Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters (original) (raw)
Abstract
Hox genes in vertebrates are clustered, and the organization of the clusters has been highly conserved during evolution. The conservation of Hox clusters has been attributed to enhancers located within and outside the Hox clusters that are essential for the coordinated “temporal” and “spatial” expression patterns of Hox genes in developing embryos. To identify evolutionarily conserved regulatory elements within and outside the Hox clusters, we obtained contiguous sequences for the conserved syntenic blocks from the seven Hox loci in fugu and carried out a systematic search for conserved noncoding sequences (CNS) in the human, mouse, and fugu Hox loci. Our analysis has uncovered unusually large conserved syntenic blocks at the HoxA and HoxD loci. The conserved syntenic blocks at the human and mouse HoxA and HoxD loci span 5.4 Mb and 4 Mb and contain 21 and 19 genes, respectively. The corresponding regions in fugu are 16- and 12-fold smaller. A large number of CNS was identified within the Hox clusters and outside the Hox clusters spread over large regions. The CNS include previously characterized enhancers and overlap with the 5′ global control regions of HoxA and HoxD clusters. Most of the CNS are likely to be control regions involved in the regulation of Hox and other genes in these loci. We propose that the regulatory elements spread across large regions on either side of Hox clusters are a major evolutionary constraint that has maintained the exceptionally long syntenic blocks at the HoxA and HoxD loci.
Keywords: conserved noncoding sequences, conserved synteny, fugu, global control region
Hox genes code for homeodomain-containing transcription factors that determine the anterior-posterior patterning of tissues along the body axis of animals. In vertebrates, Hox genes are organized into tight clusters containing up to 14 paralogs. A remarkable feature of these clusters is that the positions of genes in the cluster are colinear with their spatial and temporal expression pattern along the anterior-posterior axis of the embryo. Hox genes at the 3′ end of the cluster (see Fig. 1) express in the anterior segments of the embryo and are turned on relatively early during development, whereas genes at the 5′ end express in the posterior segments and at a later stage during development (1). More than two decades after the discovery of Hox clusters, the molecular mechanisms underlying this orchestrated expression pattern of Hox genes and the selective pressure that has maintained the clustering and colinear organization of vertebrate Hox genes still are not well understood.
Fig. 1.
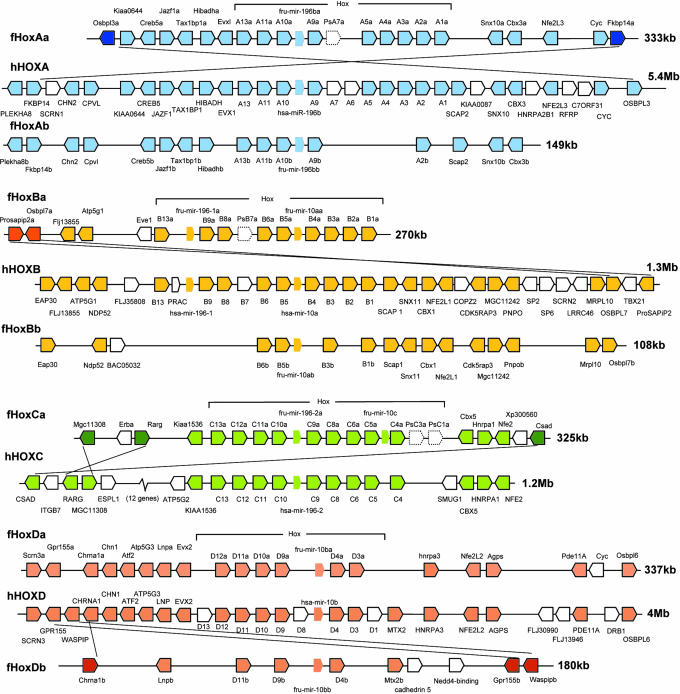
Conserved syntenic blocks at the fugu and human Hox loci. Fugu has two duplicate loci for human HoxA, HoxB, and HoxD loci. Hexagons represent genes. MicroRNA genes are shown as borderless hexagons and pseudogenes with dotted borders. Genes that are present in only fugu or human locus are shown as open hexagons. Dark colored hexagons in the fugu (linked with their human orthologs with a diagonal line) represent genes that have undergone rearrangements.
