Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition (original) (raw)
Abstract
The anaphase-promoting complex (APC) is a multisubunit E3 ubiquitin ligase that targets specific cell cycle-related proteins for degradation, regulating progression from metaphase to anaphase and exit from mitosis. The APC is regulated by binding of the coactivator proteins Cdc20p and Cdh1p, and by phosphorylation. We have developed a purification strategy that allowed us to purify the budding yeast APC to near homogeneity and identify two novel APC-associated proteins, Swm1p and Mnd2p. Using an in vitro ubiquitylation system and a native gel binding assay, we have characterized the properties of wild-type and mutant APC. We show that both the D and KEN boxes contribute to substrate recognition and that coactivator is required for substrate binding. APC lacking Apc9p or Doc1p/Apc10 have impaired E3 ligase activities. However, whereas Apc9p is required for structural stability and the incorporation of Cdc27p into the APC complex, Doc1p/Apc10 plays a specific role in substrate recognition by APC–coactivator complexes. These results imply that Doc1p/Apc10 may play a role to regulate the binding of specific substrates, similar to that of the coactivators.
Keywords: APC/coactivator binding/Doc1-Apc10/substrate recognition/ubiquitylation
Introduction
Controlled protein degradation mediated by ubiquitin-dependent proteolysis underlies the regulation of critical and diverse cellular processes ranging from the cell cycle to signal transduction and transcription events (Hershko and Ciechanover, 1998; Peters et al., 1998). The specificity of the ubiquitylation reaction is conferred by a large and varied collection of E3 ubiquitin ligases that function to facilitate the transfer of ubiquitin from activated E2 ubiquitin-conjugating proteins to lysine residues of defined substrates (Hershko et al., 1983). Formation of a ubiquitin polymer targets these substrates for proteolysis by the proteasome. Regulated cell cycle progression is dependent on both controlled protein synthesis and degradation, and reversible protein phosphorylation (Nurse, 2000). Two structurally related E3 ubiquitin ligases, the SCF and anaphase-promoting complex/cyclosome (APC), are responsible for targeting cell cycle proteins for degradation (Deshaies, 1999; Koepp et al., 1999). The APC is active during mitosis and the G1 phase of the cell cycle, and regulates both the metaphase to anaphase transition and the exit from mitosis (Zachariae and Nasmyth, 1999; Harper et al., 2002; Peters, 2002).
The APC is a complex macromolecular machine of ∼1 MDa conserved in species ranging from yeast to humans. In budding yeast, 11 core subunits have been identified (Zachariae et al., 1998b), including three tetratricopeptide repeat (TPR) subunits (Cdc16p, Cdc23p and Cdc27p) that by analogy with other TPR-containing proteins may function to mediate protein–protein interactions. The RING finger subunit Apc11p and the cullin domain-containing subunit Apc2p are conserved with two SCF subunits, and form the APC catalytic centre, with Apc11p functioning to recruit E2 to the APC (Gmachl et al., 2000; Leverson et al., 2000). However, the primary structures of the majority of APC subunits are unrelated to proteins of defined function, and little is known of their three-dimensional structures and roles. Doc1p/Apc10 is an APC subunit highly conserved from humans to the microsporidia Encephalitozoon cuniculi, and, significantly, a Doc1-homology domain occurs in several other proteins that contain other domains linked to ubiquitylation (Kominami et al., 1998; Grossberger et al., 1999). The crystal structure of the Doc1-homology domain of Doc1p/Apc10 revealed a β-jelly roll structure similar in architecture to proteins of diverse functions, but which share the common property of mediating biomolecular interactions (Wendt et al., 2001; Au et al., 2002). Conserved residues of Doc1p/Apc10 and the Doc1-homology domain map to a surface region, responsible for protein–ligand interactions in other β-jelly roll proteins, suggesting that Doc1p/Apc10 may mediate a function similar to the Doc1-homology domains of other putative E3 ligases, which may use this conserved region for biomolecular interactions. Doc1p/Apc10 plays a critical role in APC function. Hwang and Murray (1997) reported that disruption of DOC1 in budding yeast results in cells that grow only poorly at the permissive temperature (23°C). In addition, mutants of Doc1p/Apc10 in both budding and fission yeast, and in the oligosyndactylism condition of mice, cause cell cycle arrest at metaphase, and the accumulation of mitotic cyclins (Hwang and Murray, 1997; Kominami et al., 1998; Pravtcheva and Wise, 2001), observations consistent with the notion that mutants of Doc1p/Apc10 compromise the activity of the APC. Moreover, in a study of human APC, it was found that only those fractions of the APC containing Doc1p/Apc10 together with core APC subunits Cdc16 and Cdc27 were capable of ubiquitylating cyclin B, whereas other fractions containing Cdc16 and Cdc27 without Doc1p/Apc10 lacked cyclin ubiquitylation activity (Grossberger et al., 1999). It is unknown whether Doc1p/Apc10 plays a direct role in APC activity or whether it is required for an APC function independent of ubiquitin transfer, such as subcellular localization.
Association of the core APC subunits with one of two WD40 repeat-containing coactivator proteins, Cdc20 or Cdh1/Hct1, determines the timing of APC activity and dictates substrate specificity (Schwab et al., 1997; Visintin et al., 1997; Fang et al., 1998; Kramer et al., 1999). Destruction of securin/Pds1 and S-phase cyclins, mediated by the APC in complex with the coactivator Cdc20, is required for the metaphase to anaphase transition, whereas degradation of mitotic cyclins, catalysed by APCCdh1, regulates the exit from mitosis. Control of APC activity and specificity is subject to intricate regulatory mechanisms, including those exerted at the level of coactivator and reversible protein phosphorylation. For example, oscillations of APC specificity and activity during the cell cycle are determined by modulating the ability of the coactivators to bind to the APC via regulated degradation and phosphorylation of Cdc20 and Cdh1, respectively (Zachariae et al., 1998a; Harper et al., 2002). Selection of substrates for APC-catalysed ubiquitylation is dependent upon conserved destruction (D) box and/or KEN box sequence motifs located within the substrate (Glotzer et al., 1991; Pfleger and Kirschner, 2000). Recent findings demonstrating that Cdc20 and Cdh1 interact directly with substrates and that these interactions are compromised or abolished by disruption of the D box and KEN box motifs suggests that coactivators impart substrate specificity to the APC either by direct recruitment of substrates or by allowing the substrate to adopt the appropriate conformation on the APC to enable the ubiquitylation reaction to proceed (Burton and Solomon, 2001; Hilioti et al., 2001; Pfleger et al., 2001; Schwab et al., 2001).
To understand the molecular mechanisms underlying the regulation, activity and specificity of the APC, we have developed a system to purify the APC from budding yeast in quantities sufficient for structural and biochemical studies. By purifying mutant forms of the APC from yeast strains harbouring deletions of genes encoding specific APC subunits, we have begun to delineate the roles and mechanisms of individual APC subunits. Here we report the presence of two novel yeast APC-associated proteins, and demonstrate that Doc1p/Apc10 is necessary for the APC ubiquitylation activity. We have developed an APC–coactivator substrate binding assay using native gels that demonstrates, for the first time, direct binding of substrates to the APC. We show that substrate–APC interactions are dependent on coactivator and are abolished by mutations in either the substrate D or KEN box motifs. Moreover, we found that although Doc1p/Apc10 is not necessary for the association of the coactivator with the APC, it is required for the interaction of substrates with an APC–coactivator complex. This finding implicates Doc1p/Apc10 as a potential APC regulatory subunit that functions directly in APC activity by contributing to APC substrate binding.
