Extensive Chromosomal Repatterning and the Evolution of Sterility Barriers in Hybrid Sunflower Species (original) (raw)
Abstract
New species may arise via hybridization and without a change in ploidy. This process, termed homoploid hybrid speciation, is theoretically difficult because it requires the development of reproductive barriers in sympatry or parapatry. Theory suggests that isolation may arise through rapid karyotypic evolution and/or ecological divergence of hybrid neospecies. Here, we investigate the role of karyotypic change in homoploid hybrid speciation by generating detailed genetic linkage maps for three hybrid sunflower species, Helianthus anomalus, H. deserticola, and H. paradoxus, and comparing these maps to those previously generated for the parental species, H. annuus and H. petiolaris. We also conduct a quantitative trait locus (QTL) analysis of pollen fertility in a BC2 population between the parental species and assess levels of pollen and seed fertility in all cross-combinations of the hybrid and parental species. The three hybrid species are massively divergent from their parental species in karyotype; gene order differences were observed for between 9 and 11 linkage groups (of 17 total), depending on the comparison. About one-third of the karyoypic differences arose through the sorting of chromosomal rearrangements that differentiate the parental species, but the remainder appear to have arisen de novo (six breakages/six fusions in H. anomalus, four breakages/three fusions in H. deserticola, and five breakages/five fusions in H. paradoxus). QTL analyses indicate that the karyotypic differences contribute to reproductive isolation. Nine of 11 pollen viability QTL occur on rearranged chromosomes and all but one map close to a rearrangement breakpoint. Finally, pollen and seed fertility estimates for F1's between the hybrid and parental species fall below 11%, which is sufficient for evolutionary independence of the hybrid neospecies.
OVER 90% of plant and animal species differ in their karyotypes (White 1978; King 1993). Many karyotypic differences are likely to be incidental to speciation, either because they arose after reproductive isolation was complete or because they have little impact on hybrid fertility (e.g., John 1981; Coyne et al. 1991,1993) or recombination rates (Rieseberg 2001). However, some chromosomal rearrangements may trigger speciation (Levin 2002), particularly when geographical barriers to gene flow are absent. In these situations, the rearrangements may decrease gene flow sufficiently among diverging populations to allow selected differences or hybrid incompatibilities to accumulate (Rieseberg et al. 1999; Noor et al. 2001; Navarro and Barton 2003; Brown et al. 2004). Chromosomal rearrangements may impede gene flow by causing a reduction in hybrid viability or fertility (Barton and Bengtsson 1986; King 1993) and/or a reduction in recombination among the rearranged chromosomes (Noor et al. 2001; Machado et al. 2002; Navarro and Barton 2003).
It is not yet clear which of these mechanisms (reduced hybrid fertility vs. reduced recombination) most commonly contributes to chromosomal speciation, but models that treat chromosomal rearrangements as recombination modifiers (Noor et al. 2001; Rieseberg 2001) have stronger theoretical support. Mathematical analyses indicate that hybrid incompatibilities arise several times faster in regions of low recombination than in other genomic regions (Navarro and Barton 2003). However, these models are mostly applicable to inversions, which are known to greatly reduce effective recombination rates (Greenbaum and Reed 1984; Hale 1986; Navarro and Ruiz 1997; Coyne et al. 1993). In contrast, translocations and other kinds of rearrangements have smaller effects on recombination rates (although, see Davisson and Akeson 1993).
Chromosomal speciation models that rely on a reduction in hybrid fertility generally have less theoretical support than recombination modifier models because rearrangements that strongly reduce heterozygote fitness (i.e., underdominance) are unlikely to be fixed through drift, except in small, inbred populations (Hedrick 1981; Walsh 1982; Lande 1985). Weakly underdominant mutations are more easily established, but will not be very effective as reproductive barriers.
Several solutions to this paradox have been suggested. In chain or cascade models, strong isolation arises following the accumulation of weakly underdominant rearrangements (White 1978). Other models posit a scenario in which geographically isolated populations become fixed for different rearrangements (e.g., centric fissions) that have no effect on fertility on their own, but are strongly underdominant in combination (Baker and Bickham 1986). A third solution—recombinational speciation—invokes interspecific hybridization as a mechanism for karyotypic evolution; reproductive isolation is achieved via the sorting of chromosomal rearrangements that differentiate the parental species (Grant 1981) or by rearrangements induced by recombination (Templeton 1981; Rieseberg et al. 1995). Because rearrangement polymorphisms often are initiated at high frequencies in hybrid populations, establishment of a new homokaryotype through drift poses fewer theoretical hurdles than in nonhybrid populations (McCarthy et al. 1995; Pialek et al. 2001), particularly when there is ecological and spatial isolation between the hybrid derivative and its parental species (Buerkle et al. 2000).
This article focuses on the third mode of chromosomal speciation, recombinational speciation, using the annual sunflowers of the genus Helianthus as a study system. This group is well suited for the study of recombinational speciation because three of 11 species (Helianthus anomalus, Helianthus deserticola, and Helianthus paradoxus) are stabilized diploid hybrid derivatives of the same two parental species (Helianthus annuus and Helainthus petiolaris; Rieseberg 1991). Furthermore, meiotic analyses suggest that the parental and hybrid species differ significantly in chromosome structure (Chandler et al. 1986), an observation consistent with the recombinational model. Genetic map-based comparison of one of the hybrid species (H. anomalus) with that of its parents indicates that hybrid speciation was accompanied by massive karyotypic change (Rieseberg et al. 1995), as predicted by the earlier meiotic studies.
Here we extend these earlier comparative mapping studies to include the two other hybrid species, H. deserticola and H. paradoxus. We also provide a microsatellite map for H. anomalus, which provides significant additional resolution relative to the previously published RAPD map for this species (Rieseberg et al. 1995) and enables direct comparisons with the newly developed maps for H. deserticola and H. paradoxus. Microsatellite maps of the three hybrid species are compared to those recently published for the parental species (Burke et al. 2004).
In addition to the comparative mapping work, we analyze the fertility effects of the mapped chromosomal rearrangements by conducting a quantitative trait locus (QTL) analysis of pollen viability in a BC2 population between the parental species, H. annuus and H. petiolaris. Finally, we report on the pollen and seed fertility of first-generation hybrids from all combinations of crosses between the two parental species and their three hybrid-derivative species.
Results from these three data sets are used to address the following questions/assumptions that underlie the recombinational speciation model:
- Has karyotypic change accompanied the formation of the three hybrid species as required by the recombinational model and, if so, what is the nature of the karyotypic changes? That is, do the hybrid species simply possess a combination of parental rearrangements, or are there unique rearrangements in the hybrids? Also, do novel rearrangements most commonly involve chromosomes that are already rearranged between the parental species as predicted by theory (Templeton 1981)?
- Are map lengths greater in the hybrid than in parental species? Grant (1958) argues that high recombination rates will be favored during recombinational speciation.
- Are chromosomal rearrangements in Helianthus strongly underdominant as assumed by the recombinational model? Specifically, what fraction of the variance in pollen sterility maps to chromosomal rearrangements and how large are the effects of individual translocations and inversions?
- Are chromosomal sterility barriers between the hybrid and parental species strong enough to allow the hybrid species to evolve independently, even when parapatric with parental species populations (Buerkle et al. 2000)?