The colinear organization of Hox genes has been shown to be essential for the temporal progression of expression but not for the spatial expression patterns. Transgenic Hox genes inserted at ectopic locations in the genome retain spatial expression to a large extent but fail to recapitulate their temporal pattern of regulation (1). The mechanism of colinear expression of vertebrate Hox genes may require coordinated alterations in the higher-order chromatin structure from one end of the cluster to the other (2). Such a mechanism could require the clustering and colinear organization of Hox genes to be strictly maintained. Another factor that may have contributed to the clustering and colinearity of Hox genes is the existence of global enhancers that regulate several genes in a manner independent of their local enhancers but dependent on their location in the cluster. Global control regions (GCRs) have been identified on either side of the HoxD cluster. The 5′ HoxD GCR, regulating the expression of Lnp, Evx2, and posterior HoxD genes in developing digits and the central nervous system, is located ≈240 kb upstream of HoxD13 in the intergenic region between Atp5g3 and Lnp (3). The 3′ HoxD GCR mapped downstream of HoxD1, called early limb control region, is implicated in the early colinear expression of HoxD genes along the anterior-posterior axis in limb buds. Its exact location and sequence are not known (4). A 5′ long-range enhancer located upstream of HoxA has been proposed to direct expression of HoxA13 and four upstream genes (Evx1, Hibadh, Tax1bp1, and Jazf1) in the distal limb and genital bud (5). It is not known whether the 3′ region of HoxA cluster or the flanking regions of HoxB and HoxC clusters contain GCRs similar to the HoxD cluster.
Comparisons of Hox clusters from phylogenetically distant vertebrates have proved to be effective in discovering conserved regulatory sequences. Several conserved putative regulatory elements have been identified in the HoxA cluster by comparing sequences from mammals, the horn shark, and ray-finned fishes (6–10). Comparisons of subsets of Hox genes from HoxB (11, 12), HoxC (e.g., refs. 13 and 14), and HoxD (e.g., refs. 10 and 15) clusters have also identified a number of putative regulatory elements. However, a clusterwide comparison of HoxB, HoxC, or HoxD clusters has not been carried out. Three blocks of noncoding sequences in the intergenic region of Lnp and Evx2 genes at the HoxD locus have been shown to be highly conserved in mammals, chicken, fugu, and the horn shark, indicating that flanking regions of Hox clusters also contain conserved regulatory elements (16). Subsets of the 5′ GCRs of the HoxD and HoxA clusters that are located outside the Hox clusters were found to be conserved in pufferfishes (3, 5). Apart from these elements, no systematic search for conserved regulatory elements outside the Hox clusters has been attempted. We therefore decided to do a systematic search for evolutionary conserved noncoding sequences (CNS) within and outside the vertebrate Hox clusters.
In contrast to a single Hox cluster in Amphioxus (a cephalochordate), mammals contain four Hox clusters (HoxA, HoxB, HoxC, and HoxD) that have presumably evolved through two rounds of duplication of a single Hox cluster. Ray-finned fishes, on the other hand, contain up to seven Hox clusters. The additional clusters in fishes have been attributed to a whole genome duplication in the fish lineage (17). To facilitate identification of CNS within and outside Hox clusters, we obtained contiguous sequences corresponding to conserved syntenic blocks from the seven Hox loci in the pufferfish, Fugu rubripes, and identified CNS in the human, mouse, and fugu Hox loci. These CNS represent potential regulatory elements associated with Hox gene clusters and non-Hox genes present in the conserved syntenic blocks.
Results
Fugu Hox Loci and Conserved Syntenic Blocks.
We carried out an exhaustive search for Hox genes in the fugu genome assembly 3.0 and generated contiguous sequences for syntenic blocks that are conserved in human and mouse. Our analysis identified seven fugu Hox loci, HoxAa, HoxAb, HoxBa, HoxBb, HoxCa, HoxDa, and HoxDb, spanning 130–399 kb (Table 1and Fig. 1). Analysis of an earlier fugu assembly (v2.0) had identified a putative eighth locus, designated HoxAc (18). We investigated the assembly v2.0 scaffolds that were assigned to this locus and found that most of them correspond to a Tilapia bacterial artificial chromosome (BAC) sequence (GenBank accession no. AF533976). These sequences are not present in the third fugu assembly.
Table 1.
