Diverse Chromatin Remodeling Genes Antagonize the Rb-Involved SynMuv Pathways in C. elegans (original) (raw)
Abstract
In Caenorhabditis elegans, vulval cell-fate specification involves the activities of multiple signal transduction and regulatory pathways that include a receptor tyrosine kinase/Ras/mitogen-activated protein kinase pathway and synthetic multivulva (SynMuv) pathways. Many genes in the SynMuv pathways encode transcription factors including the homologs of mammalian Rb, E2F, and components of the nucleosome-remodeling deacetylase complex. To further elucidate the functions of the SynMuv genes, we performed a genome-wide RNA interference (RNAi) screen to search for genes that antagonize the SynMuv gene activities. Among those that displayed a varying degree of suppression of the SynMuv phenotype, 32 genes are potentially involved in chromatin remodeling (called SynMuv suppressor genes herein). Genetic mutations of two representative genes (zfp-1 and mes-4) were used to further characterize their positive roles in vulval induction and relationships with Ras function. Our analysis revealed antagonistic roles of the SynMuv suppressor genes and the SynMuv B genes in germline-soma distinction, RNAi, somatic transgene silencing, and tissue specific expression of pgl-1 and the lag-2/Delta genes. The opposite roles of these SynMuv B and SynMuv suppressor genes on transcriptional regulation were confirmed in somatic transgene silencing. We also report the identifications of ten new genes in the RNAi pathway and six new genes in germline silencing. Among the ten new RNAi genes, three encode homologs of proteins involved in both protein degradation and chromatin remodeling. Our findings suggest that multiple chromatin remodeling complexes are involved in regulating the expression of specific genes that play critical roles in developmental decisions.
Synopsis
In animal cells, DNA and genes are packed into a structure called chromatin. Chromatin-modifying protein complexes play a critical role in the regulation of gene expression. These complexes can alter the chemical and structural properties of the chromosome leading to either the repression or activation of gene expression. How these different complexes coordinate to regulate animal development remains to be explored. Several developmental processes in the nematode Caenorhabditis elegans present excellent model systems to study the functions of chromatin modifications. Using a genome-wide screen, the authors have identified 32 genes that encode potential chromatin-modifying proteins that antagonize the function of another set of transcription regulators including homologs of the mammalian Rb tumor suppressor and components of other chromatin-modifying complexes. The antagonistic roles of these two sets of genes have been observed in a variety of cellular and developmental processes, including organ development and expression of genes in particular tissues. This work indicates that multiple chromatin-modifying complexes are involved in maintaining proper expression of many genes that are critical for precise developmental decisions. Studies on these worm genes should shed light on the roles of the mammalian counterparts in development and related human diseases.
Introduction
In the wild-type Caenorhabditis elegans hermaphrodite, the vulva is formed from the descendants of three of six vulval precursor cells (VPCs) [1,2]. Each VPC is initially equivalent in developmental potential. An epidermal growth factor–like signal from the gonad activates the receptor tyrosine kinase (RTK)/Ras/mitogen-activated protein kinase (MAPK) signaling pathway in three central VPCs, inducing them to adopt vulval cell fates [1,2]. A large group of genes, collectively called synthetic multivulva (SynMuv) genes, antagonize this RTK/Ras/MAPK signaling activity [3,4]. The SynMuv genes fall into three subgroups: classes A, B, and C. While a single loss-of-function (lf) mutation in each class does not result in obvious vulval induction defects, a combination of mutations in two classes gives rise to a robust Muv phenotype [3,5–7]. Cloning two of the SynMuv A genes, lin-15A and lin-8, has revealed that these encode nuclear proteins, suggesting that they may play a role in transcription regulation [8–10]. Many of the class B and C SynMuv genes encode transcription factors, including those homologous to components of the mammalian RB/E2F protein complex and chromatin-remodeling complexes (e.g., the nucleosome-remodeling deacetylase [NuRD] complex and the NuA4 [nucleosomal acetyltransferase of histone H4] complex) [4,7,11–18]. Previous studies on homologous proteins indicated that many SynMuv proteins likely function via transcriptional repression. For example, the NuRD complex represses transcription through nucleosomal remodeling and deacetylase activities. Recently, Drosophila RB/E2F-containing complexes (dREAM and Myb-MuvB complexes), which contain homologs of multiple SynMuv B proteins, have been shown to act as transcriptional repressors [19–21].
Besides their redundant roles in repressing vulval induction, the SynMuv B genes are also involved in multiple cellular functions during development. These roles include regulation of cell divisions and proliferation [22,23], regulation of context-dependent transcription silencing of transgenes in somatic cells [11], RNA interference (RNAi) [24], expression of lag-2 in the intestine [18,25], and other developmental functions such as regulation of pharyngeal morphogenesis and larval development [26,27].
Chromatin remodeling plays a crucial role in regulating gene transcription. There are several ways that chromatin structure can be modulated [28]. First, the positioning of nucleosomes on DNA can be disrupted and reconfigured by ATP-dependent remodeling complexes. Second, posttranslational modifications by histone acetylases, deacetylases, and methyltransferases can generate localized distinct chromosomal domains by recruiting diverse chromatin-binding protein complexes. Third, the composition of nucleosomes can be modified by replacing major histones with variants. Most studies on chromatin modifications have been performed in yeast and mammalian cell cultures, and little is known about the functions of various chromatin-remodeling complexes during animal development. Studies of C. elegans vulval induction and other cellular and developmental processes provide us an opportunity to identify specific roles of these complexes in animal development.
To identify genes that antagonize the activity of SynMuv genes in regulating animal development, we have carried out a genome-wide RNAi screen. In this paper, we present the identification and characterization of a number of SynMuv suppressor genes that are involved in chromatin remodeling with respect to their roles in vulval induction, germline-soma distinction, germline and somatic transgene silencing, RNAi, and tissue-specific regulation of the expressions of the pgl-1 and the lag-2 genes.
Results
A Set of Chromatin Remodeling Genes Antagonize the SynMuv Gene Activities in Vulval Differentiation
A genome-wide feeding RNAi screen [29] was carried out to identify suppressors of the Muv phenotype of lin-15AB(n765). Among the 16,757 genes represented in the RNAi feeding library (out of the currently predicted 19,429 genes in C. elegans), we have identified more than 70 candidate genes. In three independent retests, RNAi of these candidate genes consistently suppressed the Muv phenotype of lin-15AB(n765). The sequence data of these identified genes were subsequently inspected for domain structures, as well as for homology to genes in Homo sapiens, Drosophila melanogaster, and Saccharomyces cerevisiae. The genes we identified include those that may function in RTK/Ras/MAPK and Notch signaling, cell fusion or migration, chromatin remodeling, dosage compensation, and other cellular processes. In this study, we focus on 32 candidate genes (called SynMuv suppressor genes herein) that are potentially involved in chromatin remodeling (Tables 1 and S1). Except for the six genes that have been recently identified as the SynMuv suppressors [24], 26 of the 32 genes were previously unknown for such a role. To investigate the allele specificity for suppression, we performed RNAi of these 32 genes in two other SynMuv mutants, lin-36(n766); lin-15A(n767) and lin-8(n111); lin-15B(n374). RNAi of all 32 genes significantly suppressed the Muv phenotype of at least one of these two strains (Table S2).
Table 1.
A Genome-Wide RNAi Screen Identified 32 SynMuv Suppressor Genes That Are Potentially Involved in Chromatin Remodeling
Among the 32 SynMuv suppressor genes, 14 encode proteins that are homologous to components of several known yeast and human chromatin remodeling complexes, such as NuA4 [30,31], SWR1 [32–34], COMPASS/MLL[35,36], ISW1[37], and SWI/SNF [38] complexes (Tables 1 and S1). Based on what are known to the chromatin remodeling complexes of their yeast homologs, these genes are further categorized into five groups. Proteins encoded by two genes in the NuA4 complex group, mrg-1 and ZK1127.3, have been shown to directly bind each other in a yeast two-hybrid assay [39], implicating that they may function in the same complex in C. elegans. The yeast homologs of GFL-1 and Y105E8A.17 proteins are shared components of the NuA4 and SWR1 complexes in yeast [40]. The SWR1 complex in S. cecrevisiae is required for depositing the histone H2A variant H2A.Z into euchromatin to maintain gene expression [32–34]. An SWR1-like complex might also be present in C. elegans and may play a role similar to that of its yeast counterpart. This hypothesis was supported by the finding of R08C7.3, a histone-variant H2A.Z, as a strong SynMuv suppressor (Table 1).