MATERIALS AND METHODS
Study system:
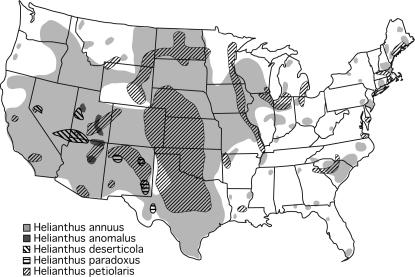
The five Helianthus species analyzed are diploid (n = 17), self-incompatible annuals, all native to North America (Figure 1). H. annuus and H. petiolaris, the parental species, are abundant in the central and western United States, but differ in habitat preference. H. annuus occurs in mesic, clay-based soils, whereas H. petiolaris is found in drier, sandier soils. The three hybrid species, in contrast, are restricted to extreme habitats in the desert Southwest. H. anomalus is found on desert sand dunes in Utah and northern Arizona, H. deserticola in more stabilized deposits in the Great Basin Desert (Nevada, Utah, Arizona), and H. paradoxus in saline desert wetlands in west Texas and New Mexico (Heiser et al. 1969; Rogers et al. 1982). Divergence times estimated from microsatellite and chloroplast DNA variation place the divergence of the parental species between 75,000 and 1 million years before present (BP) and the origin of their three hybrid derivatives between 60,000 and 200,000 years BP (Rieseberg et al. 1991; Schwarzbach and Rieseberg 2002; Welch and Rieseberg 2002; Gross et al. 2003).
Figure 1.
Present-day distributions of the two parental species, H. annuus and H. petiolaris, and their three hybrid-derivative species, H. anomalus, H. deserticola, and H. paradoxus (based on Rogers et al. 1982).
The five species appear to be strongly isolated reproductively. All five taxa occur in very different habitats and reciprocal transplant experiments demonstrate adaptive ecological divergence (Lexer et al. 2003b; Gross et al. 2004; Ludwig et al. 2004). Previous mapping studies (Rieseberg et al. 1995; Burke et al. 2004) indicate that the genomes of H. annuus, H. anomalus, and H. petiolaris are extensively rearranged and pollen viability of F1 hybrids from all three cross-combinations is <5% (Rieseberg 2000). Despite these reproductive barriers, contemporary hybrid zones are common between H. annuus and H. petiolaris (Heiser 1947; Rieseberg et al. 1998). In contrast, no natural hybrids have been reported between the three hybrid species and their parents, although hybridization has been detected between H. anomalus and H. deserticola in the Little Sahara Sand Dunes in Utah, the one location they are known to occur in parapatry.
Genetic mapping:
High-resolution genetic linkage maps were generated for each of the three hybrid species (supplementary Figures S1–S3, available at http://www.genetics.org/supplemental/). For H. anomalus, crosses were made between ANO 1497 (Mexican Water, AZ) and ANO 1506 (Hanksville, UT) (Rieseberg et al. 1995). Likewise, crosses were made between DES 1471 (Beaver Dam, AZ) and DES 1476 (Virgin, UT) for H. deserticola, and between PAR 1084 (Fort Stockton, TX) and PAR 1671 (Dexter, NM) for H. paradoxus. For each taxon, an intraspecific hybrid from the populations listed above was crossed to an inbred sunflower line (CMSHA89). This crossing design allowed us to monitor the segregation of alleles from each of the ancient hybrid species against a homogenous genetic background. A total of 54, 58, and 57 individuals were used for genetic map construction in H. anomalus, H. deserticola, and H. paradoxus, respectively.
A variety of markers were employed for mapping. For H. anomalus, this study added 241 microsatellite and 78 AFLP markers to the previously published 701 AFLP/RAPD marker map of H. anomalus (Ungerer et al. 1998), bringing the total number of markers mapped to 1019. The H. deserticola and H. paradoxus maps were generated from 120 microsatellite and 552 AFLP markers (672 markers total) and 174 microsatellite and 597 AFLP markers (771 markers total), respectively. While map positions of parental species-specific AFLP and RAPD markers were previously reported (427, 290, and 325 markers in H. anomalus, H. deserticola, and H. paradoxus, respectively; Rieseberg et al. 2003), markers informative for comparative mapping are described here for the first time.
Maps were developed with the computer program Mapmaker Macintosh V2.0 (Du Pont, Wilmington, DE). Markers were divided into groups using LOD scores of >6 and recombination limits of <0.15. Mapmaker's three-point analysis was used to determine likely orders among markers, followed by multipoint analysis to resolve any discrepancies. Marker orders were confirmed using Mapmaker's “ripple” command, and recombination values were converted to map distances using Kosambi's (1944) mapping function.
For comparisons with the parental species, we employed genetic maps previously described for H. annuus and H. petiolaris (Burke et al. 2004). The H. annuus map was constructed from four different maps for this species (Gedil et al. 2001; Burke et al. 2002; Tang et al. 2002; Yu et al. 2003) using JoinMap version 3.0 (Van Ooijen and Voorrips 2001). The integrated map includes 288 microsatellite markers that were shared across multiple H. annuus maps or that were polymorphic in one of the wild species-mapping populations reported here (Figure S4 at http://www.genetics.org/supplemental/). The map presented here is essentially identical to that provided by Burke et al. (2004), except several additional markers that are informative in interspecific comparisons have been added. Also, information on marker homology relative to the other four species is provided for all informative markers.
For H. petiolaris, an intraspecific hybrid was crossed with an inbred sunflower line (CMSHA89) and 80 progeny segregating for H. petiolaris linkage groups were employed for map construction (Rieseberg et al. 1995). The map includes 400 RAPD markers and 295 microsatellites and spans 17 linkage groups and 1592 cM. A map based on a subset of the most informative markers and information on marker homology is provided in Figure S5 (available at http://www.genetics.org/supplemental/).
Comparisons of marker orders and genetic map lengths:
Homologous genomic regions across maps were identified from the locations of presumably orthologous microsatellite, AFLP, and RAPD markers. Note that homology of the latter have previously been demonstrated by Southern hybridization and/or restriction fragment digestions (Rieseberg 1996). Rearrangements were inferred from differences among maps in the location and linear order of markers. Map lengths were compared across species by analyzing the distance separating the outermost-shared markers for each collinear segment (Burke et al. 2004), although several clearly misplaced markers were excluded from the analysis. A pairwise _t_-test was used to identify differences in mean map lengths among species (Table 1).
TABLE 1.
Comparison of map lengths between two parental and three hybrid Helianthus species
| Taxa | No. segments compared | Mean dif. (cm) | Std. error (cm) | _t_-ratio | P > |t| | | --------------------------- | --------------------- | -------------- | --------------- | ---------- | ------------------- | | H. annuus-anomalus | 23 | −20.59 | 4.81 | −4.28 | 0.0003* | | H. annuus-deserticola | 11 | −0.09 | 5.51 | −0.02 | 0.9872 | | H. annuus-petiolaris | 21 | −9.27 | 4.56 | −2.03 | 0.0557 | | H. annuus-paradoxus | 16 | −17.9 | 7.61 | −2.36 | 0.0321 | | H. anomalus-deserticola | 21 | 18.45 | 6.59 | 2.80 | 0.0110 | | H. anomalus-paradoxus | 20 | 2.52 | 4.77 | −0.53 | 0.6041 | | H. anomalus-petiolaris | 20 | 6.29 | 5.53 | 1.14 | 0.2686 | | H. deserticola-paradoxus | 13 | −5.43 | 3.26 | 1.67 | 0.1211 | | H. deserticola-petiolaris | 4 | −2.53 | 15.29 | −0.17 | 0.8793 | | H. paradoxus-petiolaris | 9 | 17.84 | 5.93 | 3.01 | 0.0168 |
QTL analyses of pollen viability:
The analysis of pollen viability employed a BC2 population of H. annuus × H. petiolaris previously described by Rieseberg et al. (2003). The population consists of 384 progeny that have been genotyped for 96 molecular markers that provide coverage of all 17 linkage groups. For each BC2 plant, fresh pollen was harvested from the first flowering head. The pollen was treated with a solution of 30% sucrose and 0.1% 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyl tetrazolium (MTT) (Chandler et al. 1986) and ∼300 grains were scored for staining indicative of pollen viability. Fully darkened grains were considered viable.