Conserved syntenic fragments at the fugu and human Hox loci
| Fugu Hox cluster | Fugu contiguous length, kb | Syntenic genes | Fugu syntenic fragment, kb | Human syntenic fragment, kb | Fold compaction of fugu locus | ||
|---|---|---|---|---|---|---|---|
| From | To | No. of genes | |||||
| HoxAa | 385.8 | Osbpl3a | Fkbp14a | 21 | 332.5 | 5,270.7 | 16 |
| HoxAb | 163.1 | Plekha8b | Cbx3b | 16 | 148.5 | 3,934.2 | 26 |
| HoxBa | 297.9 | Prosapip2a | HoxB1a | 13 | 269.7 | 1,246.5 | 5 |
| HoxBb | 130.2 | Eap30 | Osbpl7b | 15 | 107.6 | 1,212.4 | 11 |
| HoxCa | 378.8 | Mgc11308 | Csad | 16 | 325.3 | 1,182.8 | 4 |
| HoxDa | 366.8 | Scrn3a | Osbp16 | 19 | 337.4 | 4,035.7 | 12 |
| HoxDb | 398.5 | Chrna1b | Waspipb | 8 | 180.0 | 2,783.8 | 15 |
Consistent with the small size of the fugu genome (400 Mb), fugu Hox loci are 4- to 26-fold smaller compared with human loci (Table 1). The longest conserved syntenic block is found at fugu HoxDa locus, which spans 337 kb compared with 4.0 Mb of its corresponding region in human, and contains 19 coding genes and a microRNA (miRNA) gene (Fig. 1 and Table 1). The highest number of syntenic genes, however, is found at fugu HoxAa locus (Fig. 1). This locus contains 21 coding genes and a miRNA gene across 333 kb. Its human ortholog covers 5.3 Mb. Conservation of such extensive gene arrangements in the fugu and human genomes suggests that they may contain functional elements that are shared by several genes across the locus. Comparisons between paralogous fugu Hox loci show that the duplicate loci (HoxAb, HoxBb, and HoxDb) are located on shorter syntenic blocks than their original loci, indicating that, besides the loss of a large number of genes after duplication, extensive rearrangements have occurred in the flanking regions of the duplicate Hox clusters. One exception to this trend is HoxBb locus, which has retained more syntenic genes outside the Hox cluster than HoxBa locus (Fig. 1).
Rearrangements at the Fugu Hox Loci.
Although the relative positions and orientations of most of the genes are conserved at the fugu and human syntenic blocks, a few fugu genes at the ends of the fugu HoxAa, HoxBa, HoxCa, and HoxDb loci (Fig. 1) appear to have undergone intrachromosomal rearrangements. Because the positions and orientations of paralogs of some of these fugu genes are the same as their human orthologs (e.g., Fkbp14b in HoxAb locus, Osbpl7b in HoxBb locus, and Gpr155a in HoxDa locus), we infer that the rearrangements occurred in the fugu lineage after the duplication of the Hox loci in the fish lineage. Parsimonious models that can explain the rearrangements at the fugu HoxAa, HoxBa, HoxCa, and HoxDb loci are shown in Figs. 3 and 4, which are published as supporting information on the PNAS web site. This model involves intrachromosomal pairing of homologous sequences and recombination, resulting in the inversion of the loop region containing the Hox cluster and the non-Hox genes linked to the Hox cluster. In addition to the inversion of the Hox cluster, localized inversions of short segments have occurred in the flanking regions of some of these Hox clusters (Figs. 3 and 4). The maintenance of contiguous segments containing the Hox cluster and the non-Hox genes linked to the cluster despite extensive rearrangements in the flanking regions imply a strong selection against rearrangements within these syntenic blocks.
Conserved Noncoding Sequences in the Hox Loci.
The common ancestors of mammals and ray-finned fishes diverged ≈450 million years (Myr) ago, and the noncoding sequences conserved in these lineages over such a long evolutionary period are likely to represent functional elements such as RNA genes and cis-regulatory elements. Indeed, many of the noncoding elements conserved in fugu and mammals have been shown to function as tissue-specific enhancers (reviewed in refs. 19 and 20). To identify putative conserved regulatory elements in the Hox loci, we searched for CNS in the fugu, human, and mouse loci by using mlagan (21). CNS between human and mouse are typically defined by using 100-bp windows with a minimum 70% identity (22). Considering the much longer divergence period between ray-finned fishes and mammals than that between humans and rodents (≈80 Myr), we chose a lower-stringency criteria of 50-bp windows with minimum 60% identity for identifying fugu-mammal CNS. We also identified human-mouse CNS for the same regions by using the higher stringency and compared the fugu-mammal CNS with human-mouse CNS. Approximately 88% of fugu-mammal CNS were found to be part of human-mouse CNS, and another 3% partially overlap human-mouse CNS (Table 3, which is published as supporting information on the PNAS web site), indicating that most of the fugu-mammal CNS identified by using the lower stringency represent evolutionary-conserved sequences.