Results
Identification of two novel APC-associated proteins: Apc13p/Swm1p and Mnd2p
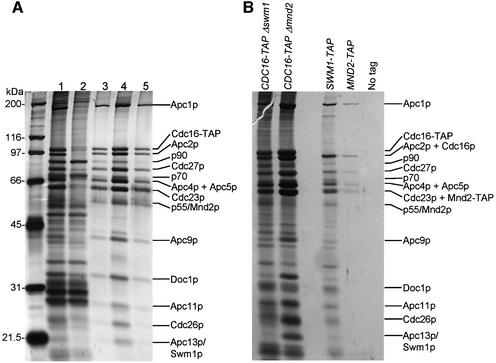
To facilitate purification of the APC, we tagged the C-terminus of the endogenous CDC16 gene in the protease-deficient Saccharomyces cerevisiae strain BJ2168 using the tandem affinity purification (TAP) tag (Rigaut et al., 1999). Using the TAP purification approach, the endogenous APC was purified to >95% homogeneity with a yield of ∼100 µg of APC from 100 g of yeast (Figure 1A). The purified proteins were confirmed as APC subunits using MALDI-TOF mass spectrometry. All 11 of the known APC subunits were present in our preparation and, in addition, two novel APC-associated proteins were identified (see below). Cdc20p and Cdh1p were not detected, consistent with the notion that these coactivators bind to the APC at substoichiometric levels. As observed by Zachariae et al. (1998b), Apc4p and Apc5p co-migrate on SDS–PAGE (Figure 1A). A protein migrating at 19 kDa was shown previously to co-purify with the yeast APC, and tentatively termed Apc13p (Zachariae et al., 1998b). Using mass spectrometry, we determined that Apc13p is Swm1p (spore wall maturation 1), a 19 kDa yeast protein required for spore wall formation, although not for vegetative growth (Ufano et al., 1999). Using two approaches, we confirmed that this APC-associated protein is Swm1p/Apc13p. First, when a TAP tag was incorporated into the endogenous SWM1 gene, all known APC subunits co-purified with Swm1p (Figure 1B). Because the major proteins that co-elute with Swm1-TAP are APC subunits, it is likely that the majority of Swm1p is associated with the APC. Secondly, Apc13p is absent from the APC purified from a CDC16-TAP yeast strain harbouring an SWM1 deletion. Significantly, Δ_swm1_ yeast are temperature sensitive, growing poorly at temperatures >30°C (data not shown), a phenotype similar to Δ_cdc26_ yeast. Another novel APC-associated protein, migrating at 55 kDa on SDS–PAGE, was identified as the 43 kDa protein encoded by the MND2 (meiotic nuclear division 2) gene, first identified in a screen for genes required for meiosis (Rabitsch et al., 2001). Purification of tagged Mnd2p and deletion of MND2 confirmed that the majority of Mnd2p associates with the APC (Figure 1B). The relatively low levels of Mnd2p and Swm1p proteins in silver-stained SDS–gels of APC purified using CDC16-TAP yeast and the low yields of APC purified from the SWM1-TAP and MND2-TAP yeast compared with the CDC16-TAP strain suggest that Mnd2p and Swm1p are associated at substoichiometric levels with the APC. The APC containing these proteins may represent subpopulations of the APC that perform yeast-specific functions differing from the APC composed of the 11 core subunits. Consistent with this notion, database searches identified Swm1p and Mnd2p homologues in related budding yeast but not in Schizosaccharomyces pombe or animal species.
Fig. 1. Purification of the intact APC and identification of subunits. (A) Analysis of the APC purification from CDC16-TAP yeast by silver-stained SDS–PAGE. Cdc16-TAP binds to IgG–Sepharose and is eluted by cleavage with TEV protease (eluate, lane 1). The eluate is bound to calmodulin Sepharose (flow through, lane 2), and eluted with EGTA (elution fractions, lanes 3–5). Proteins identified by MALDI-TOF mass spectrometry are labelled. p90 is a Cdc16p degradation product, p70 is the Hsp70 family heat shock protein SSA2, p55 is Mnd2p, and Apc13p is Swm1p. (B) Identification of Apc13p and p55. Silver-stained SDS–PAGE analysis of APC purifications from CDC16-TAP Δ_swm1_, CDC16-TAP Δ_mnd2_, SWM1-TAP, MND2-TAP and a negative-control yeast strain with no tag.
Purified APC ubiquitylates substrates with D and KEN boxes in vitro
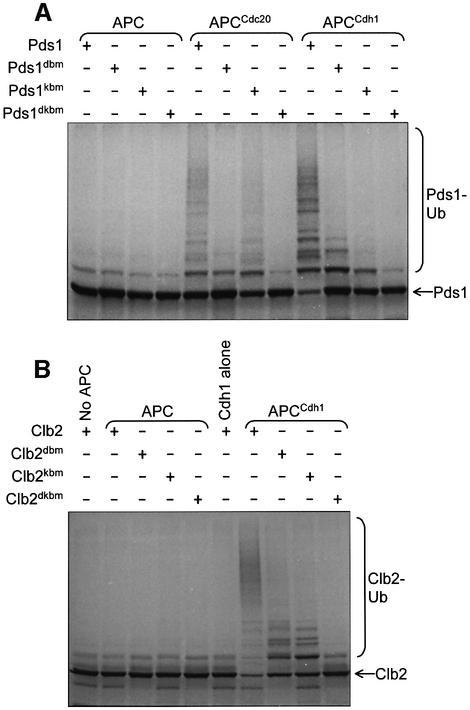
Previous studies on yeast APC have demonstrated in vitro ubiquitylation of an N-terminal D box-containing domain of sea urchin cyclin B, but activity toward endogenous yeast targets has not been demonstrated. We found that the purified yeast APC was active as an E3 ubiquitin ligase towards specific substrates, dependent upon the presence of either the Cdc20p or Cdh1p coactivator (Figure 2). Significantly, the APC catalyses the formation of polyubiquitin chains onto substrate, resulting in high molecular weight polyubiquitylated products (Figure 2). Thus, our in vitro assay reflects the in vivo function of the APC. Both APCCdc20 and APCCdh1 ubiquitylated Pds1p, whereas only APCCdh1 ubiquitylated the mitotic cyclin Clb2p (Figure 2A), and neither APCCdc20 nor APCCdh1 were active towards Cln2p, an SCF substrate (data not shown). The higher activity of APCCdh1 compared with APCCdc20 towards Pds1p (Figure 2B) may reflect more efficient processing of Cdh1p relative to Cdc20p in the reticulocyte lysate (see below). Consistent with the role of the D box and/or KEN box in mediating the substrate specificity of the APC ubiquitylation reaction, we found that mutations of the D and KEN boxes of Pds1p and Clb2p impaired the ability of the APC to ubiquitylate these substrates (Figure 3). Specifically, D box mutations have a greater effect on APCCdc20 activity, whereas KEN box mutations have a greater effect on APCCdh1 activity, findings that reflect the specificity of Cdc20p and Cdh1p towards D and KEN boxes, respectively (Pfleger and Kirschner, 2000; Burton and Solomon, 2001). Substrates with both D and KEN boxes disrupted were not ubiquitylated, consistent with the notion that the APC utilizes a bi-partite recognition motif for efficient substrate degradation (Burton and Solomon, 2001; Harper et al., 2002).