Pollen viability QTL were detected by composite interval mapping (CIM; Zeng 1994) as implemented in the program Mapmanager QTX (Manly et al. 2001). This method tests the hypothesis that a QTL is present in an interval between two adjacent markers, while at the same time controlling for the effects of segregating QTL elsewhere in the genome. Tests were performed at 1 cM steps, and five background markers were included as cofactors in each CIM model. Genome-wide threshold values for declaring the presence of QTL were determined by 1000 permutations (Churchill and Doerge 1994). One-LOD support limits for the position of each QTL were calculated from the CIM results, as were the additive effects and percentage of phenotypic variance explained (PVE) by each QTL (Table 3).
TABLE 3.
Putative QTL positions, likelihood ratios (LR), percentage variance explained (PVE), additive effects, and significance levels for the percentage viable pollen in a BC2 population of the wild sunflower species H. annuus × H. petiolaris
| Linkage group_a_ | Position (cM) | 1-LOD interval | Interval markers | LR | PVE (%) | Additive effect | P |
|---|---|---|---|---|---|---|---|
| 2_b_ | 49 | 47-49 | ORS203 | 109.3 | 26 | −0.27 | < 0.005 |
| 5_b_ | 1 | 0-4 | ORS547-ORS240 | 28.2 | 6 | −0.18 | < 0.005 |
| 6a_b_ | 3 | 0-8 | ac/ctg281-ta/gc262 | 24.2 | 5 | −0.23 | < 0.005 |
| 8a_b_ | 44 | 39-44 | ac/tg124-ac/ctg206 | 54.3 | 10 | −0.17 | < 0.005 |
| 10 | 11 | 5-14 | ORS256 | 22.1 | 7 | −0.17 | < 0.005 |
| 11 | 48 | 45-49 | ORS1146-ORS261 | 18.2 | 6 | −0.14 | < 0.05 |
| 12_b_ | 7 | 0-15 | ORS963-ORS984 | 14.7 | 12 | −0.23 | < 0.05 |
| 14_b_ | 62 | 57-64 | ac/ctg189 | 27.5 | 5 | −0.13 | < 0.005 |
| 16_b_ | 5 | 0-10 | ORS1017-ac/ctg196 | 27.4 | 5 | −0.24 | < 0.005 |
| 17a | 45 | 39-52 | ORS1097-ORS1187 | 67.1 | 14 | −0.25 | < 0.005 |
| 17b_b_ | 0 | 0-14 | ORS727-ORS845 | 25.0 | 5 | −0.13 | < 0.005 |
Mapmanager QTX also was employed to search for interaction effects or epistasis by testing all pairs of marker loci for both main effects and interaction effects for each trait (Table 4). Unlike CIM, tests were performed only at marker loci. A two-stage test for significance was employed because of the large number of comparisons that must be made for each trait and because the significance of an interaction cannot be reliably tested if there is a strong main effect. First, the total effect of the two loci had to have P ≤ 10−5. This very stringent threshold was recommended by the user manual and is almost identical to thresholds calculated from the “effective number of comparisons” (Cheverud 2000), which takes linkage among markers into account in establishing significance thresholds. Second, the interaction effect itself must have P ≤ 0.01.
TABLE 4.
Significant digenic interactions (P ≤ 1 × 10−5) and likelihood ratios (LR) for percentage of viable pollen in a BC2 population of H. annuus × H. petiolaris
| Independent interactions | Locus A | Locus B | LR total effect | LR interaction effect |
|---|---|---|---|---|
| 1 | ORS371(1) | ORS826(8a) | 27.3 | 6.7 |
| 1 | ORS371(1) | ac/ctg206(8a) | 51.4 | 8.8 |
| 2 | ORS447(2) | ORS1114(3) | 26.7 | 7.6 |
| 3 | ORS447(2) | ORS1187(17a) | 47.0 | 9.3 |
| 3 | ccgg249 (2) | ORS1187(17a) | 77.5 | 8.2 |
| 3 | ORS708(2) | ORS1187(17a) | 73.4 | 9.0 |
| 3 | ORS203(2) | ORS847(17a) | 105.3 | 7.2 |
| 4 | ORS1114(3) | ORS1108(8a) | 37.9 | 7.0 |
| 4 | ORS1114(3) | ORS826(8a) | 35.7 | 6.7 |
| 4 | ORS1114(3) | ac/ctg206(8a) | 59.7 | 7.1 |
| 5 | ORS1108(8a) | ac/ctg189(14) | 49.2 | 9.8 |
| 5 | ORS826(8a) | ac/ctg189(14) | 46.0 | 8.9 |
| 6 | ORS963(12) | ORS727(17b) | 52.6 | 6.8 |
Crossing relationships:
Pollen and seed fertility of interspecific crosses involving H. annuus, H. anomalus, and H. petiolaris have previously been reported (Rieseberg 2000). Thus, for the present study, crosses were made between these species and the two remaining hybrid taxa, H. deserticola and H. paradoxus, as well as between populations of H. deserticola and H. paradoxus. Interspecific crosses were made in both directions and replicated at least four times (56 total). Crosses employed a minimum of four individuals from one population of each species: H. annuus (Hanksville, UT, Rieseberg 1295); H. anomalus (Mexican Water, AZ, Rieseberg 1282); H. deserticola (Toquerville, UT, Rieseberg 1270), H. paradoxus (Grants, NM, Rieseberg 1370), and H. petiolaris (Glenn Canyon Recreation Area, UT, Rieseberg 1277).
Although attempts were made to propagate 3 F1's from each cross (24 per cross combination), there was significant mortality, and 4–10 F1 plants were analyzed from each cross-combination. For each F1 plant that survived to reproductive maturity, pollen was harvested from the first flowering head and tested for viability as described above. Seed set was estimated as the percentage of cross-pollinated flowers (i.e., F1 × F1) that produced achenes (one-seeded fruits). Means and standard errors were calculated for pollen viabilities and seed set for each cross combination. To explore the relationship between the number of chromosomal differences and hybrid and seed fertility, correlations were calculated for each case and the data were permuted ×1000 to obtain a null distribution for statistical testing.
RESULTS
Map lengths:
Seventeen linkage groups were recovered for each of the three hybrid species, which corresponds to their haploid chromosome number (supplementary Figures S1–S3 at http://www.genetics.org/supplemental/). Linkages were labeled according to the standard nomenclature for H. annuus (Berry et al. 1995; Gedil et al. 2001). The relationship between this nomenclature and that of Rieseberg et al. (1995, 1999) is shown in Burke et al. (2004) and Rieseberg et al. (2003). For H. anomalus, the 1019 markers spanned 1908.3 cM, with an average spacing between markers of 1.90 cM. Map lengths and average marker spacing are 1229/1.88 cM and 1420.5/1.88 cM for H. deserticola and H. paradoxus, respectively. In comparison, the lengths of the H. annuus and H. petiolaris maps are 828 and 1592 cM, respectively (Burke et al. 2004).