The vista (23) plots of mlagan alignments between seven fugu Hox loci and their orthologous human and mouse loci are shown in Figs. 5–11, which are published as supporting information on the PNAS web site. Because the numbers and extent of CNS identified between fugu-human and fugu-mouse alignments were similar, results of only fugu-human CNS will be discussed here as a representative of the fugu-mammal comparisons. Because our objective was to identify conserved regulatory elements, RNA genes in the initial set of CNS were excluded. A final set of 368 CNS comprising 39.3 kb was identified in the seven fugu Hox loci. The location and sizes of all of the 368 CNS are given in Table 3. The longest CNS is 805 bp long (79.6% identity) and is located in the intergenic region of Lnp and Evx2. Although only 1.5% of the mammalian genome code for proteins, up to 62.5% of the mouse genome has been found to be transcribed (24), indicating that a large proportion of noncoding regions are transcribed. To determine whether any of the CNS at the Hox loci are transcribed, we searched them against the human and mouse ESTs and FANTOM3 cDNAs. One hundred fifty of the 368 CNS (41%), most of them located within the Hox clusters, showed overlap with the ESTs (Table 3, column I). These CNS may represent previously undescribed noncoding exons. Based on comparisons of fugu and human genome sequences by using megablast (http://genopole.toulouse.inra.fr/blast/megablast.html), Woolfe et al. (20) have identified 1,389 conserved noncoding elements (CNEs). Only 54 of the CNS identified by us at the Hox loci overlap with these conserved noncoding elements (Table 3, column K).
Fugu HoxAa cluster contains the largest number of CNS (44 CNS totaling 4 kb), followed by HoxCa cluster (38 CNS at 3.6 kb) (Table 2). HoxDa (31 CNS at 2.8 kb) and HoxBa (35 CNS at 2.8 kb) clusters contain a similar extent of CNS. The duplicate fugu Hox clusters (HoxAb, HoxBb, and HoxDb), which have lost a large number of Hox genes after the duplication, contain far fewer CNS (4–11 CNS) than their original clusters (Table 2). Although HoxDa cluster contains the smallest number of CNS, HoxDa locus contains the highest number of CNS (111 at 16.9 kb) outside the Hox cluster (Table 2). In contrast, HoxAa locus contains only 38 CNS (3 kb) outside the Hox cluster. The extended syntenic block in the 3′ region of the HoxBb locus contains 13 CNS (1.2 kb) outside the Hox cluster (Table 2). Thus, syntenic blocks are generally associated with CNS. Comparisons of CNS between the fugu paralogous Hox loci show that 20 of the CNS have been conserved in both paralogs (Table 4, which is published as supporting information on the PNAS web site), whereas the rest (86 in HoxAa and HoxAb; 49 in HoxBa and HoxBb, and 146 in HoxDa and HoxDb) have been retained in only one of the two paralogs, indicating that a large number of CNS that were present in the ancestral vertebrate loci have undergone complementary degeneration or diversion in the fugu paralogs.
Table 2.