We also identified four factors required for protein ubiquitylation and degradation, including two of the 19S regulatory particle (RP) subunits of the 26S proteosome (RPN-10 and RPN-12) [41], an E3 ubiquitin-protein ligase (C34D4.14), and an ubiquitin protease (F30A10.10). This implicates that protein ubiquitylation and degradation are involved in vulval differentiation. However, a recent finding has brought to light a distinct nonproteolytic role of 19S RP in chromatin remodeling [42]. The yeast ortholog of C34D4.14 directly binds to the two subunits of the yeast 19S RP [43], implicating that C34D4.14 may regulate chromatin remodeling through interacting with 19S RP of the 26S proteosome. In addition, the yeast H2B ubiquitin protease Ubp8, a C. elegans homolog of F30A10.10, is required for mediating the histone H2B ubiquitylation and deubiquitylation [44]. Taken together, it suggests that these four genes may play a role in chromatin remodeling to regulate vulval differentiation in C. elegans.
Proteins encoded by 9 of the 32 SynMuv suppressor genes are predicted to have roles in chromatin remodeling based on their molecular characteristics of their homologs in yeast and/or the functional domains they have. As mentioned above, R08C7.3 encodes a histone H2 variant, H2A.Z. T02C12.3 and ZK856.9 encode the subunits of the general RNA polymerase III transcription factor TFIIIC. TFIIICs have been shown to have intrinsic histone acetyltransferase (HAT) activity to alleviate chromatin-mediated transcriptional repression of the RNA polymerase III–dependent gene in yeast [45,46]. F52B11.1 and Y53G8AR.2 encode proteins with a zinc finger-like plant homeodomain (PHD), which is found in many nuclear proteins with chromatin as their substrate [47]. B0035.4 encodes a homolog of the yeast Gim3p. Gim3p, a component of the prefoldin complex, has been shown to function redundantly with components of the SWR1 complex in yeast [33]. Mammalian ortholog of ZFP-1 binds to the GFL-1 ortholog GAS41, a component of human NuA4 complex [48]. pqn-28 encodes a C. elegans homolog of yeast Sin3p, which is well known for its function in chromatin remodeling [24,49]. In agreement with the possible involvement of histone methyltransferase in vulval development, RNAi of a S-adenosylmethionine synthetase (SAM) gene (C06E7.1) suppressed the SynMuv phenotype. There are five SAM synthetase genes, C06E7.1, C06E7.3, C49F5.1, T13A10.11, and Y105C5B.12, which encode proteins shared 99% homology. We expect that RNAi of C06E7.1 would have cross-reactions with the other four genes.
The remaining five genes, C34E10.8, F54D11.2, K08F4.2, M03C11.3, and T07E3.3, encode proteins with unknown functions. Their genetic properties (see below for detail) are more similar to those of the known chromatin factors described above than to those of other SynMuv suppressors such as genes involved in RTK/Ras/MAPK signaling, cell fusion, or cell migration (unpublished data). This suggests that these five genes may also function in chromatin remodeling. We thus characterize them together with the known chromatin factors even though it is still possible that they do not act as transcriptional regulators.
zfp-1 and mes-4 Promote Vulval Induction in SynMuv Mutants
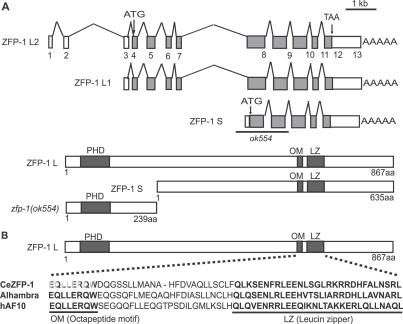
To validate the results of the RNAi screen and better characterize the functions of the SynMuv suppressor genes, we carried out further analysis of _zfp-_1 and mes-4 using genetic mutations. zfp-1 is a C. elegans ortholog of the human AF10 gene [50]. AF10 directly binds to GAS41 (a component of human NuA4 complex) and hDOT1 (a H3 K79 methyltransferase) [48,51]. ZFP-1 shares the highly conserved motifs of the AF10 protein family including the amino-terminal PHD finger, an octapeptide motif (OM), and a leucine zipper motif (LZ) in its carboxy terminus (Figure 1). The OM-LZ motifs have been shown to be required for leukemic transformation by the MLL-AF10 fusion protein [52]. The zfp-1 gene encodes three different transcripts, zfp-1 L1, zfp-1 L2, and zfp-1 S, based on expressed sequence tag data (http://www.wormbase.org; Figure 1). Both zfp-1 L1 and zfp-1 L2 are predicted to encode the larger protein (867 amino acids) bearing the PHD finger and the OM-LZ motifs. zfp-1 S encodes a shorter protein (635 amino acids) that lacks the PHD finger but retains the OM-LZ motifs. We obtained a deletion mutant affecting the zfp-1 gene from the International C. elegans Gene Knockout Consortium. This mutation, ok554, removes 1,677 base pairs from the zfp-1 locus and is predicted to generate a truncated protein that consists of the first 240 amino acids of the larger protein, resulting in the lack of the essential OM-LZ motifs (Figure 1). zfp-1(ok554) animals were viable, although they exhibited some developmental defects such as slow growth, protruding vulva, and a reduced brood size (unpublished data), which were also associated with zfp-1(RNAi). Consistent with the RNAi experiment, zfp-1(ok554) significantly reduced the Muv phenotype of lin-15AB(n765) from 100% to 62% at 19 °C, and from 98% to 19% at 18 °C (Table 2). Furthermore, zfp-1(ok554) strongly suppressed the Muv phenotype of lin-35(n745); lin-8(n111) at 19 °C (Table 2).
Figure 1. The C. elegans zfp-1 Gene.
(A) Schematic illustration of the transcripts and proteins of zfp-1 (http://www.wormbase.org). In zfp-1(ok554) mutant, the fifth and sixth exons were deleted and 2 nucleotides (CG) were inserted. Therefore, the short isoform, ZFP-1S, is unlikely to produce any proteins and the long isoform is expected to generate a truncated protein that lacks the OM-LZ motifs in the mutant.
(B) Diagram of the functional motifs of the long isoform of ZFP-1 and the sequence alignment of the OM and LZ motifs. C. elegans ZFP-1 and its homologs from human (hAF10) and Drosophila (Alhambra) are compared.
Table 2.
Genetic Interactions of zfp-1 and mes Genes with SynMuv and ras Genes

mes-2, mes-3, mes-4, and mes-6 genes were identified as maternal-effect sterile genes essential for germline viability [53]. It has been proposed that MES-2/3/6 complex, together with MES-4, are involved in X-chromosome silencing, and that MES-4 functions to exclude repressors from the autosomes [54]. Initially, we identified the RNAi of all four mes genes that suppressed the Muv phenotype of lin-15AB(n765) from the RNAi screen. However, only mes-4 could suppress the other tested SynMuv mutants by RNAi. We then carried out a genetic test using the lf mutations of mes-4 and mes-6. We confirmed that mes-4(lf), but not mes-6(lf), suppressed the Muv phenotype of all tested SynMuv mutants in the absence of the maternal MES proteins contribution (Table 2, unpublished data). MES-4 is homologous to the human MLL and the Drosophila trithorax proteins, which are histone methyltransferases maintaining active gene expression during development. In regard to a previous finding that shows MES-4 associated specifically with autosomes [54], MES-4 may positively regulate the expression of genes on autosomes by modifying chromatin, and thus antagonizing the repression function of the SynMuv genes during vulval development.
The allele-specific suppression of the Muv phenotype of lin-15AB(n765) by RNAi of mes-2, mes-3, and mes-6 genes can be explained by their roles in X-chromosome dosage compensation [55]. The Muv phenotype of lin-15AB(n765) has been previously shown to be suppressed by dosage compensation defects [56,57], which is consistent with the X-chromosomal location of the lin-15A and lin-15B genes.
Interestingly, we found mes-6(bn66) enhanced the Muv phenotype of lin-8(n111); lin-15B(n374) from 76% to 98% Muv. This result suggests that mes-6 may play a role in the SynMuv gene–related transcription repression. Such a repression function is consistent with the facts that the MES-2/MES-3/MES-6 complex keeps the X-chromosome repressed during germline development [54], and that Drosophila enhancer of zeste and extra sex combs, homologs of MES-2 and MES-6, respectively, are members of the Polycomb group of transcriptional repressors [58].