Despite the seemingly large differences in map lengths, direct comparisons based on distances between orthologous markers indicate that only the longest (H. anomalus) and shortest (H. annuus) maps differ significantly. Even this result is suspect because the integrated H. annuus map employed in these comparisons is substantially shorter than most individual maps for H. annuus (Burke et al. 2004). Thus, recombinational speciation in Helianthus does not appear to have been accompanied by significant changes in recombination rates, although note that the small size of our mapping populations may limit our ability to detect fine-scale changes.
Linkage relationships:
Reconstruction of linkage relationships was straightforward for most linkage groups; homologous segments were supported by numerous markers in the same linear order and most incongruities could be accounted for simple translocations and inversions (e.g., Figure 2). Nonetheless, in some instances individual markers were seemingly misplaced (i.e., found in the wrong position on the right linkage groups). We assumed that most of these minor discrepancies were due to mapping error since it can be difficult to order tightly linked markers, particularly when mapping populations are small. Also, the integration of multiple maps may introduce inconsistencies in marker order (Burke et al. 2004). It is also possible that some of the apparently misplaced markers represent paralogs rather than orthologs.
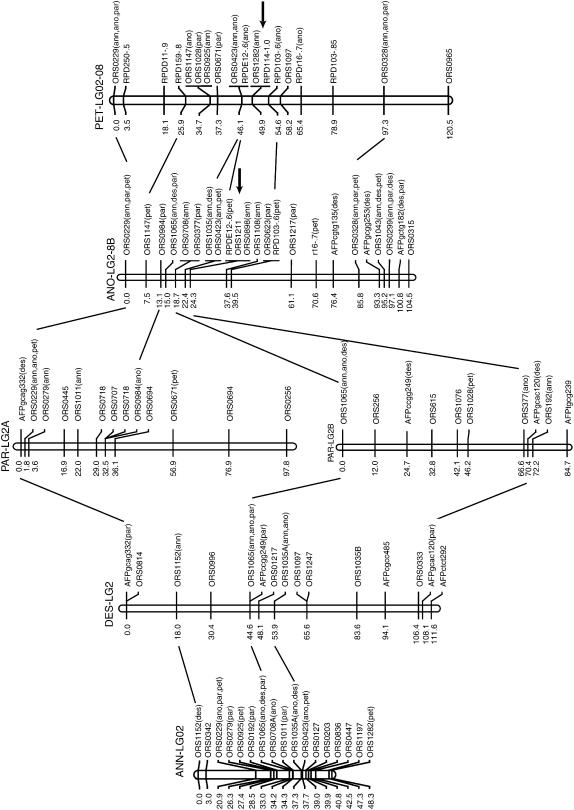
Figure 2.
Gene order relationships for a representative linkage group (LG2) between two parental sunflower species, H. annuus and H. petiolaris, and their three diploid hybrid derivatives, H. anomalus, H. deserticola, and H. paradoxus. Note the fission of LG2 in H. paradoxus and the fusion of linkages 2 and 8 in H. anomalus and H. petiolaris. Markers beginning with ORS are simple sequence repeats (SSRs), those beginning with AFP are AFLPs, and those beginning with RPD are RAPDs. Markers followed by species names in parentheses (ann, annuus; ano, anomalus; par, paradoxus; and pet, petiolaris) are informative and map to the same linkage group in the listed species. Informative loci on adjacent linkage groups are also connected by lines. Numbers to the left of each linkage group refer to genetic distance (centimorgans). Single arrows indicate the location of inferred chromosomal breakages/fusions that are necessary to account for the differences between this map and that of H. annuus (Figure S4 at http://www.genetics.org/supplemental/). Letters (A and B) following some linkage group designations indicate fragmented linkage groups, with fragments homologous to the top of the H. annuus linkage group designated A, the next highest fragment labeled B, and so forth.
Because minor differences in ordering were not uncommon, confident identification of inversions is more difficult than translocations. The three inversions recognized in this study (LG12, LG13, and LG16B; supplementary Figures S4 and S5 at http://www.genetics.org/supplemental/) were previously described by Rieseberg et al. (1995) and cause reduced introgression rates in natural hybrid zones (Rieseberg et al. 1999), so they are likely to be real. However, there was not a sufficient density of informative markers in H. deserticola and H. paradoxus to test for the presence of the inversions in these species. It is also possible that there have been multiple inversions involving LG17 because marker orders are heterogeneous across species, and this variation in marker order cannot be accounted for by a single inversion. An alternative explanation is that LG17 contains duplicated regions and the proportion of paralagous markers is unusually high.
Despite taking a conservative approach to recognizing chromosomal rearrangements, comparisons of marker locations and ordering revealed that only four linkage groups were collinear across the five species (LG01, LG07, LG09, LG10). The remaining 13 linkage groups were rearranged in one or more species (Figures 2 and 3). The two parental species, H. annuus and H. petiolaris, differ by at least 11 separate rearrangements, including eight translocations and three inversions (Rieseberg et al. 1995; Burke et al. 2004). Thus, only 6 of the 17 linkage groups were completely collinear in this comparison.
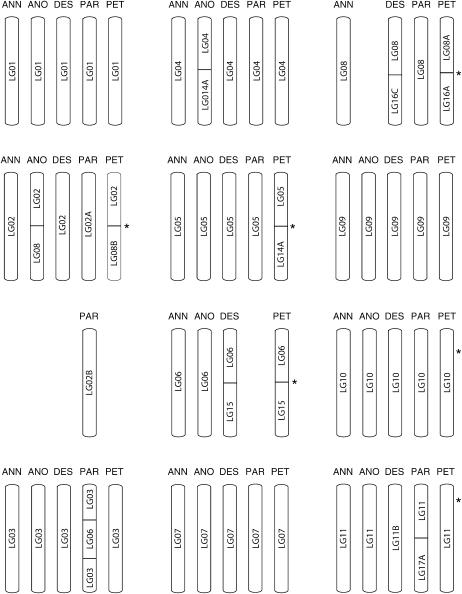
Figure 3.
Inferred chromosomal structural relationships between the parental species, H. annuus and H. petiolaris, and their three diploid hybrid derivatives, H. anomalus, H. deserticola, and H. paradoxus. Segments containing inversions are indicated by hatched lines. Note that there was insufficient marker density in H. deserticola and H. paradoxus to evaluate the presence or absence of inversions. Asterisks indicate the approximate position of pollen viability QTL (cf. Table 3).
The genomes of the three hybrid species are extensively rearranged relative to the parental species (Figure 2; Figures S1–S5 available at http://www.genetics.org/supplemental/). H. anomalus has the same linkage arrangement as both parents for six linkage groups (LG01, LG03, LG07, LG09, LG10, LG11) and as one or the other parent for an additional three linkage groups (LG05, LG06, LG12B). Note that H. anomalus has the same gene order as H. petiolaris for inverted regions in LG12 and LG13, while it has the gene order of H. annuus for the inversion in LG16B. The eight remaining linkage groups have a novel gene order in H. anomalus, and 12 chromosomal breakages/fusions are required to achieve the H. anomalus genome from its parental species. While these results are mostly consistent with Rieseberg et al. (1995), rearrangements were detected in seven rather than eight linkage groups in the previous study. Specifically, the microsatellite maps reported here imply that the fusion of LG02 and LG08 (L and M in Rieseberg et al. 1995) occurred independently in H. anomalus; previously, the fused linkage group was assumed to have been derived intact from H. petiolaris.
