Whether these elements have originated from sources external to the fungus requires additional studies.
The frequent karyotypic rearrangements in M. violaceum may provide a second explanation for the apparent inability of RIP to effectively defend against transposable elements (Perlin et al. 1997; Hood et al. 2003). In N. crassa, translocations that result in the duplication of large chromosomal regions can act as sinks for the RIP machinery and allow shorter “gene-sized” duplications to go unaffected. As suggested by Bhat and Kasbekar (2001), transposable elements may indirectly contribute to their further proliferation by promoting ectopic recombinations that overwhelm the host's genome defense.
It is also possible that RIP is no longer an active defense in M. violaceum. We may have observed the signatures of ancient RIP mutations in the background of actively proliferating lineages. In fact, RIP-like mutations are reported for some ascomycete fungi that are supposedly asexual in nature and therefore do not experience the dikaryotic stage that is required for RIP activity. It has been suggested that evidence of RIP in the asexual ascomycetes may indicate mutations that occurred either in the dikaryon of a sexual ancestor or during cryptic sex in extant populations (Neuveglise et al. 1996; Hua-Van et al. 1998; Ikeda et al. 2002).
A final possible explanation for the proliferation of transposable elements in M. violaceum is their adaptation to tolerating the RIP genome defense. Ikeda et al. (2002) suggested that the variation for preferred RIP target sites in ascomycetes contains a phylogenetic signal corresponding to the relationships between the species. Therefore, it may be possible to evaluate the changes in RIP specificity as a coevolutionary arms race between the genome defense and transposable elements. To further this hypothesis, this study showed that substitutions at the TCG target sites tended to cause synonymous changes not only on the coding strand but also on the noncoding strand of the _copia_-type retrotransposon in M. violaceum. In contrast, simulating RIP on the noncoding strands of genes not expected to be under selection by RIP tended to produce nonsynonymous changes, including housekeeping genes and even the same integrase region of the Ty1 transposable element from yeast. A broader survey of genes is needed to confirm these results. However, if the pattern observed in M. violaceum is the result of selection against the RIP-sensitive variants, there may be constant pressure to adapt the preferred RIP target site to most effectively inactivate the elements. This coevolutionary dynamic remains to be fully explored from theoretical and empirical perspectives, but may represent an important force in the interactions between molecular parasites and the host genome.
Chromosome-specific sequence variation:
Because chromosomes of fungi can be separated electrophoretically, methods used in this study provide a powerful approach to the intragenomic population structure of molecular parasites. While the transposable element sequences in M. violaceum appear hypermutated by RIP, they showed a statistically significant similarity among copies from the same chromosome. This is contrary to expectations because RIP acts most often between linked copies to diversify them due to the independent nature of mutations on the two copies involved. Furthermore, not all of the chromosome-specific variants were at RIP target sites, and some were transversion-type mutations that are not indicative of RIP. Therefore, a process other than RIP is likely responsible for the chromosomal substructuring of the variation in M. violaceum.
Gene conversion (Elder and Turner 1995) is the probable cause for the chromosome-specific patterns because it acts to homogenize repetitive sequences and does so in a linkage-dependent manner. However, Cambareri et al. (1991) suggested that the hypermutations caused by RIP may diversify repetitive sequences to the point that gene conversion is unable to act upon them. Moreover, theoretical studies suggested that gene conversion is insufficient to counter even normal mutation rates in transposable elements (Slatkin 1985). Recent evidence indicates that this is not always the case. The influence of gene conversion is seen in the MITE elements of Arabidopsis thaliana (Le et al. 2000), S2 elements of Drosophila melanogaster (Maside et al. 2003), Ty1 elements of Saccharomyces cerevisiae, and human Alu elements (Roy et al. 2000). Our sequence data from M. violaceum are consistent with gene conversion between the elements on the same chromosome. Specifically, some elements contain two different chromosome-specific mutation patterns at opposite ends of the sequence (data not shown). It remains possible that such mosaic sequences are the result of reciprocal ectopic recombinations rather than gene conversion, although this is less likely due to fitness costs normally associated with such chromosomal rearrangements. Longer DNA sequences than those used here may identify exchanged sequence “tracts” that are indicative of gene-conversion events (e.g., Sawyer 1989; Betrán et al. 1997).