The Relationship between SynMuv Suppressors and Ras
The SynMuv genes are known to act negatively on RTK/Ras/MAPK–mediated vulval induction [2]. Gain-of-function (gf) mutations in let-60/Ras cause a Muv phenotype. To investigate the relationship between identified SynMuv suppressors and the Ras pathway, we performed RNAi of each of the 32 candidate genes in let-60(n1046 gf) animals. We found that the Muv phenotype of let-60(n1046) could not be suppressed by RNAi of any of these genes (unpublished data). Consistent with the RNAi results, we observed no suppression of the Muv phenotype of let-60(n1046) by the mes-4(lf) mutation mentioned above (Table 2). These results suggest that most of the SynMuv suppressor genes may function upstream of the Ras pathway. However, it is possible that the RNAi of some of these genes were too weak to suppress the Muv phenotype of let-60(n1046) even though it is effective in suppressing the Muv phenotype of lin-15AB(n765). In contrast to mes-4(lf), zfp-1(ok554) displayed a significant suppression of the Muv phenotype of let-60(n1046) (Table 2), suggesting that zfp-1 may have an activity downstream or in parallel to let-60/Ras. This does not exclude that zfp-1 also acts with mes-4 on a target upstream of let-60/Ras. The different effects of the mes-4 and zfp-1 mutations on let-60(gf) suggest that they act at two different steps to regulate vulval induction.
SynMuv Suppressor Genes Are Required for the Somatic Expression of the Germline-Specific Gene pgl-1
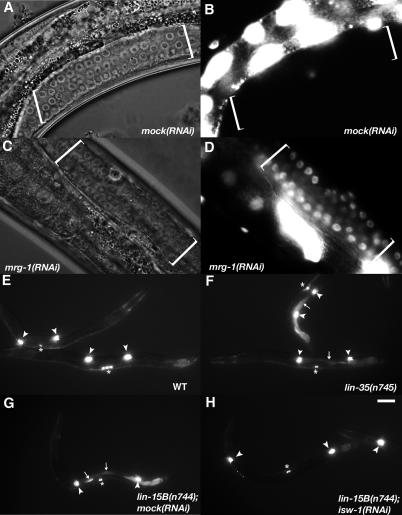
mep-1 encodes a zinc-finger protein that interacts with LET-418/Mi-2 of the NuRD complex to function in the SynMuv B pathway as well as in maintaining germline-soma distinctions in C. elegans [16]. lf mutations in mep-1 cause a first or second larval (L1/L2) lethality phenotype and ectopic expression of germline-specific genes, such as pgl-1, in somatic cells. It has also been shown that mutations in mes-2, mes-3, mes-4, and mes-6 result in a partial suppression of ectopic pgl-1 expression and a partial rescue of the larval lethality of mep-1(lf) [16]. Because mes-4 is one of the strong SynMuv suppressor genes, we investigated whether RNAi of the other SynMuv suppressor genes could rescue the mep-1 larval lethality. We found that RNAi of 12 of the 32 SynMuv suppressor genes, including mes-4, rescued the L1/L2 larval lethality of mep-1(q660) at 25 °C (Table 3, Figure 2A, and unpublished data). RNAi of isw-1, mes-4, and mrg-1 displayed the strongest rescue, resulting in 100% viable adults (Table 3, Figure 2A, and unpublished data).
Table 3.
SynMuv Suppressor Genes Play a Role in Rescuing mep(lf) Larval Lethality, Germline Transgene Silencing, RNAi and the Ectopic Expression of lag-2 in Intestine
Figure 2. The SynMuv Suppressor Genes Antagonize the SynMuv B Genes on Germline-Soma Distinction and pgl-1 Expression.
(A) The early larval arrest phenotype associated with the SynMuv B mutation mep-1(lf) was rescued by RNAi of mrg-1. The growth-arrested L1 larvae of mep-1(lf) (arrowheads) and the rescued adults of mep-1(lf) treated with mrg-1(RNAi) (arrow) are indicated. RNAi of 11 other SynMuv suppressor genes displayed a varying degree of rescue (Table 3).
(B) Quantification of relative mRNA levels of the germline specific gene pgl-1 in animals with the genotype indicated. Only three strong larval arrest rescuers, mes-4, mrg-1, and isw-1, were assayed here. rpl-26 was used as the internal reference. Mean values and ranges of the pgl-1/rpl-26 ratios based on three qRT-PCR trials are shown.
(C–K) Immunostaining using an anti–PGL-1 antibody shows that mep-1(q660) and mrg-1(RNAi) have opposite effects on ectopic expression of pgl-1 in soma. (C–E) Immunofluoresence micrographs of L1 larvae stained with an anti-PGL antibody (red). The genotype of each animal is indicated at the bottom of the corresponding column. (F–H) DAPI staining (blue) of the same animals to indicate nuclei. (I–K) The merged micrographs. White arrowheads indicate germline cells. Arrows indicate the hypodermal cells. The mep-1 mutation (D, G, and J) as well as several other SynMuv B genes (not shown, see text) caused ectopic staining of PGL-1 in somatic cells such as hypodermal cells. This ectopic staining was suppressed by mrg-1(RNAi) (E, H, and K) and RNAi of two other SynMuv suppressor genes (isw-1 and mes-4)(not shown, text). Bar: 100 μm for A, 10 μm for C-K.
We then further investigated the effects of mep-1(q660) and RNAi of three SynMuv suppressor genes on the expression of pgl-1 in L1 larvae using real-time RT-PCR. The mRNA level of pgl-1 in mep-1(q660) animals was increased by about 4-fold compared to that in wild-type (WT) animals (Figure 2B). The mRNA level in mep-1(q660) mutant was down-regulated in animals treated with RNAi of isw-1, mes-4, and mrg-1 (Figure 2B). To confirm that the down-regulation of pgl-1 resulted from the suppression of the ectopic expression in somatic cells, we immunostained mep-1(q660) animals treated with or without RNAi of isw-1, mes-4, and mrg-1 using an anti-PGL-1 antibody (gift from S. Strome). The suppression of ectopic expression of PGL-1 in the somatic cells of mep-1(q660) animals treated with RNAi of isw-1, mes-4, and mrg-1 was evident (Figure 2C–2K). These data indicate that the expression changes obtained from real-time RT-PCR are due to the ectopic expression changes in the soma. It is important to note that the disappearance of PGL-1 staining in somatic cells of mep-1(lf) animals upon inactivation of isw-1, mes-4, and mrg-1 was in accordance with the disappearance of germline cell–like somatic cells (unpublished data). This observation indicates that the soma to germline transformation associated with the mep-1(lf) mutant depends on activities of these SynMuv suppressors.
In addition, we immunostained three other SynMuv B mutants, lin-9(n112), lin-15B(n744), and lin-35(n7450), each treated with or without RNAi of isw-1, mes-4, and mrg-1. We observed ectopic expression of PGL-1 in somatic cells of all three SynMuv B mutants and the suppression of ectopic expression when animals were treated with RNAi of isw-1, mes-4, and mrg-1 (unpublished data). These results are consistent with a recent report that the ectopic expression of pgl-1 in somatic cells in a number of SynMuv B mutants and can be suppressed by mes-4(RNAi) [24]. The data here suggest that the multiple SynMuv suppressor genes are required for ectopic expression of pgl-1 in the soma, which is normally repressed by a number of SynMuv B genes, including mep-1, lin-35/Rb, lin-9, lin-15B, etc.
SynMuv Suppressor and SynMuv B Genes Play Opposite Roles in RNA Interference
During the genome-wide RNAi screen, we noticed that lin-15AB(n765) enhanced the effect of RNAi for many genes. To find out whether other SynMuv mutants are also more sensitive to RNAi than WT animals, we assayed a number of SynMuv A and B mutants by feeding RNAi of five genes, lin-1, hmr-1, dpy-13, rol-3, and unc-33, which had been previously used to test the efficiency of RNAi for the rrf-3 mutant [59]. Except for lin-36(n766), SynMuv B mutants lin-9(n112), lin-15B(n744), lin-35(n745), lin-37(n758), and tam-1(cc567) displayed an increase in RNAi sensitivity (Table S2). In particular, the lin-9(n112) mutant displayed the strongest RNAi-sensitizing effect similar to that of the rrf-3(pk1426) and eri-1(mg366) mutants, which are known as the RNAi-sensitive mutants [59,60]. In contrast, we found that the SynMuv A mutants lin-8(n111) and lin-38(n751) displayed no change in RNAi sensitivity (Table S2). These findings agree with a recent report that a number of SynMuv B genes, including lin-35/Rb, play a negative role in RNAi [24].