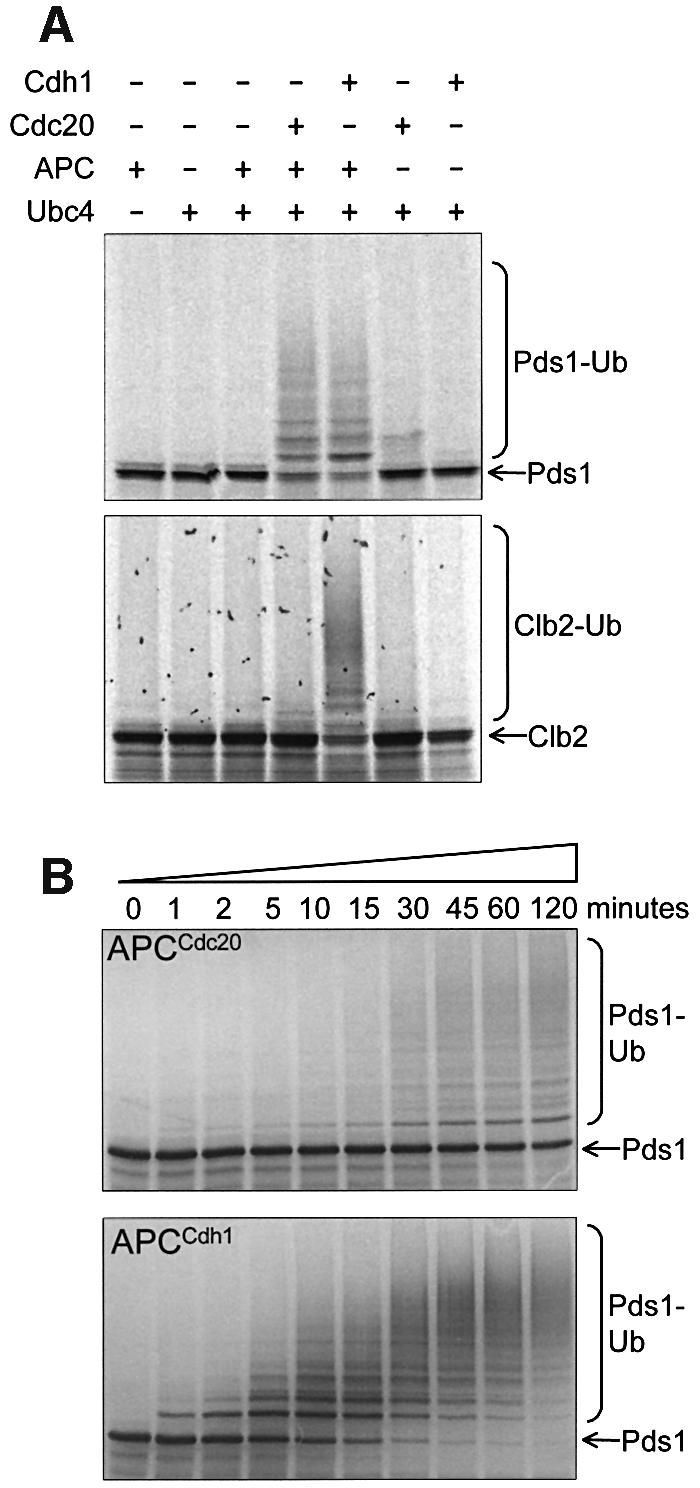
Fig. 2. In vitro activity assays of purified APC. APC was incubated with E2 (Ubc4p), ubiquitin, ATP, 35S-labelled substrate and one of the co-activators Cdc20p or Cdh1p. (A) APC ubiquitylation assays with the substrates Pds1p and Clb2p. The addition of polyubiquitin chains onto 35S-labelled Pds1p or Clb2p results in the appearance of high molecular weight smears. Where neither Cdc20p nor Cdh1p were added, an equivalent volume of reticulocyte lysate was added as a negative control. (B) Time courses of the activities of APC with Cdc20p (APCCdc20) and APC with Cdh1p (APCCdh1) towards the substrate [35S]Pds1p. Samples were taken at the indicated time points and added to gel loading buffer.
Fig. 3. Ubiquitylation of substrates containing D and KEN box mutations. APC ubiquitylation assays were performed on 35S-labelled (A) Pds1p and (B) Clb2p mutants. Pds1dbm, Pds1dkbm, Clb2dbm and Clb2dkbm contained D box mutations, and Pds1kbm, Pds1dkbm, Clb2kbm and Clb2dkbm contained KEN box mutations.
Native gel APC binding assay reveals APC–coactivator interactions
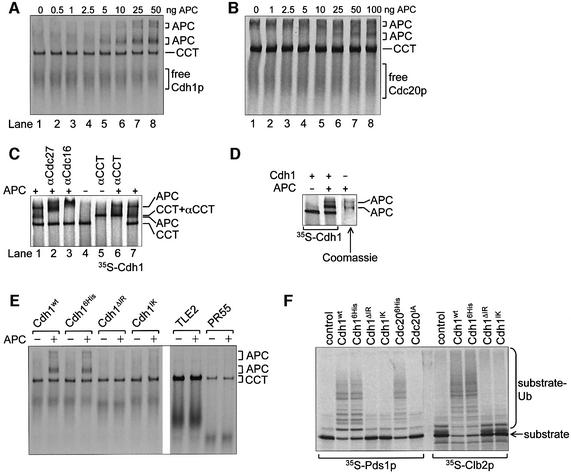
The ability of the core APC to catalyse ubiquitylation reactions depends on its ability to interact with coactivators, E2 and substrates. To understand the regulation and mechanism of these interactions, we developed an APC binding assay based on native gels and proteins produced using an in vitro transcription/translation system (IVT). First, we addressed the question of whether this assay could detect the interaction of Cdc20p and Cdh1p with purified APC. 35S-labelled Cdh1p (63 kDa) and Cdc20p (67 kDa) produced by reticulocyte lysate IVT migrate as two distinct species on native gels (lane 1, Figure 4A and B). A faster, diffuse migrating species most probably represents free Cdh1p/Cdc20p, whereas a well-resolved, slower migrating species correlates with the position of the chaperonin-containing TCP1 (CCT). The mobility of this 35S-labelled band is retarded by an antibody to CCT (Liou et al., 1998), confirming that yeast Cdh1p and Cdc20p interact with rabbit CCT in native gels (compare lanes 4 and 5 in Figure 4C; data not shown). These findings that Cdc20p and Cdh1p associate with CCT are consistent with observations that these coactivators are dependent on CCT for their correct processing in yeast (W.Zachariae and A.Camasses, unpublished data), and a recent genome-wide protein–protein interaction study in yeast showing that Cdc20p and Cdh1p, in common with numerous WD40 repeat proteins, interact with CCT subunits (Ho et al., 2002). Addition of the APC to the Cdh1p or Cdc20p IVT mixture results in the formation of a complex between the APC and coactivator (APCCdh1 or APCCdc20), which is visualized as a band above CCT on the native gel autoradiograms (Figure 4A and B). As increasing amounts of APC are added to Cdh1p or Cdc20p, a second APC–coactivator complex is observed, representing a multimer of APC (Figure 4A and B). Interestingly, previous studies have also alluded to the presence of APC multimers. For example, there is genetic evidence for multimers of budding yeast APC (Zachariae et al., 1996), and Gieffers et al. (2001) observed a slower migrating species on native gels at high concentrations of human APC. These two bands represent the APC since their mobilities in a native gel are retarded by the addition of an antibody to either of the APC subunits Cdc27p or Cdc16p (compare lane 1 with lanes 2 and 3, Figure 4C). In addition, the positions of these bands correspond to the migration position of purified APC, as determined by Coomassie Blue staining (Figure 4D).
Fig. 4. Analysis of the interactions between the APC and its coactivators. (A and B) An increasing amount of purified APC was added to IVT-produced (A) [35S]Cdh1p or (B) [35S]His6-Cdc20p, run on a 5.25% native gel and visualized by autoradiography. The positions of free Cdh1p/Cdc20p and Cdh1p/Cdc20p in complex with APC or CCT are marked. The positions of APC and CCT migration were verified using Coomassie Blue staining. Lane 1 contains no APC. (C) APC and CCT band-shifts using specific antibodies. [35S]His6-Cdh1p was run on a native gel in the presence or absence of APC and an antibody to Cdc27, Cdc16 or CCT. The band corresponding to APCCdh1 is shifted upon the addition of Cdc27 or Cdc16 antibody (lanes 2 and 3), whereas the CCT band is shifted upon the addition of a CCT antibody (lanes 5 and 6). CCT shifted with a CCT antibody runs at the same position as the APC. (D) Analysis of the migration of purified APC using Coomassie Blue-stained native gel analysis. [35S]His6-Cdh1p, in the presence or absence of APC, was analysed on the same gel to compare the positions of pure APC and the APC–[35S]Cdh1p complex. (E) Native gel analysis of Cdh1p mutants. The [35S]Cdh1p mutants were produced using IVT, run on a native gel with and without the addition of APC and analysed using autoradiography. The positions of CCT and APC are shown. Cdh1p mutants have N-terminal His6 tags. Cdh1ΔIR contains a deletion of the C-terminal IR motif, whereas Cdh1IK contains a lysine substitution of the C-terminal arginine. TLE2 and PR55 are negative control WD40 repeat-containing proteins. (F) APC ubiquitylation assays using mutant Cdh1p and Cdc20p with the substrates [35S]Clb2p and [35S]Pds1p. The control lanes contain reticulocyte lysate in place of coactivator. Cdc20IA contains an alanine substitution of the C-terminal arginine.