The same general pattern is found in H. deserticola (Figure 2; Figures S2, S4, and S5 at http://www.genetics.org/supplemental/). The species has the same linkage arrangement as both parental species for six linkages (LG01, LG03, LG04, LG07, LG09, and LG10), as H. annuus for an additional three linkage groups (LG02, LG05, and LG12), and as H. petiolaris for two linkage groups (LG06/15 and LG16C). H. deserticola has a novel gene arrangement for six linkage groups, which can be accounted for by a minimum of four chromosomal breakages and three fusions relative to the parental species.
Likewise, H. paradoxus is collinear with both parental species for six linkage groups (LG01, LG04, LG05, LG07, LG09, and LG10). It resembles H. annuus for an additional three linkage groups (LG05, LG08, and LG15) and H. petiolaris for one (LG17B). The remaining seven linkage groups in H. paradoxus are rearranged relative to both parental species and 10 breakages/fusions are required to account for the evolution of these differences.
Most of the new chromosomal breakages/fusions in the hybrid species (24 of 29) are associated with linkage groups that were already rearranged in the parental species. This is more than expected by chance, even after correcting for the smaller number of collinear than rearranged chromosomes in the parental species (χ2 = 6.86; P = 0.009; note that fusions involving both rearranged and collinear chromosomes were counted as 0.5 in both categories). Also, while the new rearrangements often involve the same linkage groups (particularly LG13, LG14, and LG16), the resulting chromosomal combinations are mostly novel (Figure 2). As a consequence, the hybrid species are as different from each other in chromosome structure as they are from the parental species (Table 2).
TABLE 2.
Percentage pollen viability (on and above the diagonal) and the number of rearranged chromosomes (below diagonal) derived from crosses within and among populations of H. annuus, H. petiolaris, and their stabilized hybrid derivatives, H. anomalus, H. deserticola, and H. paradoxus
| Taxon | H. annuus | H. anomalus | H. deserticola | H. paradoxus | H. petiolaris |
|---|---|---|---|---|---|
| H. annuus | 94.3 (0.7) | 2.6 (0.1) | 4.9 (0.9) | 9.1 (0.5) | 4.8 (0.2) |
| H. anomalus | 9 | 94.5 (0.9) | 42.9 (3.9) | 14.8 (2.8) | 3.0 (0.1) |
| H. deserticola | 9 | 10 | 90.3 (3.1) | 35.9 (3.9) | 2.5 (0.7) |
| H. paradoxus | 9 | 11 | 11 | 95.6 (1.1) | 10.9 (0.6) |
| H. petiolaris | 10 | 10 | 9 | 11 | 94.9 (0.8) |
QTL analyses of fertility:
Eleven QTL were detected for percentage viable pollen in a BC2 population of the parental species (Table 3). QTL effects were modest in magnitude when estimated in terms of percentage variance explained (PVE), averaging 9.2% (range 5–26%). However, analyses of additive effects suggest a somewhat different story, with an average reduction in the proportion of viable pollen of 19.5% (range 13–27%). This seeming discrepancy is because individuals with four or more sterility QTL often had no viable pollen (i.e., variance in pollen viability has a lower bound of zero), which appears to have resulted in an underestimate of PVE and possibly of additive effects. With respect to the latter, it is noteworthy that individuals with a single sterility QTL had a reduction in pollen viability of as much as 40%. In all instances, the H. annuus allele reduced pollen viability in the H. petiolaris genetic background.
Nine of the 11 pollen viability QTL map to rearranged linkage groups (Table 3), which is significantly more than expected by chance (χ2 = 3.97; P = 0.046), despite the preponderance of rearranged linkage groups (11 rearranged vs. 7 collinear) differentiating the parental species. Of the 9 QTL located on rearranged linkage groups, 8 map close to chromosomal breakpoints, implying that the rearrangement itself might be responsible for the observed reduction in fertility. While this observation is consistent with previous reports of complex multivalent formation in the meiosis of sunflower hybrids, it cannot be proven because gene incompatibilities may also cluster near chromosomal breakages (Noor et al. 2001; Navarro and Barton 2003). The two QTL not associated with rearranged linkage groups map to genomic regions that were previously shown to be associated with reductions in pollen viability and reduced gene flow in natural hybrid zones of H. annuus and H. petiolaris (Rieseberg et al. 1999).
Epistasis:
The genome-wide scan of digenic interactions detected 13 significant interactions (Table 4). However, many of the interactions involved linked markers from the same pair of chromosomes and may represent as few as 6 independent interactions. The clearly independent interactions involve six previously detected QTL from rearranged linkage groups (Table 3), as well as two new fertility QTL from collinear linkages (LG1 and LG3). Although multivalent formation during meiosis could generate nonadditive effects on fertility (Gardner et al. 2000), the epistasis detected here appears to be the result of interactions among genes since none of the interactions involve linkage groups that would be part of the same multivalent configuration. Also, two of the epistatic QTL derive from collinear linkages and were detected on the basis of their interaction effects rather than their additive effects.
Crossing relationships:
First-generation hybrids derived from all pairwise combinations of crosses (Table 2; Figure 4) reveal that the three hybrid species are strongly isolated from their parents. Percentages of viable pollen range from 2.5 ± 0.7% for crosses between H. deserticola and H. petiolaris to 10.9 ± 0.6% for crosses between H. paradoxus and H. petiolaris (Table 2), whereas percentage seed set (Figure 4) is lowest in crosses between H. anomalus and H. annuus (0.16 ± 0.04%) and highest for crosses between H. deserticola and H. petiolaris (2.2 ± 0.7%).
Figure 4.
Percentage seed set of first-generation hybrids between the parental species, H. annuus and H. petiolaris, and their three diploid hybrid derivatives, H. anomalus, H. deserticola, and H. paradoxus. Values are presented as mean (standard error). Line thickness is proportional to cross-compatibility. Dashed lines indicate that some individuals were completely sterile for that cross. Values for crosses among H. annuus, H. petiolaris, and H. anomalus derive from Rieseberg (2000).
The hybrid species are isolated from each other as well, but the sterility barrier is not as strong, with the pollen fertility of first-generation hybrids ranging between 14.8 ± 2.8% for crosses between H. anomalus and H. paradoxus and 42.9 ± 3.9% for crosses between H. anomalus and H. deserticola. Analyses of percentage seed set provides a similar estimate of barrier strength, with first-generation hybrids between H. anomalus and H. paradoxus producing only 2.1 ± 0.5% viable seed compared to 18.5 ± 5.5% viable seed for crosses between H. anomalus and H. deserticola.
In contrast to the interspecific crosses, all intraspecific crosses were fully fertile, with the percentage of viable pollen consistently >90% (Table 2) and the percentage seed set >80% (not shown). Lower fertility has been previously reported for some cross-combinations with H. anomalus and H. deserticola (Schwarzbach and Rieseberg 2002; Gross et al. 2003), but these involved populations that appear to have been independently derived (H. anomalus and H. deserticola appear to have multiple origins). The intraspecific crosses reported here involve populations from the same origin.