It is conceivable that processes other than gene conversion give rise to chromosome-specific patterns, but this is unlikely. Thus, it is possible that descendent copies might integrate differentially into their chromosomes-of-origin to create the similarity within chromosomes. However, transcripts of retrotransposons leave the nucleus for reverse transcription prior to integration (Havecker et al. 2004). Although some targeting of elements is known for particular chromosomal regions, such as in telomeric repeats (Anzai et al. 2001; Xie et al. 2001), the local specificity needed to explain our results has never been observed. Furthermore, the control repeat sampling of chromosome bands confirmed that the patterns were chromosome specific, as did the statistics using nonredundant sequences within a chromosome.
The sequence alterations caused by RIP and gene conversion can obscure the evolutionary history of transposable elements. First, conventional approaches to inferring phylogenies from DNA sequence data are precluded because genetic changes are in part induced by the local genomic environment. Second, because RIP tends to cause mutations at synonymous sites, it may create a false signature of purifying selection in the ratio of nonsynonymous to synonymous substitutions (i.e., _d_N/_d_S statistics). Previous studies have taken steps to avoid the complications of RIP by excluding the nucleotide combinations that are targeted for mutation (e.g., Hua-Van et al. 2001). However, it appears from the current study that gene conversion can also have a major influence upon the transposable element population even in the presence of RIP. Further studies are needed to resolve the interplay between these two homology-dependent and linkage-dependent forces, particularly as they have opposite effects upon sequence variation.
Because evolution in the genomic context of RIP and gene conversion is likely to be complicated, the dendrogram of transposable elements from M. violaceum (Figure 5) must be interpreted cautiously. Here, we suggest only that sequences found on more chromosomes than expected by chance probably represent the recently proliferating variants. The similarity in amino acids between the consensus and the most widely distributed sequences further suggests that these are active elements, particularly for the two sequences found on seven chromosomes. Finding a sequence with a base pair deletion on five chromosomes may contribute to the rising evidence of defective retrotransposons that hyperparasitize variants for which the genes are still intact (e.g., Sanz-Alferez et al. 2003; Havecker et al. 2004).
Understanding transposable elements evolution in fungi requires not only further comparative studies on a wider range of taxa, but also a theory that integrates the effects of neutral substitution and selection with the context-specific effects of RIP and gene conversion.
Acknowledgments
We thank Janis Antonovics and Louise Johnson for helpful discussions. We thank Genoscope-CNS for assistance in DNA sequencing. This research was supported by grants MCB-0129995 and DEB-0346832 from the National Science Foundation.
Footnotes
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY729078, AY729484 and AY939912, AY939913, AY939914, AY939915, AY939916, AY939917, AY939918, AY939919, AY939920, AY939921, AY939922, AY939923.
Communicating editor: D. Voytas
References
- Anderson, C., Q. Tand and J. A. Kinsey, 2001. Elimination of active Tad elements during the sexual phase of the Neurospora crassa life cycle. Fungal Genet. Biol. 33**:**49–57. [DOI] [PubMed] [Google Scholar]
- Antonovics, J., M. Hood and J. Partain, 2002. The ecology and genetics of a host shift: Microbotryum as a model system. Am. Nat. 160**:**S40–S53. [DOI] [PubMed] [Google Scholar]
- Anzai, T., H. Takahashi and H. Fujiwara, 2001. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell. Biol. 21**:**100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán, E., J. Rozas, A. Navarro and A. Barbadilla, 1997. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146**:**89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, A., and D. P. Kasbekar, 2001. Escape from repeat-induced point mutation of a gene-sized duplication in Neurospora crassa crosses that are heterozygous for a larger segment duplication. Genetics 157**:**1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, A., R. Tamuli and D. P. Kasbekar, 2004. Genetic transformation of Neurospora tetrasperma, demonstration of repeat-induced point mutation (RIP) in self-crosses and a screen for recessive RIP-defective mutants. Genetics 167**:**1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J. F. Y., 2003. The ripping yarn of the frozen genome. Curr. Biol. 13**:**R552–R553. [DOI] [PubMed] [Google Scholar]