CNS in the fugu and human Hox cluster loci
| Fugu Hox cluster loci | Fugu sequence,* kb | CNS | ||||||
|---|---|---|---|---|---|---|---|---|
| Within Hox cluster | Outside Hox cluster | Total CNS | ||||||
| No. | Size, bp | No. | Size, bp | No. | Size, bp | Avg. size, bp | ||
| HoxAa | 259.9 | 44 | 4,058 | 38 | 3,049 | 82 | 7,107 | 87 |
| HoxAb | 148.5 | 11 | 933 | 15 | 1,135 | 26 | 2,068 | 80 |
| HoxBa | 237.4 | 35 | 2,798 | 2 | 125 | 37 | 2,923 | 79 |
| HoxBb | 107.6 | 11 | 1,176 | 13 | 1,168 | 24 | 2,344 | 98 |
| HoxCa | 188.7 | 38 | 3,621 | 9 | 905 | 47 | 4,526 | 96 |
| HoxDa | 338.9 | 31 | 2,816 | 111 | 16,985 | 142 | 19,801 | 139 |
| HoxDb | 68.8 | 4 | 219 | 6 | 321 | 10 | 540 | 54 |
| Total | 368 | 39,309 |
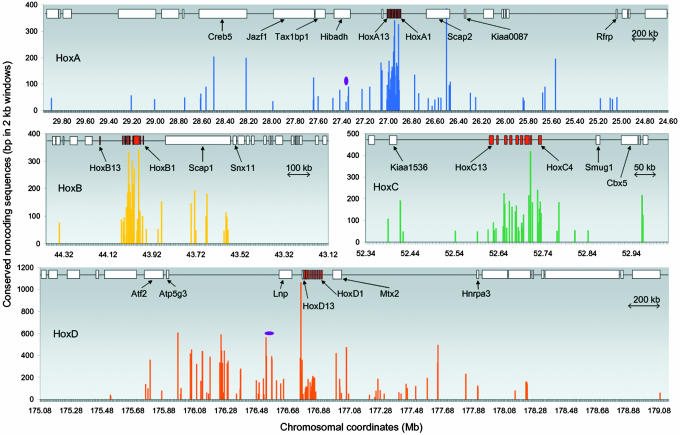
The profiles of the nonredundant set of CNS at the four human Hox loci (348 CNS comprising a total of 38 kb) are shown in Fig. 2. Human HoxD and HoxA loci stand out as they contain a large number of CNS outside the Hox clusters. Interestingly, the level of conservation of CNS outside the Hox clusters, particularly in the HoxD locus, is much higher than that within the Hox cluster indicating the importance of these regions. Some of the CNS flanking the HoxD and HoxA clusters overlap with the previously identified GCRs at these loci. A 40-kb region in the intergenic region between Atp5g3 and Lnp in the HoxD locus was shown to contain a 5′ GCR, which directs expression of posterior HoxD genes besides Lnp and Evx2. Two blocks at the extreme ends of this GCR were found to be conserved in fugu (3). Our fugu-mammal comparison has identified a remarkable cluster of 59 CNS (totaling 10 kb) in the 741-kb intergenic region between Atp5g3 and Lnp, including the two blocks that were previously identified (Fig. 2). These extensive blocks of conserved regions, which extend up to 571 kb upstream of the 40-kb 5′ GCR, may function either as independent enhancers or as a part of the 5′ GCR. Because the gene upstream of this cluster of elements is a housekeeping gene (Atp5g3, mitochondrial ATP synthase subunit 9) that expresses in a wide range of tissues, the targets of this regulatory region could be genes located downstream (Lnp, Evx2, or HoxD). The 3′ end of the HoxD cluster has been predicted to contain an early limb control region (4). We identified a cluster of 33 CNS in this region (between HoxD3 and Hnrpa3 and in the introns of Hnrpa3) (Table 3). Some of these CNS might be part of the predicted early limb control region. The 5′ region of HoxA locus contains a cluster of 12 CNS between _Tax1bp1_-Hibadh-Evx1 genes and in the introns of Tax1bp1 (Table 3 and Fig. 2), a position homologous to the 5′GCR of the HoxD cluster. A previous study had identified a putative long-range enhancer within this region associated with expression of HoxA13, Evx1, Hibadh, Tax1bp1, and Jazf1 genes (5). A highly conserved element in this region (2.25 kb in the fourth intron of Hibadh), however, was found to be inadequate to recapitulate all of the expression domains of these genes. Besides this element, fugu-mammal CNS identified in the present study include several other elements within this region (Table 3 and Fig. 2). The 3′ region of the HoxA locus contains a cluster of nine CNS in the intergenic region of HoxA1 and Scap2 and the introns of Scap2 (Table 3 and Fig. 2) at a position homologous to the 3′ GCR of HoxD locus. However, it remains to be demonstrated whether these CNS are associated with HoxA genes or the genes downstream of HoxA cluster.
Fig. 2.
Profiles of CNS at the human HoxA, HoxB, HoxC, and HoxD loci. x axis represents chromosomal coordinates, and y axis represents CNS. Genes at each locus are shown at the top as red boxes (Hox genes) or open boxes (non-Hox genes) linked with a thin line. Names of some genes are indicated. The previously identified 5′ global enhancers at HoxD and HoxA loci (3, 5) are represented by purple oval shapes.
Although the level of conservation is not as high as in HoxD and HoxA loci, HoxB and HoxC loci also contain CNS outside the Hox cluster. The 3′ flanking region of HoxB locus contains a cluster of 14 CNS between HoxB1 and Snx11 genes (Table 3 and Fig. 2). The HoxC locus contains six CNS between Kiaa1536 and HoxC13 and five CNS between HoxC4 and Smug1 (Table 3 and Fig. 2). It remains to be seen whether these CNS located outside the Hox clusters are associated with the regulation of Hox genes or non-Hox genes present in these loci.