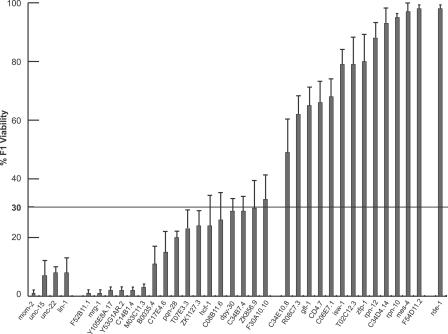
Among the 32 SynMuv suppressor genes we identified, six of these (mes-4, gfl-1, zfp-1, pqn-28, M03C11.3, and ZK1127.3) have previously been shown to play a positive role in RNAi [50,61]. Recently, these genes have been independently identified as SynMuv suppressors using a different approach [24]. To find out whether other SynMuv suppressor genes are also involved in the RNAi pathway, we co-injected WT animals with double-stranded RNA (dsRNA) of each of the 32 SynMuv suppressor genes together with dsRNA of mom-2, a gene essential for viability [50,61]. The survival of progeny indicates that RNAi of the candidate gene enables animals to overcome the lethality of mom-2(RNAi). Worms co-injected with dsRNA of a positive control gene, rde-1, and dsRNA of mom-2 showed 98% of survival, indicating strong protection from mom-2 dsRNA induced lethality by rde-1(RNAi). In contrast, co-injection with dsRNAs of unc-15, unc-22, or lin-1 that are dispensable for RNAi or injection with mom-2 dsRNA alone resulted in a low percentage of survival (Figure 3). Co-injection of dsRNAs of 13 of the 32 SynMuv suppressor genes significantly rescued the lethality caused by injection of mom-2 dsRNA (Table 3 and Figure 3), suggesting that these genes are required for robust RNAi. Except for mes-4, gfl-1, and zfp-1, which have been reported earlier as RNAi factors [50,61], the roles of ten of the 13 genes in RNAi were not previously known. These results suggest that SynMuv genes and SynMuv suppressors play opposite roles in the RNAi process.
Figure 3. Identification of Genes Required for Robust RNAi.
WT (N2) young adult hermaphrodites were co-injected with 300 ng/μl of each candidate dsRNA and 75 ng/μl of mom-2 dsRNA. rde-1(RNAi) was used as a positive control, while unc-15(RNAi), unc-22(RNAi), and lin-1(RNAi) were used as negative controls. The horizontal line indicates the cut-off for viability (>30%) that was used to define the positive genes reported in Table 3. All positive genes displayed significant differences from the negative controls with p < 0.01 (t test). The positive candidates include three genes (gfl-1, mes-4, and zfp-1) that were previously identified by Dudley et al. using the same approach [50]. Error bars represent standard error.
Notably, three genes related to protein degradation (C34D4.14, rpn-10, and rpn-12) were determined to be involved in RNAi in the above assay (Table 3 and Figure 3). It suggests that RNAi mechanism involves protein degradation and/or chromatin remodeling mediated by 19S RPs and E3 ubiquitin-protein ligase.
pgl-1 has been shown to be required for germline RNAi [62] and is ectopically expressed in the soma of SynMuv B mutants ([16,24] and this work). We asked whether a pgl-1 null mutation, ct131 [63], could suppress the RNAi-sensitizing effect of SynMuv B mutants. Three double mutants, lin-35(n745); pgl-1(ct131), pgl-1(ct131); lin-15B(n744), and lin-9(n112); pgl-1(ct131), which were treated with RNAi of rol-3, still produced obvious roller progeny similar to the SynMuv B single mutants, indicating that pgl-1 had no effect on the RNAi-sensitizing effect associated with these three SynMuv B mutants. This result suggests that pgl-1 is not necessary for RNAi sensitivity of the SynMuv B mutants.
We further investigated if lin-35(n745) up-regulated the expression of any of 11 RNAi pathway genes (dcr-1, ego-1, mut-7, mut-14, mut-16, rde-1, rde-2, rde-3, rde-4, rha-1, and rrf-1). The mRNA levels of all 11 genes were compared between WT animals and the lin-35/Rb mutant by real-time RT-PCR. No significant changes were observed for any of the 11 transcripts in the lin-35/Rb mutant (Figure S1), suggesting that effects of SynMuv B mutations on RNAi sensitivity is rather specific on a certain, unknown component(s) of the RNAi machinery.
The Antagonistic Role of the SynMuv Suppressors and the SynMuv B Genes in Somatic Transgene Silencing Was Achieved through Transcription Regulation
A number of SynMuv B genes, including tam-1, lin-35, lin-9, lin-15B, lin-51, and lin-52, are known to be involved in the modulation of context-dependent transgene silencing in somatic cells [11]. lf of any of these SynMuv B genes results in the hypersilencing of numerous highly repeated transgenes in somatic cells. To assess whether this transgene silencing is a transcriptional or a posttranscriptional event, we performed real-time RT-PCR to investigate the levels of pre-mRNA and mRNA of a transgene ccIs4251 (myo-3::Ngfp-lacZ, myo-3::Mtgfp) in tam-1(cc567) using the method developed by Grishok et al. [64]. tam-1, a SynMuv B gene, encodes a RING finger/B-Box factor that has been shown to be involved in context-dependent gene silencing [11]. We observed that the levels of gfp/lacZ pre-mRNA and mature mRNA in tam-1 animals decreased by 3-fold and 10-fold, respectively, compared to those in WT animals (Figure 4G–4H). These data indicate that transgene silencing in the tam-1 mutant likely occurs at the pre-mRNA stage and may share the same mechanism as the previously described RNAi-induced transcriptional gene silencing [64].
Figure 4. Somatic Trangene Silencing by tam-1(lf) Is Desilenced by RNAi of the SynMuv Suppressor Genes and Dicer.
(A–B) myo-3 promoter driving-GFP reporters (myo-3::Ngfp-lacZ, myo-3::Mtgfp) displayed significant transgene silencing in the somatic tissues (mainly the muscles) in the tam-1(cc567) mutant (B), but not in the WT animals (A) [11].
(C–F) RNAi of the SynMuv suppressors, mes-4 (C), mrg-1 (D), and isw-1 (E) as well as a non-SynMuv suppressor gene, dcr-1 (F), restored GFP expression in the tam-1(cc567) mutant. dcr-1 encodes the C. elegans Dicer protein that is required for RNAi [81,82]. Bar: 100 μm.
(G–H) The relative levels of gfp/lacZ of a transgene ccIs4251 (myo-3::Ngfp-lacZ, myo-3::Mtgfp) [11] were measured by qRT-PCR in various genetic background indicated. The level of ama-1 mRNA, encoding an RNA PolII, was used as the internal reference. Mean values and ranges of the lacZ/ama-1 ratios based on three qRT-PCR trials are shown.
We next examined whether RNAi of the SynMuv suppressors could revert the somatic transgene silencing in the tam-1(cc567) mutant. Among the 32 SynMuv suppressor genes, we found that RNAi of mes-4, isw-1, mrg-1, zfp-1, and C06E7.1 prevented transgene silencing in somatic tissues in the tam-1(cc567) mutant (Figure 4A–4E and unpublished data). RNAi of dcr-1, which encodes the only dicer protein in C. elegans, was also found to be capable of suppressing the somatic transgene silencing in the tam-1 mutant (Figure 4F). To quantify the observed effects, we performed real-time RT-PCR to measure the pre-mRNA and mature mRNA levels of the transgene following injection RNAi of dcr-1, mes-4, isw-1, and mrg-1 in the tam-1 (cc567) mutant. The levels of both pre-mRNA and mRNA of the transgene, myo-3::gfp/lacZ, were significantly increased following RNAi of these four genes (Figure 4G–4H). These results suggest that these SynMuv suppressor genes are required for somatic transgene silencing in the tam-1 mutant and regulating transgene expression at the transcription level as the SynMuv B genes do.
SynMuv Suppressor Genes Are Required for Germline Transgene Silencing and May Function in Germline Maintenance
In C. elegans, multiple-copy transgenes are commonly silenced in the germline. It depends on the four mes genes (mes-2, mes-3, mes-4, and mes-6 [65]), his-24 [66], the SynMuv B gene hpl-2 [67], and a number of genes that function in the RNAi pathway and germline cosuppression [61,62].
We investigated whether the SynMuv suppressor genes are required for germline transgene silencing using the marker transgene let-858::GFP (PD7271), which is silenced in the germline but expressed in somatic tissues [65]. RNAi of eight of the 32 SynMuv suppressor genes resulted in desilencing of the transgenes in the germline (Table 3 and Figure 5A–5D), suggesting that they are required for inhibiting the expression of transgenes in the germline. These included six genes that have not been previously reported for such function: mrg-1, C08B11.6, F30A10.10, F52B11.1, F54D11.2, and K08F4.2. The molecular functions of the latter two genes are not known. However, as their genetic behaviors are more close to the potential chromatin remodeling genes, we propose that F54D11.2 and K08F4.2 may play a role related to chromatin remodeling.
Figure 5. The SynMuv Suppressor Genes Are Required for Germline Transgene Silencing in WT Animals and for the lag-2::gfp Ectopic Expression in lin-15B(n744).