Pollen and seed fertility are highly correlated across crosses (r = 0.97; P ≪ 0.001), but the direction of the cross did not have a significant effect on fertility in any of the 10 cross-combinations. Thus, interactions between the cytoplasmic and nuclear genes do not appear to be important contributors to the evolution of sterility barriers between these sunflower species. This differs from recent reports emphasizing the ubiquity of asymmetric reproductive barriers in angiosperms (Tiffin et al. 2001; Levin 2003). Also, there was not a significant correlation between the total number of rearrangements that differentiate the species and pollen (r = 0.49; P = 0.09) or seed fertility (r = 0.33; P = 0.21). However, this should not be viewed as evidence that chromosomal rearrangements do not affect fertility, because there is very little variance in the number of rearranged chromosomes differentiating the species (Table 2). Nonetheless, the fact that the hybrid species differ by a similar number of rearranged chromosomes as do the parental species, yet have weaker sterility barriers, does imply that genic incompatibilities are significant contributors to sterility as well. This argument is further buttressed by the fact that the two hybrid species that are most similar in gene composition, H. anomalus and H. deserticola (Rieseberg et al. 2003), are also most interfertile (Table 2; Figure 4), despite differing by 10 rearranged chromosomes.
DISCUSSION
Homoploid hybrid speciation:
For new homoploid hybrid species to become established, they must somehow become reproductively isolated from their parental species. This is a difficult step. Unlike allopolyploidy where genome doubling provides instantaneous isolation, there is no simple means by which a homoploid hybrid can become isolated. Verbal models suggest that a hybrid neospecies might become isolated through rapid karyotypic evolution (i.e., the recombinational speciation model; Stebbins 1957; Grant 1981), ecological divergence (Grant 1981; Templeton 1981), and/or spatial isolation, perhaps mediated by hybrid founder events (Charlesworth 1995). Simulation models confirmed the feasibility of these various scenarios (McCarthy et al. 1995; Buerkle et al. 2000), but show that while ecological divergence alone was sufficient for a new hybrid species to arise, it was unlikely to evolve independently without significant karyotypic change and/or spatial isolation. Thus, an important empirical question is whether rapid karyotypic evolution often accompanies the origin of homoploid hybrid species.
This study provides important confirmation of the recombinational speciation model, at least in Helianthus. The three hybrid sunflower species differ from their parents by a minimum of nine rearranged chromosomes (Table 2; Figure 3). About one-third of the differences in karyotype have arisen from the simple sorting of chromosomal rearrangements that differentiate the parental species, whereas the remainder have arisen de novo (six breakages/six fusions in H. anomalus; four breakages/three fusions in H. deserticola, and five breakages/five fusions in H. paradoxus). The fact that a significant overproportion of new rearrangements involved linkages that were already rearranged in the parental species, as opposed to those that are collinear, implies that chromosomal breakage in hybrids may be a consequence of meiotic abnormalities involving rearranged linkage groups (Templeton 1981; Rieseberg et al. 1995). However, it is also possible that these linkage groups are susceptible to chromosomal mutation for other reasons.
Despite the consistent association between chromosomal repatterning and homoploid hybrid speciation in Helianthus, chromosomal change is not universally associated with this mode of speciation. Of ∼11 confirmed examples of homoploid hybrid speciation (Gross and Rieseberg 2005), karyotypic change has been reported in six cases––the three Helianthus examples plus Argyranthemum sundingii (Brochmann et al. 2000; Borgen et al. 2003), Iris nelsonii (Randolph 1966; Arnold 1993), and Stephanomeria diegensis (Gallez and Gottlieb 1982). Also, some karyotypic races of house mice are speculated to have arisen through hybridization (Pialek et al. 2001). However, only in Helianthus has there been detailed analyses of the hybrid species' genomes.
Even less is known about karyotypic evolution in the other five homoploid hybrid taxa: Daphnia mendota (Taylor et al. 1996), Gila seminuda (Demarais et al. 1992), Paeonia spp. (Sang et al. 1995,1997; Sang and Zhang 1999), Penstemon clevelandii (Wolfe et al. 1998), and Pinus densata (Wang et al. 2001). Habitat and/or pollinator isolation is considered to be the predominant form of isolation in each case, but as far as we are aware the only formal karyotypic analysis has been conducted in Pinus (Yang 1987); P. densata was shown to have the same karyotype as one of its parental species (Yang 1987; Wang et al. 2001).
Chromosomal rearrangements and sterility:
An underlying assumption of the recombinational model that has not previously been verified is that the karyotypic changes accompanying speciation are underdominant and create a sterility barrier with the parental species. The QTL analyses reported here indicate that pollen sterility QTL in Helianthus are significantly more likely to occur on rearranged than on collinear linkage groups and that sterility QTL in rearranged chromosomes almost always map near chromosomal breakpoints. Chromosomal rearrangements in Helianthus have previously been shown to generate multivalent configurations in meiois (Heiser 1947; Chandler et al. 1986). Thus, the rearrangements themselves appear to be at least partially responsible for the fertility reductions observed in hybrids.
Interspecific incompatibilities among genes also contribute to hybrid sterility in Helianthus. The best evidence for this is that, as previously reported by Rieseberg et al. (1999), several sterility QTL map to collinear rather than to rearranged linkage groups. Also, because none of the interactions involve linkage groups that would be part of the same multivalent configuration, the epistasis detected here is likely to be the result of interactions among genes. This is consistent with theory and empirical evidence, which indicate that genic incompatibilities are likely to cluster within or near chromosomal rearrangements (Noor et al. 2001; Navarro and Barton 2003; Brown et al. 2004). Finally, crosses among the hybrid species, which are similar in gene composition (Rieseberg et al. 2003), but not in karyotype, were more fertile than crosses between the parental species, despite differing by similar numbers of chromosomal rearrangements.
It is difficult to evaluate the relative contributions of genic incompatibilities vs. chromosomal rearrangements to hybrid sterility. A crude estimate can be gleaned by analyzing the genomic distribution of sterility QTL; nine of 11 sterility QTL map to rearranged linkage groups and account for 87% of the phenotypic variance for pollen sterility. However, at least some of the sterility mapping to rearranged linkage groups must be caused by genes. If we assume an even distribution of sterility QTL across the genome, then ∼35% of the reduction in pollen viability may be attributed to genes and 65% to chromosomal rearrangements. Even this is likely to be an underestimate of the contribution of genes to reduced pollen viability, because genic incompatibilities are likely to be recessive (Orr and Presgraves 2000; Fishman and Willis 2001), epistatic effects on sterility are not considered, and genic incompatibilities may accumulate disproportionately on rearranged chromosomes (Noor et al. 2001).
Reproductive barrier strength and the independent evolution of hybrid neospecies:
Computer simulation models indicate that in the absence of a strong sterility barrier, a new hybrid lineage is unlikely to evolve independently from its parental species (Buerkle et al. 2000). Instead, the hybrid population is likely to represent a steep step in a cline between the parental populations. Evolutionary independence can be achieved if there are strong ecological and sterility barriers (i.e., hybrid fertility < 0.1). This requirement does appear to be met in Helianthus. All pollen and seed fertility estimates for F1's between the hybrid species and their parents fall below 11%, and we have previously documented the occurrence of strong ecological selection in the habitats of all three hybrid species (Lexer et al. 2003a,b; Gross et al. 2004; Ludwig et al. 2004).