The CNS identified by us include several previously characterized functional regulatory elements. In particular, they include HB1 elements in the introns of HoxA4, HoxA7, HoxB4, and HoxD4 and upstream region of HoxD9, retinoic acid response elements in flanking regions of HoxA3, HoxA4, HoxB3, HoxB4, HoxC4, and HoxD4, the neural enhancer within the 5′ GCR of HoxD cluster (3), and the central nervous system enhancer located 5′ to the HoxA cluster (5) (Table 3, column L). It is likely that the other CNS identified in this study represent previously undescribed regulatory elements associated with Hox genes and other genes present in these loci. However, this hypothesis needs to be confirmed by functional assays in transgenic systems.
Transcription Factor Binding Sites (TFBS) Within CNS.
To identify transcription factors that bind to the CNS at the Hox loci, we searched the human CNS against the transfac (BiobaseWolfenbüttel, Germany) database by using tess (www.cbil.upenn.edu/tess). Our TFBS analysis identified 12,292 binding sites spread among 368 CNS (data not shown). Of these binding sites, 2,478 were found on sequences that are 100% identical in human, mouse, and fugu. The names and locations of these TFBS are shown in Table 3 (column M). A list of the most abundant of these TFBS is given in Table 5, which is published as supporting information on the PNAS web site. These transcription factors are likely to be involved in the regulation of Hox and non-Hox genes in the Hox loci analyzed by us. Among these TFBS, sites for Cdx-1, USF, E12, IUF-1, Pbx-1, and c-Jun were found to be significantly enriched (P < 8 × 10−3) in these conserved sequences as compared with TFBS predicted on random noncoding sequences in the human genome (see footnote to Table 5). Cdx-1 and USF are known regulators of Hox genes (25, 26), and because our loci contain a large number of Hox genes, many of the binding sites identified for these factors might be functional. The TFBS predicted on the CNS constitute targets for assaying the potential functions of the associated CNS.
Discussion
Chromosomal segments that have escaped rearrangements over a long evolutionary period reflect a strong selective pressure that has preserved the integrity of the cluster of genes located on the segment. Vertebrate Hox clusters typify such conserved chromosomal segments. The relative order and orientation of Hox genes in each cluster have been highly conserved during vertebrate evolution. In the present study, we have identified large syntenic blocks extending far beyond the Hox clusters that have been conserved in fugu and mammals, indicating that the flanking regions of Hox clusters are also under selective pressure. The syntenic blocks at the human and mouse HoxA and HoxD loci are exceptionally large, spanning 5.4 Mb and 4 Mb, respectively. Genomewide comparisons of fugu and human had indicated that the conserved segments in the two genomes were restricted to short fragments with few syntenic genes on them (27). Nevertheless, longer syntenic regions have been identified at several loci for which contiguous sequences are available (28–30). The longest syntenic block identified so far is at the fugu and human Shh locus. The order and orientation of 16 genes spread across 4 Mb in the human Shh loci are totally conserved in fugu, although the syntenic block in rodents is restricted to only 12 genes (30). Approximately 25 conserved noncoding elements were identified in this locus, and a large number of them were found to exhibit enhancer activity in transgenic assays. It was proposed that association of these elements with several genes in this locus is responsible for maintaining the large syntenic block (30). The conserved syntenic blocks at the human HoxA and HoxD loci contain 97 and 149 CNS, respectively. These CNS include previously characterized enhancer elements, including a subset of elements associated with the 5′ global enhancers of the HoxD and HoxA clusters (3, 5). We propose that most of these CNS are likely to be regulatory elements, and the distribution of these CNS over large regions may be responsible for the maintenance of the unusually large syntenic blocks at these loci.
The distribution of potential regulatory elements over a large region in the mammalian genome poses a major challenge in testing their function. For example, our analysis has identified 59 putative regulatory elements, including subsets of the HoxD 5′ GCR spread across a 741-kb intergenic region of Atp5g3 and Lnp. Functional assay of this control region should ideally include all of the 59 elements, but it is not easy to obtain a mammalian genomic clone containing all these elements. The compact intergenic regions of fugu offer an attractive alternative for testing the function of such clusters of CNS. The intergenic region between Atp5g3 and Lnp in the fugu genome is only 74 kb, and the locus from Atp5g3 to HoxD3a is 131 kb. A fugu BAC containing this region will be invaluable for testing the function of these elements. Although the fugu clone may lack mammal-specific elements, transgenic experiments in zebrafish and mice should provide useful insights into the role of this cluster of CNS in the regulation of Hox and other genes in this locus.