- Cambareri, E. B., M. J. Singer and E. U. Selker, 1991. Recurrence of repeat-induced point mutation (RIP) in _Neurospora crassa._Genetics 127**:**699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., A. Lapid and D. Canada, 1992. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster 1. Element frequencies and distribution. Genet. Res. 60**:**103–114. [DOI] [PubMed] [Google Scholar]
- Clutterbuck, A. J., 2004. MATE transposable elements in Aspergillus nidulans: evidence of repeat-induced point mutation. Fungal Genet. Biol. 41**:**308–316. [DOI] [PubMed] [Google Scholar]
- Daboussi, M. J., J. M. Daviere, S. Graziani and T. Langin, 2002. Evolution of the Fot1 transposons in the genus Fusarium: discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol. 19**:**510–520. [DOI] [PubMed] [Google Scholar]
- Díez, J., T. Béguiristain, F. Le Tacon, J. M. Casacuterta and D. Tagu, 2003. Identification of Ty1-copia retrotransposons in three ectomycorrhizal basidiomycetes: evolutionary relationships and use as molecular markers. Curr. Genet. 43**:**34–44. [DOI] [PubMed] [Google Scholar]
- Elder, J. F., and B. J. Turner, 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 70**:**297–320. [DOI] [PubMed] [Google Scholar]
- Favre, D., 1992. Improved phenol-based method for the isolation of DNA fragments from low melting temperature agarose gels. Biotechniques 13**:**22–26. [PubMed] [Google Scholar]
- Galagan, J. E., and E. U. Selker, 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20**:**417–423. [DOI] [PubMed] [Google Scholar]
- Garber, E. D., and M. Ruddat, 1994. Genetics of Ustilago violacea XXXII. Genetic evidence for transposable elements. Theor. Appl. Genet. 89**:**838–846. [DOI] [PubMed] [Google Scholar]
- Garber, E. D., and M. Ruddat, 1998. Genetics of Ustilago violacea XXXIV. Genetic evidence for a transposable element functioning during mitosis and two transposable elements functioning during meiosis. Int. J. Plant Sci. 159**:**1018–1022. [Google Scholar]
- Garber, E. D., and M. Ruddat, 2000. Genetics of Ustilago violacea XXXV. Transposition in haploid and diploid sporidia and germinating teliospores. Int. J. Plant Sci. 161**:**227–231. [DOI] [PubMed] [Google Scholar]
- Garber, E. D., and M. Ruddat, 2002. Transmission genetics of Microbotryum violaceum (Ustilago violacea): a case history. Adv. Appl. Microbiol. 51**:**107–127. [DOI] [PubMed] [Google Scholar]
- Goodwin, T. J. D., and T. M. Poulter, 2001. The diversity of retrotransposons in the yeast _Cryptococcus neoformans._Yeast 18**:**865–880. [DOI] [PubMed] [Google Scholar]
- Hamann, A., F. Feller and H. D. Osiewacz, 2000. The degenerate DNA transposon Pat and repeat-induced point mutation (RIP) in _Podospora anserina._Mol. Gen. Genet. 263**:**1061–1069. [DOI] [PubMed] [Google Scholar]
- Havecker, E. R., X. Gao and D. Voytas, 2004. The diversity of LTP retrotransposons. Genome Biol. 5**:**225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E., 2002. Dimorphic mating-type chromosomes in the fungus _Microbotryum violaceum._Genetics 160**:**457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E., 2005. Repetitive DNA in the automictic fungus _Microbotryum violaceum._Genetica 124**:**1–10. [DOI] [PubMed] [Google Scholar]
- Hood, M. E., and J. Antonovics, 2004. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity. Genetics 166**:**1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E., J. Antonovics and H. Heishman, 2003. Karyotypic similarity identifies multiple host-shifts of a pathogenic fungus in natural populations. Infect. Genet. Evol. 2**:**167–172. [DOI] [PubMed] [Google Scholar]
- Hood, M. E., J. Antonovics and B. Koskella, 2004. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics 168**:**141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van, A., F. Héricourt, P. Capy, M. J. Daboussi and T. Langin, 1998. Three highly divergent subfamilies of the impala transposable element coexist in the genome of the fungus _Fusariumoxysporum._Mol. Gen. Genet. 259**:**354–362. [DOI] [PubMed] [Google Scholar]