The native gel assay reveals that lower amounts of Cdc20p bind to the APC than Cdh1p (Figure 4A and 4B), with most of the Cdc20p remaining bound to CCT. This suggests that yeast Cdc20p is processed poorly by rabbit CCT, leaving very little free Cdc20p to interact with the APC, and explaining the reduced ubiquitin ligase activity of APCCdc20 relative to APCCdh1 (Figure 2B). Phosphorylation of Cdc20p and/or APC, or additional factors may be required for efficient APC–Cdc20p interactions. Because the IVT-produced Cdc20p and Cdh1p are required for the ubiquitin ligase activity of the APC, the interaction of these coactivators with the APC detected in the native gel assay is likely to represent a physiologically relevant interaction. In support of this notion, other WD40 repeat proteins, such as the transcriptional repressor TLE2 and the PR55 regulatory subunit of PP2A, synthesized using IVT, were shown to bind to CCT but did not interact with the APC (Figure 4E) (Valpuesta et al., 2002). APC coactivators (Cdc20, Cdh1 and Ama1) from diverse species share an invariant C-terminal IR motif outside of the conserved WD40 domain (Schwab et al., 2001). Deletion of the IR motif or substitution of a lysine for the arginine residue of the motif does not affect the behaviour of Cdh1p in native gels. However, these mutants are unable to activate the APC (Figure 4F), correlating with their inability to bind the APC (Figure 4E). Mutation of the C-terminal arginine of Cdc20p also abolished its ability to bind to and activate the APC (Figure 4F; data not shown).
Interaction of the APC with substrates requires coactivator
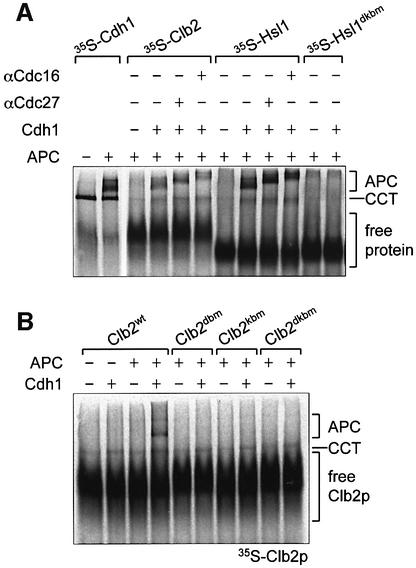
Recent studies demonstrating direct interaction between defined substrates and purified Cdc20p and Cdh1p suggest that coactivators function to activate the APC by means of substrate recruitment to the complex (Burton and Solomon, 2001; Hilioti et al., 2001; Pfleger et al., 2001; Schwab et al., 2001). However, to date, there has been no definitive demonstration that the interactions of substrates with the APC are dependent on the coactivator. To investigate substrate–APC interactions, we used our native gel assay to examine the migration of Clb2p and a D- and KEN box-containing domain of Hsl1p (Hsl1p667–872) in the presence and absence of APC. Hsl1p667–872 is a Swe1p inhibitor and a well characterized APCCdh1/Cdc20 substrate (Burton and Solomon 2001). Using the native gel assay, we visualized directly the association of Clb2p and Hsl1p667–872 with the APC and, moreover, determined that these interactions are dependent on the presence of IVT-produced Cdh1p (Figure 5A). The Clb2p–APCCdh1 and Hsl1p–APCCdh1 interactions are likely to be physiologically relevant because they are abolished by mutations in either the D or KEN box motifs of Clb2p or Hsl1p667–872 (Figure 5). The two slowly migrating bands, which appear upon the addition of APCCdh1 to 35S-labelled substrate, correspond to APCCdh1–substrate complexes because their migrations in a native gel match those of APCCdh1 (Figure 5), but are retarded by the addition of either anti-Cdc16p or anti-Cdc27p antibodies (Figure 5A) in an identical manner to shifts in APCCdh1 (Figure 4C). Interactions between APC and Pds1p were not detectable in the native gel assay, presumably reflecting the lower affinity of APCCdh1 for Pds1p.
Fig. 5. Analysis of APC-substrate interactions. (A) 35S-labelled Clb2p, His6-Hsl1667–872 or His6-Hsl1667–872 dkbm were mixed with APC in the presence or absence of His6-Cdh1p, run on a native gel and analysed by autoradiography. Antibodies to Cdc27p or Cdc16p, which retard the migration of the APC, were added to some samples. [35S]His6-Cdh1 with or without APC was included as a marker for the position of APC. (B) 35S-labelled Clb2p (Clb2wt), Clb2p with a D box mutation (Clb2dbm), Clb2p with a KEN box mutation (Clb2kbm) or Clb2p with both D and KEN box mutations (Clb2dkbm) were mixed with APC. Samples were run on a native gel in the presence or absence of His6-Cdh1p, then analysed by autoradiography.
Role of APC subunits Apc9p, Cdc26p, Swm1p and Mnd2p in the ubiquitylation reaction
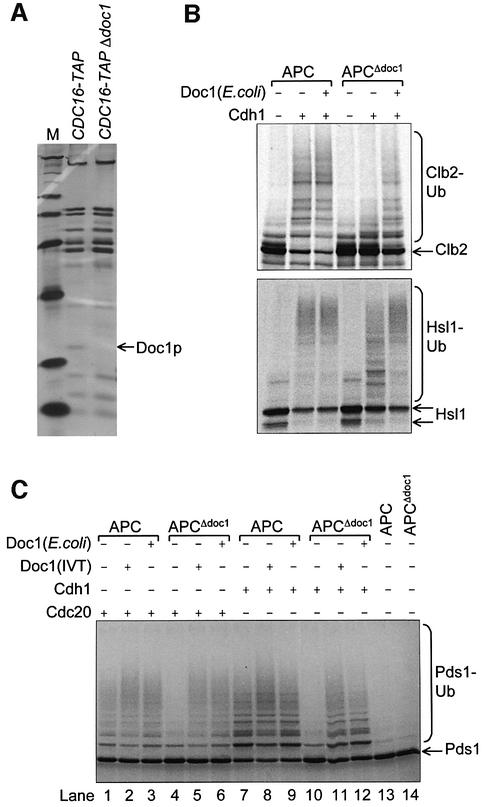
The in vitro ubiquitylation and binding assays described above provide model systems to delineate the roles of individual APC subunits by assessing the properties of APC purified from yeast strains harbouring deletions of one of the five non-essential APC genes (DOC1, APC9, CDC26, SWM1 or MND2). Yeast strains with deletion of DOC1, CDC26 or SWM1 are temperature sensitive, but sufficient quantities of yeast cultures for purification of APCΔdoc1, APCΔcdc26 and APCΔswm1 were obtained by growth at low temperature. Yeast strains with deletion of APC9 or MND2 were not temperature sensitive.
APCΔapc9 contains no Apc9p, and also has significantly reduced amounts of Cdc27p but normal levels of all other subunits (Figure 6A), in agreement with results of Zachariae et al. (1998b), indicating that the budding yeast-specific protein Apc9p mediates the incorporation of Cdc27p during APC assembly. APCΔapc9 has >10-fold less activity than wild-type APC assessed using both APC coactivators (Figure 6B). Addition of exogenous Apc9p and/or Cdc27p does not significantly recover ubiquitylation activity (data not shown). The APC binding assay showed that Cdh1p is unable to bind to APCΔapc9, indicating that one role of Apc9p/Cdc27p is to facilitate APC–coactivator interactions (Figure 6C). The stoichiometries of APC subunits appear normal in APC purified from Δ_cdc26_ yeast (Figure 6A), in contrast to findings of Zachariae et al. (1998b) who reported a reduction in the relative amounts of Cdc16p, Cdc27p and Apc9p in their preparations of APCΔcdc26. These differences in APC subunit composition may result from differences in the temperature of yeast growth (25°C in this study compared with 37°C used by Zachariae et al., 1998b). Purified APCΔcdc26 has wild-type ubiquitylation activity that is not affected by readdition of Cdc26p (Figure 6B; data not shown). This finding would be consistent with a model in which Cdc26p plays a role in assembly of the APC at high temperatures but does not play a direct role in the ubiquitylation reaction. We found that the wild-type APC had very low in vitro ubiquitylation activity at 37°C (data not shown) and, therefore, we were unable to examine further the role of Cdc26p at high temperatures. Finally, APC purified from Δ_swm1_ and Δ_mnd2_ yeast have normal subunit compositions (Figure 1B) and ubiquitylation activities (data not shown).