Materials and Methods
Sequences and Annotation.
We analyzed the third fugu assembly (www.fugu-sg.org) to identify scaffolds containing Hox gene sequences (Table 6, which is published as supporting information on the PNAS web site) by tblastn (www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html) search by using known fugu, zebrafish, and human Hox protein sequences. A contig map for each locus was generated based on cosmid and BAC end-sequence linkage information obtained by searching the scaffold sequences against the cosmid and BAC end sequences. Contiguous sequences extending on both sides of Hox clusters were obtained by filling gaps within and between scaffolds until two or more genes that were unrelated to genes in the orthologous human Hox loci were encountered. Gaps were filled by sequencing cosmid or BAC DNA directly or by sequencing PCR products amplified from genomic DNA. Protein-coding genes were annotated by blastx search of the fugu sequence against the nr protein database at the National Center for Biological Information (NCBI) and followed by tblastn search of the most similar full-length protein sequence in the nr database against the fugu sequence. For some non-Hox genes with ambiguous exon boundaries, gene structures were predicted by using genewise (www.ebi.ac.uk/Wise2). Genomic sequences and annotation details for the human (v30.35c; NCBI 35) and mouse (v25.33; NCBI m33) Hox cluster loci were extracted from ensembl (www.ensembl.org). Human, mouse, and rat multiple alignments (Human hg16, mouse mm4, and rat rn3) were obtained from http://pipeline.lbl.gov/downloads.shtml. Human-mouse CNS that fulfilled the criteria of ≥70% identity across 100-bp windows were extracted, and coordinates were taken with respect to the NCBI 35 (hg17) assembly.
Identification and Analysis of CNS.
Repetitive sequences in the human, mouse, and fugu loci were masked by using repeatmasker repbase (Update September 8, 2005; Genetic Information Research Institute, Mountain View, CA). Multiple alignments were generated by using mlagan (21) and visualized by using vista (23). All mlagan alignments and vista visualization were carried out on a Compaq (Palo Alto, CA) DEC Alpha server, and all scripts were written in perl (O'Reilly Media, Sebastopol, CA). CNS between fugu and mammals were identified by using a criterion of ≥60% identity across 50 bp windows.
The initial set of fugu-mammal CNS were blastx searched against the NCBI nr protein database (cutoff 1 × 10−4) to identify coding exons that were missed during the annotation. This search resulted in a refinement of the exon annotation of some fugu genes (e.g., Atf2 and Hnrpa3). The revised set of CNS was then blastn searched against rfam (www.sanger.ac.uk/Software/Rfam) and mirbase (http://microrna.sanger.ac.uk/sequences) to identify and eliminate RNA genes. The fugu-mammal CNS were searched against the fugu-human conserved noncoding elements (26) and human and mouse ESTs and FANTOM3 cDNAs by using the cutoff of 1 × 10−4 and minimum 50% coverage to determine the extent of overlap. Finally, CNS between paralogous fugu loci were compared by using blastn.
Identification of TFBS.
The CNS were submitted to tess (www.cbil.upenn.edu/tess) for the identification of putative TFBS. A combined search (by using filtered string-matching and searching against weight matrices) was carried out with vertebrate transcription factors and a maximum allowable string mismatch of 10%. Only string matches with L a/ value equal to 2, and matrix matches with _S_c > 0.9 and _S_m > 0.8 were retained.
Supplementary Material
Supporting Information
Acknowledgments
This project was funded by Singapore's Agency for Science, Technology, and Research. A.P.L. is supported by the A*STAR Graduate Scholarship. B.V. is an adjunct staff of the Department of Pediatrics, Yong Loo Lin School of Medicine, National University of Singapore.
Abbreviations
BAC
bacterial artificial chromosome
CNS
conserved noncoding sequences
GCR
global control region
NCBI
National Center for Biological Information
TFBS
transcription factor binding site.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The fugu Hox loci sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ481663–DQ481669).