(A–D) DIC (A) and (C) and GFP fluorescence (B) and (D) images of hermaphrodite germline from transgenic strain PD7271 that contains the multicopy let-858::gfp reporters [75]. Brackets indicate the region of germ cell nuclei. The transgene was silenced in WT, but desilenced in animals treated with mrg-1(RNAi). RNAi of seven other SynMuv suppressors also displayed a similar effect (Table 3). Bar: 10 μm.
(E–H) GFP fluorescence images of mid-L4 larvae carrying the lag-2::gfp transgene. (E) WT animals display a strong expression of the transgene in distal tip cells (arrowheads) and the vulva (asterisks). (F–G) a strong ectopic expression of the transgene in the intestine (arrows) is seen in two SynMuv mutants. (H) RNAi of isw-1 suppressed the ectopic expression of lag-2::gfp in the intestine of the lin-15B(n744) mutant, but not its expression in distal tip cells and vulval cells. RNAi of 14 other SynMuv suppressors also displayed a similar effect (Table 3). Bar: 100 μm.
It has been reported previously that mutations in the known germline silencing genes (the four mes genes, his-24, and hpl-2) cause sterile phenotypes and disrupt germline maintenance [65–67]. We observed that RNAi of the two new germline silencing genes, mrg-1 and F30A10.10, displayed a completely sterile progeny phenotype, suggesting that they might be required for germline maintenance. The sterility of mrg-1(RNAi) has been reported and shown to be the consequence of a defect in mitotic proliferation of primordial germ cells [68,69].
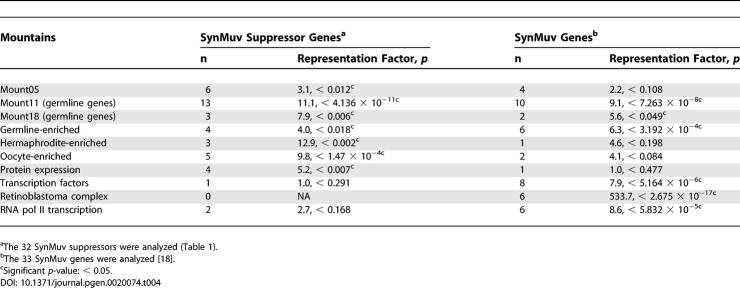
SynMuv Suppressor Genes Are Enriched in Germline Gene Clusters
The germline functions of the SynMuv suppressor genes are consistent with the distribution of most of these genes in germline gene clusters. Using TopoExpression Mountains devised by Kim et al. [70], we found that seven “mountains” were overrepresented (p < 0.05) by 32 SynMuv suppressor genes, whereas six “mountains” were overrepresented by the 33 SynMuv genes (Table 4). More strikingly, two germline-related mountains (mount 11 and mount 18), as well as the so-called “germline-enriched group,” were overrepresented by both the SynMuv genes and the SynMuv suppressor genes. Specifically, 16 SynMuv suppressor genes, dpy-30, gfl-1, isw-1, mes-4, mrg-1, pqn-28, rpn-12, C08B11.6, C14B1.4, C17E4.6, C34E10.8, C46A5.9, CD4.7, F30A10.10, K08F4.2, M03C11.3, R08C7.3, B0035.4, and ZK856.9, are in either mount 11 or the germline-enriched group [70]. RNAi of half of these genes, isw-1, mes-4, mrg-1, pqn-28, rpn-12, F30A10.10, M03C11.3, and R08C7.3, displayed a sterile phenotype.
Table 4.
Representations of TOPO Expression Mountains by the SynMuv Suppressor Genes and the SynMuv Genes
SynMuv Suppressor Genes Are Required for the Ectopic Expression of the lag-2 Gene in SynMuv B Mutants
lag-2 encodes a Delta_-_like ligand for the Notch receptors GLP-1 and LIN-12 that mediate multiple cell–cell interaction events in C. elegans [71]. In WT animals, lag-2 is expressed in distal tip cells of the gonad to signal GLP-1 and controls germline proliferation. It has been known that a number of SynMuv genes transcriptionally repress the lag-2::gfp expression in the intestinal and epidermal cells [18,25]. Consistent with these previous reports in which RNAi of the SynMuv genes was being used, we found that a genetic mutation in lin-15B produced a strong ectopic lag-2::gfp expression in the intestinal cells (Figure 5E–5H and Table 3). To investigate whether the SynMuv suppressor genes are required for the ectopic expression of lag-2 in the intestine of SynMuv B mutant, we performed RNAi of each of the 32 SynMuv suppressor genes on the lin-15B mutant carrying the lag-2::gfp transgene and scored for ectopic lag-2::gfp expression (Figure 5E–5H and Table 3). We observed that RNAi of 15 of the 32 SynMuv suppressors suppressed the ectopic reporter gene expression in the intestine. These data indicate that these SynMuv suppressors are required for the ectopic expression of lag-2 in the intestine and may function as transcription activators. These results are consistent with the hypothesis that both the SynMuv suppressor genes and the SynMuv B genes act antagonistically in transcription regulation of common target genes.
Discussion
Chromatin Remodeling Genes Play Dual Roles in Vulval Development, Germline-Soma Distinction, and RNAi
A number of SynMuv B proteins are homologous to yeast, Drosophila, and/or human proteins that have been implicated in histone modification, nucleosome remodeling, and transcription repression. Some SynMuv B proteins are homologs of components of the NuRD complex that is well known to play a major role in transcription repression [4,13–15,17]. Recently, the Drosophila Myb-MuvB (also known as dREAM) complex, which contains homologs of the C. elegans SynMuvB gene products (lin-35/Rb, hda-1, lin-52, lin-61, lin-9, efl-1, and DPL-1) has been isolated as a transcription repressor complex from embryonic extracts [19,20]. Thus, it has been proposed that chromatin remodeling plays a repressive role on inductive signaling in vuval development in C. elegans [7,15]. Our findings revealed that chromatin remodeling plays a dual role in vulval development, both repressive and active. Taking the biochemical characterizations of their mammalian and yeast counterparts into consideration, the SynMuv suppressor genes we identified have links to transcription activation. Histone acetylation carried out by the NuA4 complex and/or the subunits of TFIIIC, the incorporation of H2A.Z into the euchromatin carried out by the SWR1 complex, and the histone H3 K4 tri-methylation carried out by the COMPASS and ISW1 complexes, are generally associated with transcription activation. Therefore, we propose that the SynMuv suppressor genes compete with the SynMuv genes at the level of transcription regulation on target genes, promoting ectopic vulval induction in SynMuv mutants. The antagonistic roles of chromatin remodeling carried out by the SynMuv genes and the SynMuv suppressor genes are also involved in germline-soma distinction, RNAi, and other developmental events.
Two Different NuA4-Like Complexes May Exist in C. elegans
The NuA4 complex is a well characterized S. cerevisiae HAT complex that acetylates the N-terminus of nucleosomal histones H2A and H4 and functions as a transcription coactivator [31]. The components of the human NuA4 complex correspond to a combination of two distinct complexes in yeast, namely NuA4 and SWR1 [31]. In this study, five SynMuv suppressor genes that we identified (C34B7.4, gfl-1, mrg-1, Y105E8A.17, and ZK1127.3) encode proteins homologous to components of the yeast NuA4 complex (Tables 1 and S1). C34B7.4 encodes a MYST family HAT enzyme that is a key component of the NuA4 complex in other species. In addition to gfl-1, and Y105E8A.17, three other SynMuv suppressor genes, C08B11.6, C17E4.6, and CD4.7, encode proteins homologous to components of the yeast SWR1 complex (Tables 1 and S1). The human homologs of six of these eight genes, C17E4.6, C34B7.4, gfl-1, mrg-1, Y105E8A.17, and ZK1127.3, encode components of the human NuA4 complex. Furthermore, it has been shown that the MRG-1 and ZK1127.3 proteins directly bind to each other in the yeast two-hybrid assay [39], implicating that these two proteins may function in the same complex in C. elegans. In fact, the human ortholog of MRG-1 directly binds to MRGB, a human ortholog of ZK1127.3 [30]. Thus, these observations allow us to consider that there is a NuA4-like complex in C. elegans that plays a similar transcription activation role to the yeast and human NuA4 complexes. Our findings that C34B7.4, GFL-1, MRG-1, ZK1127.3, and C08B11.6 are required for the ectopic expression of lag-2 in the intestine of SynMuv B mutants (Table 3) further support the hypothesis that these proteins may act as transcription activators.