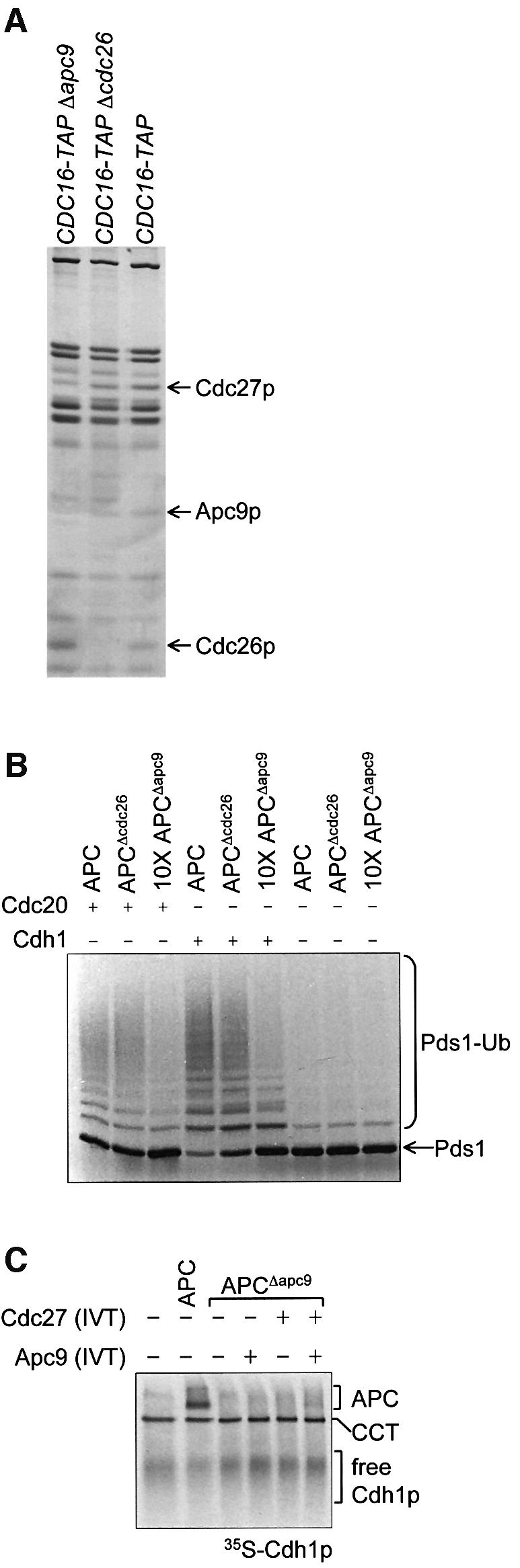
Fig. 6. Purifications and ubiquitylation activities of APCΔapc9 and APCΔcdc26. (A) Silver-stained SDS–PAGE analysis of APC purifications from CDC16-TAP (wild-type), CDC16-TAP Δ_apc9_ and CDC16-TAP Δ_cdc26_ yeast strains. Arrows indicate the positions of deleted or missing subunits. (B) Ubiquitylation activities of APCΔapc9 and APCΔcdc26. Ten-fold more APCΔapc9 was added because it had no apparent activity at lower concentrations. (C) Native gel analysis of Cdh1p binding to APCΔapc9. [35S]His6-Cdh1p was mixed with APC or APCΔapc9, with or without the readdition of Apc9p and/or Cdc27p produced in IVT.
Doc1p is required for E3 ligase activity of the APC
APC purified from the Δ_doc1_ strain (APCΔdoc1) lacks the Doc1p subunit, but contains all other APC subunits at apparently normal stoichiometries (Figure 7A), suggesting that Doc1p is not required for the assembly of the core APC subunits. However, strikingly, APCΔdoc1 is almost completely inactive as an E3 ubiquitin ligase assayed using either Clb2p or Pds1p as a substrate (Figure 7B and C). The readdition to APCΔdoc1 of IVT-produced Doc1p or purified Doc1p overexpressed in Escherichia coli completely restores wild-type ubiquitylation activity (Figure 7B and C). APCΔdoc1 is partially able to ubiquitylate the substrate Hsl1p667–872; however, the readdition of Doc1p substantially increases the amount of ubiquitylated product as well as the length of the polyubiquitin chains (Figure 7B). These results demonstrate that Doc1p plays an important role in promoting the ubiquitylation activity of both APCCdc20 and APCCdh1. Since addition of Doc1p also enhances wild-type APC activity (Figure 7C, compare lane 1 with lane 2, and lane 7 with lane 8), Doc1p may represent a substoichiometric subunit of purified APC.
Fig. 7. Purification and ubiquitylation activity of APCΔdoc1. (A) Silver-stained SDS–PAGE analysis of APC purifications from CDC16-TAP (wild-type) and CDC16-TAP Δ_doc1_ yeast strains. An arrow indicates the position of the deleted Doc1p subunit. M = molecular weight markers. (B and C) Ubiquitylation activity of APCΔdoc1 with the substrates (B) [35S]Clb2, [35S]His6-Hsl1667–872 and (C) [35S]Pds1p. Doc1p produced in IVT or Doc1p purified from E.coli was added where indicated. The lower Hsl1 band is most probably an internal initiation product that does not contain the N-terminal tag.
Doc1p mediates the interaction of substrates with APC–coactivator
Our finding that the compromised E3 ligase activity of APCΔdoc1 can be restored to that of wild-type APC by simple addition of bacterially expressed and purified Doc1p indicated that Doc1p is capable of interacting with the core APC subunits in a potentially reversible and regulated manner. Doc1p may promote APC ubiquitylation activity by one or a combination of mechanisms including: (i) functioning in the E3 ubiquitin ligase catalytic reaction; or (ii) promoting the association of coactivators, the E2-conjugating subunit or substrates with the APC. Such effects could be exerted either directly, or indirectly as a result of Doc1p-induced allosteric changes of the APC. We used the native gel APC binding assay to determine if Doc1p is necessary for the association of coactivators or substrates with the APC. First, we showed that, as expected, IVT-produced Doc1p binds to APCΔdoc1 (Figure 8A). Interestingly, similarly to the coactivators, Doc1p interacts with CCT. Next we found that the binding of both Cdh1p and Cdc20p to APCΔdoc1 is indistinguishable from wild-type APC (Figure 8B and C), indicating that Doc1p is not required for the interaction of the APC with coactivators. As described above, substrates interact with APCCdh1 but not with APC alone (Figure 5). We therefore tested whether Clb2p and Hsl1p667–872 could interact with an APCΔdoc1–Cdh1p complex. Figure 8D shows that Clb2p and Hsl1p667–872 fail to interact with APCΔdoc1–Cdh1p, demonstrating that loss of Doc1p from the APC results in a form of the complex that is incapable of interacting with substrate, explaining the loss of ubiquitylation activity of APCΔdoc1–Cdh1p. Significantly, addition of purified Doc1p to APCΔdoc1–Cdh1p restores the ability of the APC to bind Clb2p and Hsl1p667–872 to the same level as wild-type APC (Figure 8D). These results reveal that Doc1p participates in the formation of a high affinity substrate-binding site on the APC coactivator complex and suggest that Doc1 could function as a potential regulator of APC substrate recognition.