- Hua-Van, A., T. Langin and M.-J. Daboussi, 2001. Evolutionary history of the impala transposon in _Fusarium oxysporum._Mol. Biol. Evol. 18**:**1959–1969. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., H. Nakayashiki, T. Kataoka, H. Tamba, Y. Hashimoto_et al._, 2002. Repeat-induced point mutation RIP in Magnaporthe grisea: implications for its sexual cycle in the natural field environment. Mol. Microbiol. 45**:**1355–1364. [DOI] [PubMed] [Google Scholar]
- Jordan, I. K., and J. F. McDonald, 1999. Tempo and mode of Ty element evolution in _Saccharomyces cerevisiae_Genetics 151**:**1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, I. K., L. V. Matyunina and J. F. McDonald, 1999. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc. Natl. Acad. Sci. USA 96**:**12621–12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V. V., and J. Jurka, 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98**:**8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempken, F., and U. Kück, 1998. Transposons in filamentous fungi—facts and perspectives. BioEssays 20**:**652–659. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115**:**49–63. [DOI] [PubMed] [Google Scholar]
- Le, Q. H., S. Wright, Z. Yu and T. Bureau, 2000. Transposon diversity in _Arabidopsis thaliana._Proc. Natl. Acad. Sci. USA 97**:**7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovsin, N., F. Gubensek and D. Kordis, 2001. Evolutionary dynamics in a novel L2 clade of non-LTR retrotransposons in deuterostomia. Mol. Biol. Evol. 18**:**2213–2224. [DOI] [PubMed] [Google Scholar]
- Maside, X., C. Bartolome and B. Charlesworth, 2003. Inferences on the evolutionary history of the S-element family of _Drosophila melanogaster._Mol. Biol. Evol. 20**:**1183–1187. [DOI] [PubMed] [Google Scholar]
- Neuveglise, C., J. Sarfati, J. P. Latge and S. Paris, 1996. Afutl, a retrotransposon-like element from _Aspergillus fumigatus._Nucleic Acids Res. 24**:**1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin, M. H., C. Hughes, J. Welch, S. Akkaraju, D. Steinecker_et al._, 1997. Molecular approaches to differentiate subpopulations or formae speciales of the fungal phytopathogen _Microbotryum violaceum._Int. J. Plant Sci. 158**:**568–574. [Google Scholar]
- Peterson-Burch, B. D., and D. F. Voytas, 2002. Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol. Biol. Evol. 19**:**1832–1845. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., Y. T. Aminetzach, J. C. Davis, D. Bensasson and A. E. Hirsh, 2003. Size matters: non-LTR retrotransposable elements and ectopic recombination in _Drosophila._Mol. Biol. Evol. 20**:**880–892. [DOI] [PubMed] [Google Scholar]
- Poulter, R. T. M., T. J. D. Goodwin and M. I. Bulter, 2003. Vertebrate helentrons and other novel _Helitrons._Gene 313**:**201–212. [DOI] [PubMed] [Google Scholar]
- Roy, A. M., M. L. Carroll, S. V. Nguyen, A. H. Salem, M. Oldridge_et al._, 2000. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 10**:**1485–1495. [DOI] [PubMed] [Google Scholar]
- Sanz-Alferez, S., P. SanMiguel, Y. K. Jin, P. S. Springer and J. L. Bennetzen, 2003. Structure and evolution of the Cinful retrotransposon family of maize. Genome 46**:**745–752. [DOI] [PubMed] [Google Scholar]
- Sawyer, S., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6**:**526–538. [DOI] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000 Arlequin: a software for population genetics data analysis, Ver 2. Genetics and Biometry Lab, Department of Anthropology, University of Geneva, Geneva.
- Selker, E. U., 2002. Repeat-induced gene silencing in fungi. Adv. Genet. 46**:**439–450. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., 1985. Genetic differentiation of transposable elements under mutation and unbiased gene conversion. Genetics 110**:**145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters, M. K., T. A. Randall, B. S. Margolin, E. U. Selker and D. R. Stadler, 1999. Action of repeat-induced point mutation on both strands of a duplex and on tandem duplications of various sizes in _Neurospora._Genetics 153**:**705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöstemeyer, J., and A. Kreibich, 2002. Repetitive DNA elements in fungi Mycota: impact on genomic architecture and evolution. Curr. Genet. 41**:**189–198. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., N. Agrawal and T. E. Bureau, 2003. Effects of recombination rate and gene density on transposable element distributions in _Arabidopsis thaliana._Genome Res. 13**:**1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W. W., X. Gai, Y. Zhu, D. C. Zappulla, R. Sternglanz_et al._, 2001. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21**:**6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]