Intriguingly, it has recently been shown that strong lf mutations in three SynMuv C genes results in the Muv phenotype in SynMuv A or SynMuv B mutant backgrounds [7]. These three genes, epc-1, mys-1, and trr-1, also encode components of the NuA4 complex. MYS-1 may function as the MYST family HAT enzyme in a NuA4-like complex with EPC-1 and TRR-1, as suggested in a previous report [7]. The SynMuv phenotype indicates a repressive role of these genes in vulval induction. Consistent with this, epc-1, mys-1, and trr-1 have been shown to repress the ectopic expression of lag-2 in the intestine [18]. Therefore, the NuA4 complex containing these components appears to function differently from the above-proposed NuA4-like complex containing SynMuv suppressors that promote vulval induction. A repressive role of the NuA4 complex has also been observed in mammalian cells [72], suggesting that transcriptional repression by NuA4 complexes may be conserved between C. elegans and mammals. These results suggest that there may be two distinct NuA4-like complexes with opposing functions. An alternative explanation is that there is only one NuA4 complex with both positive and negative roles on transcription regulation in development. However, such a scenario is unlikely for the following reasons. In SynMuv A or SynMuv B mutants, RNAi of NuA4 components identified as SynMuv suppressors did not cause a Muv phenotype [7] (unpublished data), indicating that these genes do not have the same repressive roles as that of the SynMuv C genes. Reciprocally, RNAi of the three SynMuv C genes that are NuA4 components failed to suppress the SynMuv phenotype of lin-15AB(n765) (unpublished data), suggesting that these SynMuv C genes do not have the activation roles associated with the SynMuv suppressor genes. Biochemical purification and characterization of these complexes will elucidate whether there are two different NuA4-like complexes and what their actual functions are in C. elegans.
The Role of Chromatin Remodeling in Transgene Silencing and RNAi
A number of SynMuv B genes are required for repetitive transgene expression in somatic cells. In contrast, we found that dcr-1, an RNAi pathway gene, as well as four SynMuv suppressor genes (mes-4, isw-1, mrg-1, and zfp-1), are required for the somatic transgene silencing at the transcriptional level. Since SynMuv B proteins are likely transcription repressors and SynMuv suppressor proteins are likely transcription activators, somatic transgene expression may be inhibited by factors or functions that are targets of these two classes of genes with opposite roles.
It has been known that tandem repeats form heterochromatin to silence gene expression [73]. Recent studies have revealed that components of the RNAi machinery are associated with the formation of heterochromatin [74]. Kelly and colleagues [75] have shown that histone H3 lysine 9 (H3K9) di- and trimethylation, a modification associated with heterochromatin and recognized by the heterochromatin protein HP1, is found on extrachromosomal transgenic fragments in C. elegans. It is consistent that the C. elegans HP1 homolog, hpl-2, and RNAi pathway genes are required for both germline transgene silencing and somatic transgene silencing associated with RNAi transcriptional gene silencing and tam-1(lf) [64,67] (unpublished data). It suggests that heterochromatin formation on extrachromosomal transgenes might be the major cause for transgene silencing in C. elegans.
We propose that SynMuv B genes and SynMuv suppressors play opposite regulatory roles on repetitive transgene silencing through regulating heterochromatin formation. One simple explanation for the hypersilencing associated with SynMuv B mutants is that the SynMuv B genes repress the expression of target genes involved in heterochromatin formation, while the SynMuv suppressor genes activate the expression of these target genes. In addition, the phenomenon of RNAi enhancement associated with the SynMuv B mutations may also be explained by the up-regulation of target genes in RNAi pathway in the SynMuv B mutants. We have so far been unable to identify the potential target genes. However, our efforts excluded the possibility of hpl-2 and 11 RNAi pathway genes acting as the transcriptional targets of lin-35/Rb to regulate transgene silencing and RNAi, since the mRNA levels of these genes were not changed in lin-35(n745) mutants (Figure S1). To test such a hypothesis, an extensive genome-wide expression pattern analysis of SynMuv B and SynMuv suppressor genes may be necessary.
Roles of SIN3 and SAM Synthetase in Vulval Differentiation Opposing to that of SynMuvB Protein
Identification of pqn-28, the C. elegans ortholog of the yeast/human SIN3 protein, as a SynMuv suppressor was unexpected because SIN3 generally functions as a transcription corepressor together with a histone deacetylase (HDAC). hda-1, encoding a worm HDAC, has been shown to repress the ectopic vulval induction [13,25], while we found in this study that pqn-28 promotes ectopic vulval induction. Since there are several more genes in C. elegans genome that encode HDAC superfamily proteins, one simple explanation for this intriguing finding is that PQN-28 may interact with one or more of these HDAC proteins and function as a transcription activator. There have been genetic and genomic evidences that SIN3 may activate as well as repress transcription [49]. One alternative explanation is that PQN-28 may interact with transcription activators to regulate gene transcription independent of HDAC function in C. elegans.
Another interesting finding is that the gene encodes a SAM synthetase that also behaves as a SynMuv suppressor. In addition to suppressing the SynMuv phenotype, RNAi of this gene also rescued the larval lethality of mep-1(lf) and reverted the transgene silencing in the soma (Table 3). SAM can serve as a substrate for multiple histone methyltrasferases, including histone H3 lysine 9 (H3K9) and histone H3 lysine 4 (H3K4) methyltransferases. H3K9 methylation is known to be involved in transcription repression, while H3K4 methylation is known to be involved in transcription activation [28]. Therefore, it is possible that SAM synthetase is involved in both gene activation and repression. However, our genetic assay only revealed the SynMuv suppressor role that is likely involved transcription activation. One explanation is that the cellular concentration of SAM may be more critical to the H3K4 methylation than to the H3K9 methylation, with regard to its specific functions.
Functional Distinctions among Different SynMuv Suppressor Genes
All 32 candidate genes were identified by suppression of the SynMuv phenotype, which indicates a common function for these genes in vulval induction. Although we expect that many of them act on one or more common targets for the function, not all of the suppressors behave the same. For example, zfp-1(lf) and mes-4(lf) have different genetic interactions with let-60/Ras(gf) mutations, indicating that these two genes do not act on the same targets for vulval functions. The functional differences among these SynMuv suppressor genes were also indicated by the differential roles of several cellular processes examined. For example, we showed that isw-1 was required for RNAi and the ectopic expression of the pgl-1 and lag-2 genes, but was not essential for germline transgene silencing (Table 3). Three SynMuv suppressor genes in the protein degradation group (C34D4.14, rpn-10, and rpn-12) were determined to be involved in RNAi. However, RNAi of any of these three genes had no effect on rescuing the mep-1(lf) larval lethal phenotype, desilencing transgenes in the germline, or suppressing the ectopic expression of lag-2 in the intestine (Table 3). Overall, these results implicate that there are functional distinctions among different chromatin remodeling factors even though they share some common functions.
Conclusive Remarks
In this study, we identified and characterized a large number of potential chromatin remodeling genes that antagonize the functions of the SynMuv B pathways in several developmental and cellular processes. Our genetic analysis also implicated potential chromatin remodeling functions for several genes that do not reveal a connection to chromatin remodeling by structure alone. While each of these factors is implicated to play regulatory roles on many target genes, the study underscores the involvement of multiple chromatin remodeling complexes in regulating specific gene expression for respective functions. The precise mechanisms by which these chromatin-remodeling factors collaborate and coordinate to regulate gene expression and cellular functions remain to be investigated. Identifying target genes that are responsible for specific developmental or cellular functions, such as RNAi, vulval induction, and transgene silencing, would be the key to the success of further studies. As most of the SynMuv suppressor genes are conserved in mammals, genetic characterization of these genes in C. elegans should shed light on the role of the mammalian chromatin remodeling factors in development and related human diseases.
Materials and Methods
Genetic mutations and strains.
Information regarding the following mutations can be found at http://www.wormbase.org. LGI: mes-3(bn35), dpy-5(e61), lin-35(n745), and sDP2(l,f). LGII: mes-2(bn11), unc-4(e120), lin-8(n111), lin-38(n751), rrf-3(pk1426), and mnC1. LGIII: lin-36(n766), lin-37(n758), lin-9(n112), and zfp-1(ok554). LGIV: eri-1(mg366), mep-1(q660), pgl-1(ct131), dpy-20(e1282), let-60(n1046), mes-6(bn66), and nT1(IV;V). LGV: mes-4(bn23) and dpy-11(e224). LGX: lin-15A(n767), lin-15B(n374), lin-15B(n744), and lin-15AB(n765). nT1(qIs51) was used as dominant green balancer chromosomes. PD4251 (ccIs4251; dpy-20(e1282)), PD6249 (ccIs4251; tam-1(cc567), JK2868 (unc-119(ed3); qIs56(lag-2::gfp, unc-119(+)), and PD7271 (pha-1(e12123ts); ccEx7271) are described in previous reports [11,75–77]. The Bristol strain N2 was used as the WT strain.
Genome-wide RNAi screen.