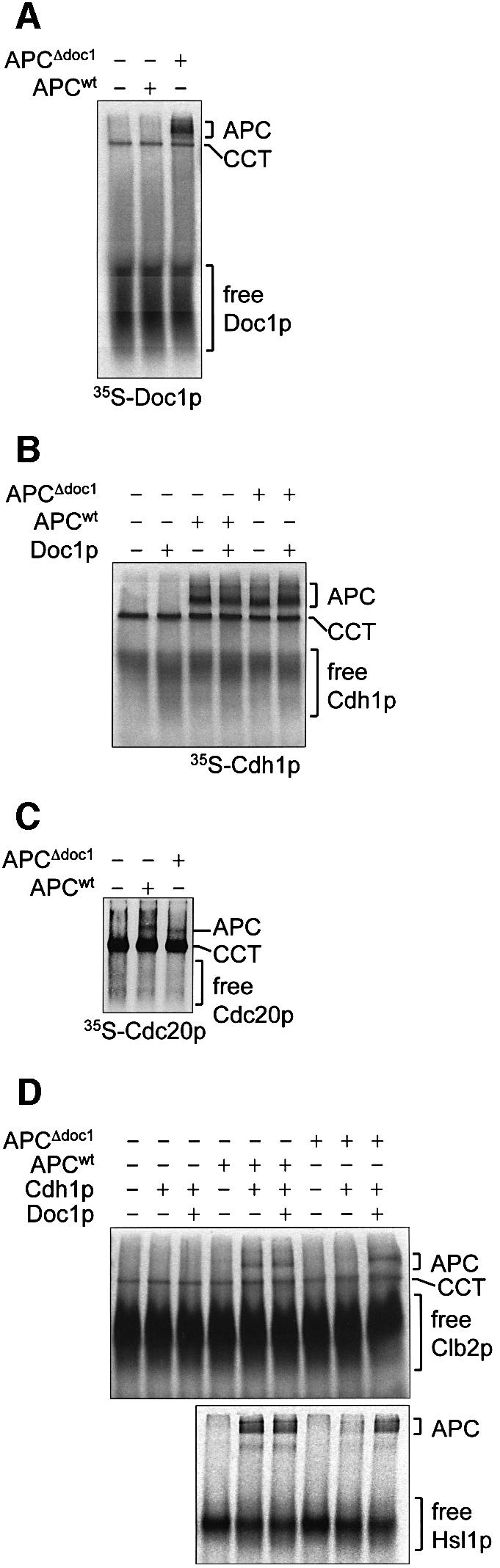
Fig. 8. Binding studies on APCΔdoc1. Native gels were used to analyse protein–protein interactions between (A) [35S]His6-Doc1p and APC or APCΔdoc1; (B) [35S]His6-Cdh1p and APC or APCΔdoc1, in the presence or absence of Doc1p purified from E.coli; (C) [35S]His6-Cdc20p and APC or APCΔdoc1; (D) [35S]Clb2p or [35S]His6-Hsl1667–872 and APC or APCΔdoc1, in the presence or absence of Doc1p purified from E.coli and His6-Cdh1p. APCΔdoc1 sometimes runs slightly faster than APC (C).
Discussion
In this study, we have demonstrated that S.cerevisiae APC purified using a TAP tag is activated as an E3 ubiquitin ligase by the coactivators Cdc20p and Cdh1p. In our assay, the APC mediates the physiologically relevant formation of polyubiquitin chains onto a range of yeast substrates; Clb2p, Hsl1p and Pds1p. We have identified two novel non-essential yeast APC-associated proteins, Swm1p and Mnd2p, that are likely to be components of a subpopulation of APC responsible for specific meiotic and/or yeast functions. It has been suggested that such subpopulations are also present in Drosophila (Huang and Raff, 2002). The in vitro ubiquitylation assay revealed that Pds1p is a substrate of both APCCdc20 and APCCdh1, but only APCCdh1 is active towards the mitotic cyclin Clb2p. Recently, APCCdc20 was shown to be responsible for initiating the degradation of the high levels of Clb2p present early in mitosis, but APCCdh1 is more important in ubiquitylating the lower levels of Clb2p late in mitosis (Yeong et al., 2000; Wasch and Cross, 2002). Our in vitro ubiquitylation results may reflect these higher in vivo activities of APCCdh1 towards low levels of Clb2p compared with APCCdc20. Substrates are targeted for ubiquitylation by the APC via their D box and KEN box motifs mediated by coactivators (Schwab et al., 1997; Visintin et al., 1997; Fang et al., 1998; Kramer et al., 1999). The selectivity of Cdc20 for a substrate is determined primarily by the D box, whereas the KEN box is more important for Cdh1 selectivity, although both motifs contribute to efficient APC-dependent ubiquitylation (Pfleger and Kirschner, 2000; Burton and Solomon, 2001; Harper et al., 2002). The activities of the budding yeast APC in the in vitro ubiquitylation assays towards Pds1p and Clb2p with D and KEN box mutations (Figure 3) are consistent with these preferences.
We have developed a native gel APC binding assay to explore the properties of coactivator and substrate interactions with wild-type and mutant forms of the APC deficient in individual APC subunits. Using the APC binding assay, we reveal for the first time that the association of two substrates, Clb2 and Hsl1, with the APC is dependent on coactivator, thus explaining the ability of Cdc20/Cdh1 to activate the APC (Figure 5). Interactions of Clb2 and Hsl1 with APCCdh1 are abolished by disruptions of either their D or KEN boxes, reflecting the requirement for these motifs for APC-mediated ubiquitylation reactions (Figures 3 and 5). The conserved C box, located N-terminal to the WD40 domain of coactivators, is necessary for APC–Cdh1 interactions (Schwab et al., 2001). Here, the native gel APC binding assay demonstrated that the invariant C-terminal Ile–Arg motif of Cdc20p and Cdh1p is also required for coactivator interactions with the APC, and disruption of this motif is associated with loss of ubiquitylation activity (Figure 4).
Using the in vitro ubiquitylation and APC-binding assays, we have delineated the roles of Apc9p and Doc1p/Apc10 in the activities of the APC. While both APCΔapc9 and APCΔdoc1 have severely reduced E3 ligase activities, only APCΔdoc1 activity can be restored by the readdition of the missing subunit (Figures 6 and 7). APCΔapc9 appears to contain a structural defect causing the loss of at least one other subunit (Cdc27p) and an inability to bind coactivator. In contrast, the absence of Doc1p in APCΔdoc1 does not alter the composition of the remaining APC subunits, suggesting that Doc1p is not involved in the assembly of the core APC, but is likely to be located on the external surface of the APC. Our results are consistent with numerous genetic and biochemical data suggesting that Doc1p/Apc10 is required for the function of the APC to promote degradation of key cell cycle regulatory proteins and to mediate cell cycle progression (Hwang and Murray, 1997; Kominami et al., 1998; Grossberger et al., 1999; Pravtcheva and Wise, 2001). Consistent with our analysis of the APC subunits of APCΔdoc1, the composition of APC subunits is normal in APC isolated from budding and fission yeast with temperature-sensitive Doc1p/Apc10 mutants, suggesting that Doc1p/Apc10 mutants did not inactivate the APC by destabilizing the complex (Hwang and Murray, 1997; Kominami et al., 1998; Grossberger et al., 1999).
With the APC binding assay, we characterized the nature of the defect caused by the loss of Doc1p from the APC. APCΔdoc1 binds Cdh1p and Cdc20p normally; however, whereas a wild-type APCCdh1 complex interacts with Clb2p and Hsl1p, the specific loss of Doc1p eliminates APCCdh1 interactions with both of these substrates (Figure 8). Significantly, readdition of purified Doc1p to APCΔdoc1 restores APC–substrate interactions and E3 ligase activity. These results suggest that Doc1p mediates the E3 ligase activity of the APC by contributing to substrate recognition. The failure of an APCΔdoc1–Cdh1p complex to recruit substrate, even though free Cdh1p has been shown to bind substrate (Burton and Solomon, 2001; Hilioti et al., 2001; Pfleger et al., 2001; Schwab et al., 2001), could be explained in a number of ways. First, the substrate-binding sites of Cdh1p are blocked in APCΔdoc1 but are revealed by a Doc1p-induced conformational change of the APC. Alternatively, in addition to the coactivator, APC subunits are required for high affinity APC–substrate interactions. Doc1p may contribute directly to the substrate-binding site, or it may induce a conformational change of the APC that exposes these sites. Doc1p/Apc10 may have functions reminiscent of the accessory proteins necessary for substrate recruitment to the SCFSkp2 ubiquitin ligase (Ganoth et al., 2001; Harper et al., 2001; Spruck et al., 2001) where Skp2 association with its substrate p27 is dependent on Cks1 that directly interacts with Skp2. We propose that at physiological levels of substrate concentrations, free coactivators and substrates may not interact. Interactions between substrate and coactivator may be dependent on the prior association of the coactivator with the APC. This requirement for the coactivator to be bound to the APC before coactivator–substrate interactions can take place prevents substrate from being sequestered by free coactivator. In humans and yeast, Doc1p/Apc10 is probably not a constitutive subunit of the APC (Hwang and Murray, 1997; Kominami et al., 1998; Grossberger et al., 1999), suggesting a potential for Doc1p to play a regulatory role in the APC ubiquitylation reaction.