References
- 1.Kmita M., Duboule D. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 2.Chambeyron S., Da Silva N. R., Lawson K. A., Bickmore W. A. Development (Cambridge, U.K.) 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 3.Spitz F., Gonzalez F., Duboule D. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 4.Zakany J., Kmita M., Duboule D. Science. 2004;304:1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]
- 5.Lehoczky J. A., Williams M. E., Innis J. W. Evol. Dev. 2004;6:423–430. doi: 10.1111/j.1525-142X.2004.04050.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim C.-B., Amemiya C., Bailey W., Kawasaki K., Mezey J., Miller W., Minoshima S., Shimizu N., Wagner G., Ruddle F. Proc. Natl. Acad. Sci. USA. 2000;97:1655–1660. doi: 10.1073/pnas.030539697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu C.-H., Amemiya C., Dewar K., Kim C.-B., Ruddle F. H., Wagner G. P. Proc. Natl. Acad. Sci. USA. 2002;99:5492–5497. doi: 10.1073/pnas.052709899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santini S., Boore J. L., Meyer A. Genome Res. 2003;13:1111–1122. doi: 10.1101/gr.700503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu C.-H., Dewar K., Wagner G. P., Takahashi K., Ruddle F., Ledje C., Bartsch P., Scemama J. L., Stellwag E., Fried C., et al. Genome Res. 2004;14:11–17. doi: 10.1101/gr.1712904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prohaska S. J., Fried C., Flamm C., Wagner G. P., Stadler P. F. Mol. Phylogenet. Evol. 2004;31:581–604. doi: 10.1016/j.ympev.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Marshall H., Studer M., Popperl H., Aparicio S., Kuroiwa A., Brenner S., Krumlauf R. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 12.Hadrys T., Prince V., Hunter M., Baker R., Rinkwitz S. J. Exp. Zool. 2004;302:147–164. doi: 10.1002/jez.b.20012. [DOI] [PubMed] [Google Scholar]
- 13.Geada A. M., Coletta P. L., Sharpe P. T. Mamm. Genome. 1996;7:81–84. doi: 10.1007/s003359900021. [DOI] [PubMed] [Google Scholar]
- 14.Wang W. C., Anand S., Powell D. R., Pawashe A. B., Amemiya C. T., Shashikant C. S. J. Exp. Zool. 2004;302:436–445. doi: 10.1002/jez.b.21009. [DOI] [PubMed] [Google Scholar]
- 15.Nolte C., Amores A., Nagy Kovacs E., Postlethwait J., Featherstone M. Mech. Dev. 2003;120:325–335. doi: 10.1016/s0925-4773(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 16.Sabarinadh C., Subramanian S., Tripathi A., Mishra R. K. BMC Genomics. 2004;5:75. doi: 10.1186/1471-2164-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 18.Amores A., Suzuki T., Yan Y. L., Pomeroy J., Singer A., Amemiya C., Postlethwait J. H. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatesh B., Yap W. H. BioEssays. 2005;27:100–107. doi: 10.1002/bies.20134. [DOI] [PubMed] [Google Scholar]
- 20.Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., Smith S. F., North P., Callaway H., Kelly K., et al. PloS Biol. 2005 doi: 10.1371/journal.pbio.0030007. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brudno M., Do C. B., Cooper G. M., Kim M. F., Davydov E., Green E. D., Sidow A., Batzoglou S. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loots G. G., Locksley R. M., Blankespoor C. M., Wang Z. E., Miller W., Rubin E. M., Frazer K. A. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 23.Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 25.Haerry T. E., Gehring W. J. Proc. Natl. Acad. Sci. USA. 1996;93:13884–13889. doi: 10.1073/pnas.93.24.13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J., Giannola D. M., Zhang Y., Rivera A. J., Emerson S. G. Blood. 2003;102:2420–2427. doi: 10.1182/blood-2003-01-0251. [DOI] [PubMed] [Google Scholar]
- 27.Aparicio S., Chapman J., Stupka E., Putnam N., Chia J. M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., et al. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesh B., Gilligan P., Brenner S. FEBS Lett. 2000;476:3–7. doi: 10.1016/s0014-5793(00)01659-8. [DOI] [PubMed] [Google Scholar]
- 29.Smith S. F., Snell P., Gruetzner F., Bench A. J., Haaf T., Metcalfe J. A., Green A. R., Elgar G. Genome Res. 2002;12:776–784. doi: 10.1101/gr.221802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goode D. K., Snell P., Smith S. F., Cooke J. E., Elgar G. (2005) Genomics. 2005;86:172–181. doi: 10.1016/j.ygeno.2005.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information