In conclusion, the requirements of the recombinational speciation model do appear to be fulfilled in Helianthus. The hybrid species are strongly divergent karytoypically, the karyotypic differences contribute to reproductive isolation, and barrier strength is sufficient to achieve evolutionary independence in parapatry. However, it remains to be determined whether homoploid hybrid species in other groups of plants and animals satisfy the recombinational mode. It might be, for example, that some of the more weakly isolated hybrid taxa will exhibit little genetic isolation from their parental species (i.e., only slightly reduced gene flow) when tested and are better viewed as introgresssive races. More generally, there is a need to discriminate between the effects of chromosomal rearrangements and linked genic incompatibilities in both plants and animals. This seemingly requires a laborious combination of fine-mapping and map-based cloning, although some inferences may be made by studying patterns of interfertility in different hybrid classes (e.g., Fishman and Willis 2001).
Acknowledgments
We thank M.-J. Kim for technical assistance and D. Rosenthal and L. Donovan for providing pollen from BC2 plants. This work was supported by grants from the National Science Foundation (DEB-9806290 and DEB-0314654 to L.H.R.), the National Institutes of Health (GM059065 to L.H.R.), and the United States Department of Agriculture (00-35300-9244 and 03-35300-13104 to J.M.B., 98-35300-6166 to S.J.K., and 00-52100-9609 to L.H.R. and S.J.K.).
References
- Arnold, M. L., 1993. Iris nelsonii (Iridaceae)—Origin and genetic composition of a homoploid hybrid species. Amer. J. Bot. 80**:** 577–583. [DOI] [PubMed] [Google Scholar]
- Baker, R. J., and J. W. Bickham, 1986. Speciation by monobrachial centric fusions. Proc. Natl. Acad. Sci. USA 83**:** 8245–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N., and B. O. Bengtsson, 1986. The barrier to genetic exchange between hybridizing populations. Heredity 57**:** 357–376. [DOI] [PubMed] [Google Scholar]
- Berry, S. T., A. J. Leon, C. C. Hanfrey, P. Challis and A. Burkholz et al., 1995. Molecular marker analysis of Helianthus annuus L. 2. Construction of an RFLP linkage map for cultivated sunflower. Theor. Appl. Genet. 91**:** 195–199. [DOI] [PubMed] [Google Scholar]
- Borgen, L., I. Leitch and A. Santos-Guerra, 2003. Genome organization in diploid hybrid species of Argyranthemum (Asteraceae) in the Canary Islands. Bot. J. Linn. Soc. 141**:** 491–501. [Google Scholar]
- Brochmann, C., L. Borgen and O. E. Stabbetorp, 2000. Multiple diploid hybrid speciation of the Canary Island endemic Argyranthemum sundingii (Asteraceae). Plant Syst. Evol. 220**:** 77–92. [Google Scholar]
- Brown, K. M., L. M. Burk, L. M. Henagan and M. A. F. Noor, 2004. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution 58**:** 1856–1860. [DOI] [PubMed] [Google Scholar]
- Buerkle, C. A., R. J. Morris, M. A. Asmussen and L. H. Rieseberg, 2000. The likelihood of homoploid hybrid speciation. Heredity 84**:** 441–451. [DOI] [PubMed] [Google Scholar]
- Burke, J. M., S. Tang, S. J. Knapp and L. H. Rieseberg, 2002. Genetic analysis of sunflower domestication. Genetics 161**:** 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. M., Z. Lai, M. Salmaso, T. Nakazato, S. Tang et al., 2004. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics 167**:** 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, J. M., C. C. Jan and B. H. Beard, 1986. Chromosomal differentiation among the annual Helianthus species. Syst. Bot. 11**:** 354–371. [Google Scholar]
- Charlesworth, D., 1995. Evolution under the microscope. Current Biol. 5**:** 835–836. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., 2000. Detecting epistasis among quantitative trait loci, pp. 58–81 in Epistasis and the Evolutionary Process, edited by J. Price, B. Brodie and M. J. Wade. Oxford University Press, New York.
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138**:** 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., S. Aulard and A. Berry, 1991. Lack of underdominance in a naturally-occurring pericentric inversion in Drosophila melanogaster and its implications for chromosome evolution. Genetics 129**:** 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., W. Meyers, A. P. Crittenden and P. Sniegowski, 1993. The fertility effects of pericentric inversion in Drosophila melanogaster. Genetics 134**:** 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson, M. T., and E. C. Akeson, 1993. Recombination suppression by heterozygous Robertsonian chromosomes in the mouse. Genetics 133**:** 649–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarais, B. D., T. E. Dowling, M. E. Douglas, W. L. Minckley and P. C. Marsh, 1992. Origin of Gila seminuda (Teleostei: Cyprinidae) through introgressive hybridization: Implications for evolution and conservation. Proc. Natl. Acad. Sci. USA 89**:** 2747–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallez, G. P., and L. D. Gottlieb, 1982. Genetic evidence for the hybrid origin of the diploid plant Stephanomeria diegensis. Evolution 36**:** 1158–1167. [DOI] [PubMed] [Google Scholar]
- Gardner, K., A. Buerkle, J. Whitton and L. H. Rieseberg, 2000. Inferring epistasis in wild sunflower hybrid zones, pp. 264–279 in Epistasis and the Evolutionary Process, edited by J. Price, B. Brodie and M. J. Wade. Oxford University Press, New York.
- Gedil, M. A., C. Wye, S. T. Berry, B. Segers, J. Peleman et al., 2001. An integrated RFLP-AFLP linkage map for cultivated sunflower. Genome 44**:** 213–221. [PubMed] [Google Scholar]
- Grant, V., 1958. The regulation of recombination in plants. Cold Spring Harbor Symp. Quant. Biol. 23**:** 337–363. [DOI] [PubMed] [Google Scholar]
- Grant, V., 1981. Plant Speciation. Columbia University Press, New York.
- Greenbaum, I. F., and M. J. Reed, 1984. Evidence for heterosynaptic pairing of the inverted segment in pericentric-inversion heterozygotes of the deer mouse (Peromyscus-maniculatus). Cytogenet. Cell Genet. 38**:** 106–111. [DOI] [PubMed] [Google Scholar]
- Gross, B. L., and L. H. Rieseberg, 2005. The ecological genetics of homoploid hybrid speciation. J. Hered. 96**:** 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, B. L., A. E. Schwarzbach and L. H. Rieseberg, 2003. Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae). Amer. J. Bot. 90**:** 1708–1719. [DOI] [PubMed] [Google Scholar]
- Gross, B. L., N. C. Kane, C. Lexer, F. Ludwig, D. Rosenthal et al., 2004. Reconstructing the origin of Helianthus deserticola: survival and selection on the desert floor. Am. Nat. 164**:** 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, D. W., 1986. Heterosynapsis and suppression of chiasmata within heterozygous pericentric inversions of the Sitka deer mouse. Chromosoma 94**:** 425–432. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 1981. The establishment of chromosomal variants. Evolution 35**:** 322–332. [DOI] [PubMed] [Google Scholar]
- Heiser, C. B., 1947. Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution 1**:** 249–262. [Google Scholar]
- Heiser, C. B., D. M. Smith, S. B. Clevenger and W. C. Martin, 1969. The North American sunflowers (Helianthus). Mem. Torrey Bot. Club 22**:** 1–213. [Google Scholar]
- Fishman, L., and J. H. Willis, 2001. Evidence for Dobzhansky-Muller incompatibilites contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution 55**:** 1932–1942. [DOI] [PubMed] [Google Scholar]
- John, B., 1981. Chromosomal change and evolutionary change: a critique, pp. 23–51 in Evolution and Speciation, edited by W. R. Atchley and D. S. Woodruff. Cambridge University Press, Cambridge, UK.