Since submission of our manuscript, Carroll and Morgan (2002) reported the finding, using a sea urchin cyclin substrate, that Doc1p is required for the E3 ligase activity of budding yeast APC. Specifically, at physiological substrate concentrations, defects in the processivity and rate of the ubiquitylation reaction were observed, although we note that Lys48-linked ubiquitin polymers were not formed in this assay, possibly because of the artificial substrate used. Kinetic analysis of APCΔdoc1 indicated that whereas affinities for E2 and Cdh1p are normal, defects in APC activity can be explained by a >30-fold increased _K_m for the cyclin substrate. In principle, the increased _K_m may result from a combination of decreased catalytic efficiency (_k_cat) and reduced APC–substrate affinities. Our finding that the Doc1p subunit is required for binding of physiological substrates (Clb2p and Hsl1p) to budding yeast APCCdh1 indicates that the increased _K_m for cyclin substrate observed by Carroll and Morgan, and hence reduced rates of E3 ligase activity, can be explained by decreased APC–substrate affinities. Loss of processivity observed by these authors is consistent with such reduced APC–substrate affinities caused by increased dissociation rate constants and, therefore, reduced APC–substrate retention times. However, Carroll and Morgan (2002) do not demonstrate that the physiologically relevant formation of a polyubiquitin chain (as seen in our assay, Figure 2) is processive, and further experiments are required to establish this.
Detailed understanding of the mechanism of APC–coactivator substrate interactions requires structural studies of such complexes. However, if Doc1p/Apc10 directly participates in the substrate-binding site of the APC, it might be expected that free Doc1p/Apc10 may interact with substrates and be detected in a binding assay and, importantly, it suggests the interesting possibility that a Doc1 recognition motif would be present within APC substrates. Future studies will be directed towards addressing these questions. In conclusion, our study demonstrates that Doc1p contributes to APC–substrate recognition, independently of coactivator, and implicates Doc1p as a potential APC regulatory subunit.
Materials and methods
PCR-based gene targeting and yeast strains
The TAP tag was created using recursive PCR (Prodromou and Pearl, 1992) for optimal S.cerevisiae codon usage, and was inserted into Pac_I–_Asc_I sites of the pFA6a-kanMX6 vector (Wach et al., 1998). PCR products derived from this pFA6a-TAP-kanMX6 vector and containing flanking genomic regions of the gene to be tagged were transformed into the BJ2168 S.cerevisiae strain (Mata leu2 trp1 ura3-52 pep4-3 prc1-407 prb1-1122; Jones, 1977; Zubenko et al., 1980) using the lithium acetate method (Wach et al., 1998). Tagged strains include CDC16-TAP, SWM1-TAP and MND2-TAP. To generate the deletion mutants CDC16-TAP Δ_swm1, CDC16-TAP Δ_mnd2_, CDC16-TAP Δ_doc1_, CDC16-TAP Δ_cdc26_ and CDC16-TAP Δ_apc9_, the open reading frames (ORFs) were replaced with the hygromycin B kinase gene using a PCR product derived from the pAG32c vector (Goldstein and McCusker, 1999). Transformants were selected on YPD plates containing 300 µg/ml G418 and/or 200 µg/ml hygromycin B. Sequences of oligonucleotides and the TAP tag can be provided upon request.
APC purification
Yeast cultures (10–15 l) were grown in YPD at 25 or 30°C and 200 r.p.m. to an OD600 nm of ∼1.0. APC was purified using the TAP method, essentially as described (Rigaut et al., 1999; see also Supplementary data available at The EMBO Journal Online). The final APC storage buffer was 10 mM Tris–HCl pH 8.0, 150 mM NaCl, 10% (w/v) glycerol, 3 mM dithiothreitol (DTT), 1 mM magnesium acetate, 0.1% (v/v) Igepal CA-630, 2 mM EGTA. Samples were analysed by SDS–PAGE on 8% polyacrylamide gels, and silver stained.
Mass spectrometry
Protein bands excised from Coomassie Blue-stained 8 or 15% gels were treated with 12 ng/µl trypsin (Promega) at 4°C overnight (Rosenfeld et al., 1992; Hellman et al., 1995). Tryptic peptide products were resolved using an α-cyano-4-hydroxycinammic acid matrix and MALDI-TOF mass spectrometry (ABI Voyager-DE STR). Database searching of the monoisotopic peptide masses was performed using the MS-Fit algorithm.
Plasmids and reagents
All reagents were constructed using S.cerevisiae ORFs. The His6 tag of pRSET-His6-CLB2 was removed to create pRSET-CLB2. pRSET-PDS1 was made by cloning the PDS1 cDNA into the _Nde_I–_Bam_HI site of pRSET-A. D and KEN box mutants were made using Quickchange site-directed mutagenesis (Stratagene). The PDS1 D box was changed from R85xxL88 to A85xxA88, and the KEN box was changed from K8E9N10 to K8A9A10. The CLB2 D box was changed from R25xxL29xxxxN33 to A25xxA29xxxxA33, and the KEN box was mutated from K100E101N102 to A100A101A102. MBP–Hsl1667–872 (wild-type) and MBP–Hsl1667–872mDB/mKB (containing D and KEN box mutations) were obtained from Mark J.Solomon. Hsl1667–872 and Hsl1667–872mDB/mKB were subcloned into the _Bam_HI–_Pst_I sites of pRSET-A to make His6-Hsl1667–872 and His6-Hsl1667–872 dkbm. The CDC20 and CDH1 cDNAs were cloned into the _Eco_RI–_Sal_I sites of pET28c for pET28–His6-Cdc20 and pET28–His6-Cdh1. PCR mutagenesis was performed to make the following mutations: R610A in pET22-His6-Cdc20IA, deletion of I565 and R566 in pET22-His6-Cdh1ΔIR, R566K in pET22-His6-Cdh1IK, and removal of the His6 tag in pET22-Cdh1wt. The cDNAs for CDC27, APC9 and DOC1 were cloned into pET28 vectors, and the UBC4 ORF was cloned into pET15b, all with N-terminal His6 tags. Plasmids containing the WD40 repeats of human TLE2 (pET17b-TLE2, residues 416–743) and PR55/Bα (pT7T3D-Pac-PR55/Bα) were provided by Laura Pickles and Brian Hemmings, respectively. His6-Ubc4p and His6-Doc1p were overexpressed and purified from E.coli. The affinity-purified Cdc27 polyclonal antibody was made using a peptide corresponding to the N-terminus of Cdc27p (amino acids 4–21).
Ubiquitylation assays
Substrates (Pds1p and Clb2p), activators (His6-Cdc20p and His6-Cdh1p), APC subunits (Apc9p, Cdc27p and Doc1p) and control non-APC-binding WD40 repeat proteins (TLE2 and PR55) were prepared using TNT T7 Quick coupled in vitro transcription/translation (Promega). Ubiquityl ation assays were performed for 45 min at room temperature in 10 µl reaction volumes with 40 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.6 mM DTT, 2.7 mM ATP, 6.6 µg of ubiquitin (Affiniti), 500 ng of His6-Ubc4p, 200 ng of ubiquitin aldehyde (Affiniti), 2 µM LLnL (_N_-acetyl-Leu-Leu-Norleu-aldehyde; Sigma), 1 µl of 35S-labelled substrate, 2 µl of Cdc20p or Cdh1p or reticulocyte lysate (negative control) and 10 ng of APC. The addition of exogenous E1 was not necessary since E1 is abundant in reticulocyte lysate. Reactions were analysed by 8% SDS–PAGE.
Native gel assays
APC (50 ng in 1–2 µl) was mixed with 2 µl of 35S-labelled IVT-produced protein and 0.7 µl of 100 mM CaCl2, and brought up to a final volume of 14 µl with binding buffer (10 mM Tris pH 8.0, 150 mM NaCl, 3 mM DTT, 1 mM magnesium acetate, 2 mM EGTA). For some reactions, 2 µl of unlabelled IVT-produced His6-Cdc20p, His6-Cdh1p or Cdh1p was added, or 2 µl of reticulocyte lysate was used as a negative control. For antibody band shifts, 2 µl of αCCT (23C; Liou et al., 1998), αCdc27 or αCdc16 (provided by Jane Endicott) was added. Samples were incubated at room temperature for 15 min, 1 µl of native gel loading buffer [125 mM Tris pH 8.0, 84% (v/v) glycerol] was added and the entire reaction was loaded onto a 5.25% non-denaturing polyacrylamide gel (Liou and Willison, 1997). Gels were fixed and stained with Coomassie Blue, dried and exposed to film.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank B.Hemmings, X.Leng, B.Panaretou, L.Pickles, M.J.Solomon and J.A.Endicott for reagents, C.Richardson for designing oligonucleotides, and W.Zachariae and A.Camasses for communicating results prior to publication. This work was supported by grants from HFSP (D.B. and J.W.H.), by CR-UK (D.B. and K.R.W.) and by NIH grant AG11085 to J.W.H. L.P. was supported by the ICR and the BACR.