For the genome-wide screen, RNAi clones from the RNAi library [29] were inoculated in 96-well plates with 200 μl of Luria broth medium containing 50 μg/ml ampicillin and 15 μg/ml tetracycline per well, and cultured for 16 h at 37 °C. These RNAi clones were transferred to a new 96-well plate with 200 μl of Luria broth medium supplemented with 50 μg/ml ampicillin per well and again cultured for 16 h at 37 °C. NGM plates (6-cm) supplemented with 0.4 mM IPTG and 50 μg/ml ampicillin were seeded with these cultures, and were incubated for 24 h at room temperature and then spotted with four to eight synchronized L1 larvae of lin-15AB(n765). The plates were incubated for 7 d at 19 °C and then were scored for the Muv phenotype. Three rounds of retest were performed to determine the final list of positive suppressors. For the retests, RNAi clones were inoculated into 5 ml of Luria broth medium containing 50 μg/ml ampicillin and were cultured for 16 h at 37 °C. Each bacterial culture (~400 μl) was dispensed onto NGM plates supplemented with 0.4 mM IPTG and 50 μg/ml ampicillin. For all clones that suppressed the Muv phenotype of lin-15AB(n765), the dsRNA-encoding DNA inserts were sequenced. Several clones were found to contain genes that were different from those listed in the library. All RNAi screen were done at 19 °C except those specified.
Other RNAi experiments.
An RNAi construct for mes-2 gene is not present in the available RNAi library [29]. To make the RNAi feeding construct for mes-2, 1.5 kb of mes-2 genomic DNA was PCR-amplified from genomic DNA and subsequently cloned into pPD129.36 (gift from A. Fire). The construct was transformed into Escherichia coli HT115(DE3). Injection RNAi was carried out as described previously [76]. Worms were injected with 200 ng/ml dsRNA of the candidate gene and were allowed to recover for 12–16 h before single worms were placed for egg-laying.
The co-injection experiments were conducted at 20 °C, as described previously [50,61]. DNA templates were PCR-amplified from plasmids corresponding to each RNAi clone with T7 promoters. dsRNA were synthesized in a single reaction using T7 polymerase-based transcription kit (Ambion Megascript Kit; Ambion, Austin, Texas, United States). Concentrations of dsRNAs were determined using a spectrophotometer. The quality and size of the dsRNAs were assessed by gel eletrophoresis. WT young hermaphrodites were co-injected with the mixture of 300 ng/μl of the dsRNA of candidate gene and 75 ng/μl of mom-2 dsRNA. Worms were allowed to recover for 12–16 h before single worms per plate were placed for consecutive 48-h egg laying. Then, the total number of eggs was scored upon removal of P0 worms from the plates. The viability of the progeny were assayed after 24 h and expressed as the (number of hatched larvae) / (total number of eggs).
Characterization of zfp-1(ok554).
The zfp-1(ok554) lesion was determined by sequencing genomic DNA and found to be identical to that reported by the C. elegans Gene Knockout Consortium (http://celeganskoconsortium.omrf.org) for the corresponding gene, F54F2.2. Primers outside of the deletion (attcaatcagcctgtggagg and tgctgctgctttctcgttta) and a primer inside of the deletion (gagggtccgcaagcatatgc) were used to confirm there is no additional copy of zfp-1 in the genome and to distinguish the homozygotes from the heterozygotes when building double or triple mutants with mutations in other genes.
Vulval induction assay.
VPC (P3p-P8p) cell fates in L4 hermaphrodites were scored under Nomarski optics as described previously [78]. Scores of 1, 0.5, and 0 were assigned to cells that fully, partially, or did not adopt vulval cell fates, respectively. In WT worms, three VPCs (P5.p, P6.p, and P7.p) fully adopted the vulval fate and the other three VPCs fused with epidermal syncytium, giving a score of 3.0. A score of more than 3.0 indicates ectopic VPC induction.
Quantitative real-time RT-PCR (qRT-PCR).
Total RNA was isolated from worms using TRI Reagent (MRC, Inc., Cincinnati, Ohio, United States) followed by purification and DNase I treatment. Random-primed cDNA was prepared using Superscript III reverse transcriptase (Invitrogen, Carlsbad, California, United States). Reactions were run in triplicate with 2XSYBR Green Jumpstart Taq ReadyMix (Sigma, St. Louis, Missouri, United States) on the Rotor Gene 3000 (Corbett Research, Cambridge, United Kingdom). Relative fold changes were calculated using the 2−ΔΔCt method. For pgl-1, we picked ~200 non-green L1/L2 stage worms (mep-1(q660) homozygote) from mep-1(q660)/nT1(qIs51) mothers treated with or without RNAi of the SynMuv suppressor genes. Primers for real-time PCR were designed to span an exon-exon junction. The rpl-26 gene encoding the ribosome large subunit was used as an internal control for data normalization. For measuring the expression of gfp/lacZ, we picked ~200 L4-young adult stage worms from the strains PD4251, PD6249, and the progeny of the PD6249 injected with dsRNA of dcr-1, isw-1, mes-4, or mrg-1. RNA polymerase II large subunit AMA-1 gene was used as an internal control for data normalization. Primers for ama-1 mRNA, mature mRNA of gfp/lacZ, and pre-mRNA of gfp/lacZ, were designed by Grishok et al. [64]. For measuring the mRNA levels of hpl-2 and 11 RNAi pathway genes (see Figure S1) in lin-35(n745), synchronized L1 larvae are used. The ama-1 was used as the internal control for data normlization. The data shown are representative of three experiments with independent worm growths and RNA isolations.
Assay for the early larval arrest of mep-1(q660) rescue.
The mep-1(q660) mutant is temperature sensitive: at 25 °C, nearly 100% of mep-1 homozygotes derived from a heterozygous mother are arrested as young larvae (L1 or L2) [79]. The RNAi plates were seeded with 12–18 synchronized L1 larvae of mep-1(q660) IV/nT1(qIs51) and cultured at 25 °C. qIs51 is a genomic insertion containing three markers: myo-2::GFP expressed in the pharynx throughout development, pes-10::GFP expressed in the embryo, and F22B7.9::GFP expressed in the intestine [79]. Any mep-1(q660) homozygote hermaphrodite (nongreen animals) in the progeny were transferred onto the corresponding new RNAi plates. The larval progeny were scored four days later for larval stages according to the cell divisions of three VPCs (P5.p–P7.p) and the body size under Nomarski optics.
Assay for somatic transgene silencing.
The restoration of green fluorescent protein (GFP) fluorescence in the strain PD6249 (ccIs4251; tam-1(cc567)) [11] was assayed for the GFP expression intensity by fluorescent microscopy with Nomarski optics, following feeding RNAi of all the candidates. The integrated array (ccIs4251) contains three markers: myo-3 promoter driving mitochondrial targeted GFP, myo-3 promoter driving a nuclear-targeted GFP/LacZ fusion gene, and a dpy-20(+) gene [76]. The RNAi plates were seeded with 12–16 synchronized L1 larvae and cultured for 6 days at 20 °C. The GFP expression in the F1 progeny at L4 to young adult stages was examined.
Assay for germline transgene silencing.
Strain PD7271 (gift from W. Kelly) carries a multicopy extrachromosomal transgene array of let-858::GFP, of which the expression is silenced in the germline, but is expressed in most somatic lineages [75]. Synchronized L1 larvae of the strain PD7271 (12–18 larvae) were placed onto the candidate RNAi plates. The restoration of GFP expression in the germline was assayed in adults of the F1 progeny.
Assay for lag-2::gfp ectopic expression in the intestine.
The ectopic expression of lag-2::gfp in lin-15B(n744) was assayed as described previously [25]. In brief, mid-L4 worms were mounted on 3% agar pads and were scored for the ectopic expression in the intestine cells under fluorescence microscope with Nomarski optics.
Immunofluorescence staining.
PGL-1 antibody staining was conducted using L1 larvae that were fixed as described by Finney and Ruvkun [80]. Affinity purified polyclonal anti-PGL-1 antibody (gift from S. Strome) was used at 1:2000 dilution.
Bioinformatics.
Analysis of the distributions of SynMuv and SynMuv suppressor genes in the TOPO expression mountains was performed using a program in http://workhorse.stanford.edu/cgi-bin/gl/gl_mod.cgi [70].
Supporting Information
Figure S1. Quantification of Relative mRNA Levels of hpl-2, and 11 RNAi Pathway Genes in lin-35(n745) Mutant.
rpl-26 was used as the internal reference. Mean values and ranges of the pgl-1/rpl-26 ratios based on three qRT-PCR trials are shown.
(655 KB EPS)
Table S1. The 32 SynMuv Suppressor Genes and Their Homologs in Other Species.