- King, M., 1993. Species Evolution. Cambridge University Press, Cambridge, UK.
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12**:** 172–175. [Google Scholar]
- Lande, R., 1985. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54**:** 323–332. [DOI] [PubMed] [Google Scholar]
- Levin, D. A., 2002. The Role of Chromosomal Change in Plant Evolution. Oxford University Press, Oxford.
- Levin, D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28**:** 5–11. [Google Scholar]
- Lexer, C., M. Welch, J. L. Durphy and L. H. Rieseberg, 2003. a Natural selection for salt tolerance QTL in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a homoploid hybrid species. Mol. Ecol. 12**:** 1225–1235. [DOI] [PubMed] [Google Scholar]
- Lexer, C., M. E. Welch, O. Raymond and L. H. Rieseberg, 2003. b The origin of ecological divergence in Helianthus paradoxus (Asteraceae): selection on transgressive characters in a novel hybrid habitat. Evolution 57**:** 1989–2000. [DOI] [PubMed] [Google Scholar]
- Ludwig, F., D. Rosenthal, J. A. Johnston, N. Kane, B. L. Gross et al., 2004. Selection on leaf ecophysiological traits in a hybrid Helianthus species and early generation hybrids in a desert dune habitat. Evolution 58**:** 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19**:** 472–488. [DOI] [PubMed] [Google Scholar]
- Manly, K. F., R. H. Cudmore and J. M. Meer, 2001. Map manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12**:** 930–932. [DOI] [PubMed] [Google Scholar]
- McCarthy, E. M., M. A. Asmussen and W. W. Anderson, 1995. A theoretical assessment of recombinational speciation. Heredity 74**:** 502–509. [Google Scholar]
- Navarro, A., and N. Barton, 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57**:** 447–459. [DOI] [PubMed] [Google Scholar]
- Navarro, A., and A. Ruiz, 1997. On the fertility effects of pericentric inversions. Genetics 147**:** 931–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci and J. Reiland, 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98**:** 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and D. C. Presgraves, 2000. Speciation by postzygotic isolation: forces, genes and molecules. BioEssays 22**:** 1085–1094. [DOI] [PubMed] [Google Scholar]
- Pialek, J., H. C. Hauffe, K. M. Rodriguez-Clark and J. B. Searle, 2001. Raciation and speciation in house mice from the Alps: the role of chromosomes. Mol. Ecol. 10**:** 613–625. [DOI] [PubMed] [Google Scholar]
- Randolph, L. F., 1966. Iris nelsonii, a new species of Louisiana iris of hybrid origin. Baileya 14**:** 143–169. [Google Scholar]
- Rieseberg, L. H., 1991. Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am. J. Bot. 78**:** 1218–1237. [Google Scholar]
- Rieseberg, L. H., 1996. Homology among RAPD fragments in interspecific comparisons. Mol. Ecol. 5**:** 99–105. [Google Scholar]
- Rieseberg, L. H., 2000. Crossing relationships among ancient and experimental sunflower hybrid lineages. Evolution 54**:** 859–865. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16**:** 351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., S. M. Beckstrom-Sternberg, A. Liston and D. M. Arias, 1991. Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus (Asteraceae). Syst. Bot. 16**:** 50–76. [Google Scholar]
- Rieseberg, L. H., C. Van Fossen and A. M. Desrochers, 1995. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature 375**:** 313–316. [Google Scholar]
- Rieseberg, L. H., S. Baird and A. Desrochers, 1998. Patterns of mating in wild sunflower hybrid zones. Evolution 52**:** 713–726. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152**:** 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. D. Livingstone et al., 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301**:** 1211–1216. [DOI] [PubMed] [Google Scholar]
- Rogers, C. E., T. E. Thompson and G. J. Seiler, 1982. Sunflower Species of the United States. National Sunflower Association, Fargo, ND.
- Sang, T, and D. Zhang, 1999. Reconstructing hybrid speciation using sequences of low copy nuclear genes: Hybrid origins of five Paeonia species based on Adh gene phylogenies. Syst. Bot. 24**:** 148–163. [Google Scholar]
- Sang, T., D. J. Crawford and T. F. Stuessy, 1995. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: Implications for biogeography and concerted evolution. Proc. Natl. Acad. Sci. USA 92**:** 6813–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, T., D. J. Crawford and T. F. Stuessy, 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Amer. J. Bot. 84**:** 1120–1136. [PubMed] [Google Scholar]
- Schwarzbach, A. E., and L. H. Rieseberg, 2002. Likely multiple origins of a diploid hybrid sunflower species. Mol. Ecol. 11**:** 1703–1717. [DOI] [PubMed] [Google Scholar]
- Stebbins, G. L., 1957. The hybrid origin of microspecies in the Elymus glaucus complex. Cytologia Suppl. 36**:** 336–340. [Google Scholar]
- Tang, S., J. K. Yu, M. B. Slabaugh, D. K. Shintani and S. J. Knapp, 2002. Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105**:** 1124–1136. [DOI] [PubMed] [Google Scholar]
- Taylor, D. J., P. D. Hebert and J. K. Colbourne, 1996. Phylogenetics and evolution of the Daphnia logispina group (Crustacea) based on 12S rDNA sequence and allozyme variation. Mol. Phylogenet. Evol. 5**:** 495–510. [DOI] [PubMed] [Google Scholar]
- Templeton, A. R., 1981. Mechanisms of speciation–a population genetic approach. Annu. Rev. Ecol. Syst. 12**:** 23–48. [Google Scholar]
- Tiffin, P., M. S. Olson and L. C. Moyle, 2001. Asymmetrical crossing barriers in angiosperms. Proc. R. Soc. Lond. Ser B 268**:** 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M. C., S. Baird, J. Pan and L. H. Rieseberg, 1998. Rapid hybrid speciation in wild sunflowers. Proc. Natl. Acad. Sci. USA 95**:** 11757–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap 3.0: Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Walsh, J. B., 1982. Rate of accumulation of reproductive isolation by chromosomal rearrangements. Am. Nat. 120**:** 510–532. [Google Scholar]
- Wang, X.-R., A. E. Szmidt and O. Savolainen, 2001. Genetic composition and diploid hybrid speciation of a high mountain pine, Pinus densata, native to the Tibetan plateau. Genetics 159**:** 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M. E., and L. H. Rieseberg, 2002. Patterns of genetic variation suggest a single, ancient origin for the diploid hybrid species Helianthus paradoxus. Evolution 56**:** 2126–2137. [DOI] [PubMed] [Google Scholar]
- White, M. J. D., 1978. Modes of Speciation. W. H. Freeman, San Francisco.
- Wolfe, A. D., Q.-Y. Xiang and S. K. Kephart, 1998. Diploid hybrid speciation in Penstemon (Scrophulariaceae). Proc. Natl. Acad. Sci. USA 95**:** 5112–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.-Y., 1987. Nuclear type analysis of Pinus densata. J. Northwest. Coll. For. 2**:** 51–54 (in Chinese). [Google Scholar]
- Yu, J. K., S. Tang, M. B. Slabaugh, A. Heesacker, G. Cole et al., 2003. Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43**:** 367–387. [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136**:** 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]