Note added in proof
Yoon et al. (2002) recently reported the identification of Mnd2p and Swm1p as subunits of S.cerevisiae APC.
Yoon,H.J., Feoktistova,A., Wolfe,B.A., Jennings,J.L., Link,A.J. and Gould,K.L. (2002) Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr. Biol., 12, 2048–2054.
References
- Au S.W., Leng,X., Harper,J.W. and Barford,D. (2002) Implications for the ubiquitination reaction of the anaphase-promoting complex from the crystal structure of the Doc1/Apc10 subunit. J. Mol. Biol., 316, 955–968. [DOI] [PubMed] [Google Scholar]
- Burton J.L. and Solomon,M.J. (2001) D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev., 15, 2381–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W. and Morgan,D.O. (2002) The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat. Cell Biol., 4, 880–887. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell, 2, 163–171. [DOI] [PubMed] [Google Scholar]
- Ganoth D., Bornstein,G., Ko,T.K., Larsen,B., Tyers,M., Pagano,M. and Hershko,A. (2001) The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat. Cell Biol., 3, 321–324. [DOI] [PubMed] [Google Scholar]
- Gieffers C., Dube,P., Harris,J.R., Stark,H. and Peters,J.M. (2001) Three-dimensional structure of the anaphase-promoting complex. Mol. Cell, 7, 907–913. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray,A.W. and Kirschner,M.W. (1991) Cyclin is degraded by the ubiquitin pathway. Nature, 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Gmachl M., Gieffers,C., Podtelejnikov,A.V., Mann,M. and Peters,J.M. (2000) The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 97, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L. and McCusker,J.H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast, 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Grossberger R., Gieffers,C., Zachariae,W., Podtelejnikov,A.V., Schleiffer,A., Nasmyth,K., Mann,M. and Peters,J.M. (1999) Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J. Biol. Chem., 274, 14500–14507. [DOI] [PubMed] [Google Scholar]
- Harper J.W. (2001) Protein destruction: adapting roles for Cks proteins. Curr. Biol., 11, R431–R435. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Burton,J.L. and Solomon,M.J. (2002) The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev., 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- Hellman U., Wernstedt,C., Gonez,J. and Heldin,C.H. (1995) Improvement of an ‘in-gel’ digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem., 224, 451–455. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hershko A., Heller,H., Elias,S. and Ciechanover,A. (1983) Components of ubiquitin-protein ligase system. Resolution, affinity purification and role in protein breakdown. J. Biol. Chem., 258, 8206–8214. [PubMed] [Google Scholar]
- Hilioti Z., Chung,Y.S., Mochizuki,Y., Hardy,C.F. and Cohen-Fix,O. (2001) The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol., 11, 1347–1352. [DOI] [PubMed] [Google Scholar]
- Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Huang J.Y. and Raff,J.W. (2002) The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci., 115, 2847–2856. [DOI] [PubMed] [Google Scholar]
- Hwang L.H. and Murray,A.W. (1997) A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol. Biol. Cell, 8, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.W. (1977) Proteinase mutants of Saccharomyces cerevisiae. Genetics, 85, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D.M., Harper,J.W. and Elledge,S.J. (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell, 97, 431–434. [DOI] [PubMed] [Google Scholar]
- Kominami K., Seth-Smith,H. and Toda,T. (1998) Apc10 and Ste9/Srw1, two regulators of the APC-cyclosome, as well as the CDK inhibitor Rum1 are required for G1 cell-cycle arrest in fission yeast. EMBO J., 17, 5388–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer,N., Podtelejnikov,A.V., Mann,M. and Peters,J.M. (1999) Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell, 11, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson J.D., Joazeiro,C.A., Page,A.M., Huang,H., Hieter,P. and Hunter,T. (2000) The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol. Biol. Cell, 11, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou A.K. and Willison,K.R. (1997) Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J., 16, 4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou A.K., McCormack,E.A. and Willison,K.R. (1998) The chaperonin containing TCP-1 (CCT) displays a single-ring mediated disassembly and reassembly cycle. Biol. Chem., 379, 311–319. [DOI] [PubMed] [Google Scholar]
- Nurse P. (2000) A long twentieth century of the cell cycle and beyond. Cell, 100, 71–78. [DOI] [PubMed] [Google Scholar]
- Peters J.M., King,R.W. and Deshaies,R.J. (1998) Cell cycle control by ubiquitin-dependent proteolysis. In Peters,J.M., Harris,J.R. and Finey,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, NY, pp. 345–387.
- Peters J.M. (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell, 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Pfleger C.M. and Kirschner,M.W. (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev., 14, 655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., Lee,E. and Kirschner,M.W. (2001) Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev., 15, 2396–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravtcheva D.D. and Wise,T.L. (2001) Disruption of Apc10/Doc1 in three alleles of oligosyndactylism. Genomics, 72, 78–87. [DOI] [PubMed] [Google Scholar]
- Prodromou C. and Pearl,L.H. (1992) Recursive PCR: a novel technique for total gene synthesis. Protein Eng., 5, 827–829. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P. et al. (2001) A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol., 11, 1001–1009. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Seraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J., Capdevielle,J., Guillemot,J.C. and Ferrara,P. (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem., 203, 173–179. [DOI] [PubMed] [Google Scholar]
- Schwab M., Lutum,A.S. and Seufert,W. (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell, 90, 683–693. [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner,M., Mocker,D. and Seufert,W. (2001) Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J., 20, 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck C., Strohmaier,H., Watson,M., Smith,A.P., Ryan,A., Krek,T.W. and Reed,S.I. (2001) A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol. Cell, 7, 639–650. [DOI] [PubMed] [Google Scholar]
- Ufano S., San-Segundo,P., del Rey,F. and Vazquez de Aldana,C.R. (1999) SWM1, a developmentally regulated gene, is required for spore wall assembly in Saccharomyces cerevisiae. Mol. Cell Biol., 19, 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valpuesta J.M., Martin-Benito,J., Gomez-Puertas,P., Carrascosa,J.L. and Willison,K.R. (2002) Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 529, 11–16. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz,S. and Amon,A. (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Rebischung,C., Steiner,S., Pokorni,K., te Heesen,S. and Philippsen,P. (1998) PCR-based gene targeting in S.cerevisiae. In Brown,A.J.P. and Tuite,M. (eds), Methods in Microbiology, Vol. 26: Yeast Gene Analysis. Academic Press, London, UK, pp. 67–82.
- Wasch R. and Cross,F.R. (2002) APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature, 418, 556–562. [DOI] [PubMed] [Google Scholar]
- Wendt K.S., Vodermaier,H.C., Jacob,U., Gieffers,C., Gmachl,M., Peters,J.M. Huber,R. and Sondermann,P. (2001) Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat. Struct. Biol., 8, 84–88. [DOI] [PubMed] [Google Scholar]
- Yeong F.M., Lim,H.H., Padmashree,C.G. and Surana,U. (2000) Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell, 5, 501–511. [DOI] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shin,T.H., Galova,M., Obermaier,B. and Nasmyth,K. (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science, 274, 1201–1204. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab,M., Nasmyth,K. and Seufert,W. (1998a) Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science, 282, 1721–1724. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shevchenko,A., Andrews,P.D., Ciosk,R., Galova,. M, Stark,M.J., Mann,M. and Nasmyth,K. (1998b) Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science, 279, 1216–1219. [DOI] [PubMed] [Google Scholar]
- Zubenko G.S., Mitchell,A.P. and Jones,E.W. (1980) Mapping of the proteinase b structural gene PRB1, in Saccharomyces cerevisiae and identification of nonsense alleles within the locus. Genetics, 96, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]