(38 KB DOC)
Table S2. The Muv Suppression Observed in Two SynMuv Mutants upon RNAi of Each of the 32 Candidate Genes.
(38 KB DOC)
Table S3. The SynMuv B Mutants Enhanced Effect of Feeding RNAi.
(25 KB DOC)
Acknowledgments
We are indebted to Keiko Hirono and Matt Sieber for their assistance in this project and Erik Anderson and Bob Horvitz for communicating unpublished results. We thank Susan Strome for the anti-PGL antibody and the mes genes mutants, Bob Barstead of the C. elegans Knockout Consortium for zfp-1(ok554) allele, Bill Kelly for strain PD7271, Judith Kimble for mep-1(q660) and JK2868, Andy Fire for tam-1(cc567) and strain PD4251, C. elegans Genetic Center for a number of strains, Wade Johnson for help on qPCR analysis, Lois Edgar, Yo Suzuki, Morgan Tucker, Wade Johnson, Jennifer Blanchette and Aileen Sewell for comments on the manuscript, and members of the Han/Wood laboratories for discussion during the course of this study.
Abbreviations
dsRNA
double-stranded RNA
gf
gain of function
GFP
green fluorescent protein
HAT
histone acetyltransferase
HDAC
histone deacetylase complex
L1/L2
first or second larval
lf
loss of function
LZ
leucine zipper motif
MAPK
mitogen-activated protein kinase
NuA4
nucleosomal acetyltransferase of histone H4
NuRD
nucleosome-remodeling deacetylase
OM
octapeptide motif
PHD
plant homeodomain
RNAi
RNA interference
RP
regulatory particle
RTK
receptor tyrosine kinase
SAM
S-adenosylmethionine synthetase
SynMuv
synthetic multivulva
VPC
vulval precursor cell
WT
wild type
Footnotes
Author contributions. MC and MH conceived and designed the experiments. MC performed the experiments and analyzed the data. EBK provided assistance on RNAi screen. MC and MH wrote the paper.
Competing interests. The authors have declared that no competing interests exist.
Funding. This work was supported by a grant from the NIH (GM47869) and by HHMI.
References
- Greenwald I. Development of the vulva. In: Riddle DL, Blumental T, Meyer BJ, Priess JR, editors. C. elegans II. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 519–541. [PubMed] [Google Scholar]
- Sternberg P. Vulval development. 2005. Available: http://www.wormbook.org/chapters/www_vulvaldev/vulvaldev.html. Accessed 19 April 2006. [DOI] [PMC free article] [PubMed]
- Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans . Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans . Nature. 1987;326:259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- Clark SG, Lu X, Horvitz HR. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Tzou P, Sternberg PW. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison EM, Harrison MM, Walhout AJ, Vidal M, Horvitz B. lin-8, which antagonizes C. elegans Ras-mediated vulval induction, encodes a novel nuclear protein that interacts with the LIN-35 Rb protein. Genetics. 2005;171:1017–1031. doi: 10.1534/genetics.104.034173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Liu J, Kostas SA, Chang C, Sternberg PW, et al. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans . Genes Dev. 1999;13:2958–2970. doi: 10.1101/gad.13.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Greenwald I. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics. 2000;155:1127–1137. doi: 10.1093/genetics/155.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F, Ahringer J. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans . Curr Biol. 2000;10:223–226. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, et al. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol Cell. 2001;7:461–473. doi: 10.1016/s1097-2765(01)00194-0. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, et al. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans . Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Ceol CJ, Schwartz HT, Horvitz HR. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans . Genetics. 2003;164:135–151. doi: 10.1093/genetics/164.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans . EMBO J. 2005;24:2613–2623. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS. synMuv verite—Myb comes into focus. Genes Dev. 2004;18:2837–2844. doi: 10.1101/gad.1274804. [DOI] [PubMed] [Google Scholar]
- Boxem M, van den Heuvel S. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol. 2002;12:906–911. doi: 10.1016/s0960-9822(02)00844-8. [DOI] [PubMed] [Google Scholar]
- Fay DS, Keenan S, Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–517. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, et al. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol Cell Biol. 2002;22:3024–3034. doi: 10.1128/MCB.22.9.3024-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Large E, Han M, Darland M. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans . Development. 2003;130:3319–3330. doi: 10.1242/dev.00561. [DOI] [PubMed] [Google Scholar]
- Cui M, Fay DS, Han M. lin-35/Rb cooperates with the SWI/SNF complex to control Caenorhabditis elegans larval development. Genetics. 2004;167:1177–1185. doi: 10.1534/genetics.103.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling complexes: Strength in diversity, precision through specialization. Curr Opin Genet Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:e131. doi: 10.1371/journal.pbio.0020131. DOI: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol Cell Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, et al. A map of the interactome network of the metazoan C. elegans . Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, et al. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Iwasaki H, Inoue H, Takahashi K. Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference. Biol Chem. 2002;383:1263–1266. doi: 10.1515/BC.2002.140. [DOI] [PubMed] [Google Scholar]
- Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, et al. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu TK, Wang Z, Roeder RG. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YJ, Kundu TK, Wang Z, Kovelman R, Roeder RG. The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol Cell Biol. 1999;19:7697–7704. doi: 10.1128/mcb.19.11.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Debernardi S, Bassini A, Jones LK, Chaplin T, Linder B, et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99:275–281. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: A flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Dudley NR, Labbe JC, Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc Natl Acad Sci U S A. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, et al. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans . Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y, Bender L, Wang W, Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans . Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin C, Holdeman R, Strome S. The phenotype of mes-2, mes-3, mes-4, and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics. 1998;148:167–185. doi: 10.1093/genetics/148.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely PM, Wood WB. Genetic analysis of X-chromosome dosage compensation in Caenorhabditis elegans . Genetics. 1987;117:25–41. doi: 10.1093/genetics/117.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DR, Meyer BJ. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 1994;137:999–1018. doi: 10.1093/genetics/137.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Fan Y, Strome S. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development. 1998;125:2469–2478. doi: 10.1242/dev.125.13.2469. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Wang J, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans . Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, et al. Functional genomic analysis of RNA interference in C. elegans . Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, et al. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans . Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans . Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans . Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik MA, Schulze E. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans . Development. 2001;128:1069–1080. doi: 10.1242/dev.128.7.1069. [DOI] [PubMed] [Google Scholar]
- Couteau F, Guerry F, Muller F, Palladino F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 2002;3:235–241. doi: 10.1093/embo-reports/kvf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Takasaki T, Nakajima N, Kawano T, Shimura Y, et al. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans . Mech Dev. 2002;114:61–69. doi: 10.1016/s0925-4773(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Olgun A, Aleksenko T, Pereira-Smith OM, Vassilatis DK. Functional analysis of MRG-1: The ortholog of human MRG15 in Caenorhabditis elegans . J Gerontol A Biol Sci Med Sci. 2005;60:543–548. doi: 10.1093/gerona/60.5.543. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, et al. A gene expression map for Caenorhabditis elegans . Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans . Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J Biol Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- Martienssen R, Lippman Z, May B, Ronemus M, Vaughn M. Transposons, tandem repeats, and the silencing of imprinted genes. Cold Spring Harb Symp Quant Biol. 2004;69:371–379. doi: 10.1101/sqb.2004.69.371. [DOI] [PubMed] [Google Scholar]
- Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee MH, Kim SK, et al. X-chromosome silencing in the germline of C. elegans . Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Schvarzstein M, Morphy KM, Blelloch R, Spence AM, et al. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans . Development. 2004;131:4333–4343. doi: 10.1242/dev.01288. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. Pattern formation during vulval development in C. elegans . Cell. 1986;44:761–772. doi: 10.1016/0092-8674(86)90842-1. [DOI] [PubMed] [Google Scholar]
- Belfiore M, Mathies LD, Pugnale P, Moulder G, Barstead R, et al. The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3′ untranslated region in Caenorhabditis elegans . RNA. 2002;8:725–739. doi: 10.1017/s1355838202028595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans . Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans . Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Quantification of Relative mRNA Levels of hpl-2, and 11 RNAi Pathway Genes in lin-35(n745) Mutant.
rpl-26 was used as the internal reference. Mean values and ranges of the pgl-1/rpl-26 ratios based on three qRT-PCR trials are shown.
(655 KB EPS)
Table S1. The 32 SynMuv Suppressor Genes and Their Homologs in Other Species.
(38 KB DOC)
Table S2. The Muv Suppression Observed in Two SynMuv Mutants upon RNAi of Each of the 32 Candidate Genes.
(38 KB DOC)
Table S3. The SynMuv B Mutants Enhanced Effect of Feeding RNAi.
(25 KB DOC)