Genetic Variation in the CCL18-CCL3-CCL4 Chemokine Gene Cluster Influences HIV Type 1 Transmission and AIDS Disease Progression (original) (raw)
Abstract
CCL3 (MIP-1α), CCL4 (MIP-1β), and CCL18 (DC-CK1/PARC/AMAC-1) are potent chemoattractants produced by macrophages, natural killer cells, fibroblasts, mast cells, CD4+ T cells, and CD8+ T cells. CCL3 and CCL4 are natural ligands for the primary human immunodeficiency virus type 1 (HIV-1) coreceptor CCR5 and are also known to activate and enhance the cytotoxicity of natural killer cells. Genomic DNAs from >3,000 participants enrolled in five United States–based natural-history cohorts with acquired immunodeficiency syndrome (AIDS) were genotyped for 21 single-nucleotide polymorphisms (SNPs) in a 47-kb interval on chromosome 17q12 containing the genes CCL3, CCL4, and CCL18. All 21 SNPs were polymorphic in African Americans (AAs), whereas 7 of the 21 had minor-allele frequencies <0.01 in European Americans (EAs). Substantial linkage disequilibrium was observed in a 37-kb interval containing 17 SNPs where many pairwise _D_′ values exceeded 0.70 in both racial groups, but particularly in EAs. Four and three haplotype blocks were observed in AAs and EAs, respectively. Blocks were strongly correlated with each other, and common haplotype diversity within blocks was limited. Two significant associations are reported that replicate an earlier study. First, among AA members of the AIDS Link to the Intravenous Experience cohort of injection drug users, frequencies of three correlated SNPs covering 2,231 bp in CCL3 were significantly elevated among highly exposed, persistently HIV-1–uninfected individuals compared with HIV-1–infected seroconvertors (_P_=.02–.03). Second, seven highly correlated SNPs spanning 36 kb and containing all three genes were significantly associated with more-rapid disease progression among EAs enrolled in the Multicenter AIDS Cohort Study cohort (_P_=.01–.02). These results reiterate the importance of chemokine gene variation in HIV-1/AIDS pathogenesis and emphasize that localized linkage disequilibrium makes the identification of causal mutations difficult.
The heterogeneity among individuals in their susceptibility to HIV type 1 (HIV-1) infection and the variable rates of disease progression among HIV-1–infected individuals are the results of many factors, including viral virulence, environmental conditions, age, and host genetics. Host antiviral immunity can act at different levels, including the inhibition of viral attachment, entry, or assembly, and may include various responses to viral infection, such as cytotoxic T-cell response, the generation of noncytolytic factors such as cytokines, and antibody production.1
AIDS restriction genes have been defined in which allelic variation has been shown to influence infection or disease progression. These include genes encoding chemokine receptors, chemokines, human leukocyte antigen, and cytokines and genes involved with innate or acquired immunity.2 Chemokines became prominent in HIV-1 research when the culture supernatants of CD8+ T cells demonstrated HIV-1 suppressor activity,3,4 and this activity was shown to be attributable to three CC chemokines—namely, RANTES (CCL5), MIP-1α (CCL3), and MIP-1β (CCL4).5 Chemokines are a group of low-molecular-weight, basic, heparin-binding proteins that mediate an array of homeostatic and inflammatory processes. The amino acid sequences contain four conserved cysteine residues, and these molecules signal through receptors belonging to the G-protein–coupled seven-transmembrane domain receptor family.6 Over 50 chemokine genes are assigned to two major subgroups: the α (or C×C) and the β (or CC) subgroups. Sixteen members of the CC gene family have been assigned to chromosome 17q12, and many of these genes exhibit extreme sequence conservation, suggesting recent duplication and selection for conserved function in their evolutionary histories.7,8
The CCL3 and CCL4 chemokine genes, along with CCL18, are located within 40 kb of each other. SNPs in the CCL3 gene were evaluated in two previous HIV-1 epidemiological studies. Four CCL3 SNPs were not significantly associated with HIV-1 infection in a Japanese population,9 whereas a haplotype containing two CCL3 intron 1 SNPs was associated with resistance to HIV-1 acquisition in African Americans (AAs) and with accelerated progression to AIDS in European Americans (EAs).10 Given the obvious role played by CCL3 and CCL4 in HIV-1/AIDS pathogenesis and the availability of high-density SNP maps for population-based association analyses, we examined, in the present study, variation in 21 SNPs spanning 47 kb of genomic DNA containing the CCL18, CCL3, and CCL4 genes in >3,000 participants enrolled in five United States–based longitudinal HIV-1/AIDS cohorts. Two primary questions were addressed in the study. First, what is the extent and nature of SNP and haplotype variation in and around these three genes in two racial groups? Second, do genetic variants exist in these genes that influence HIV-1 infection and/or AIDS disease progression?
Material and Methods
Participants and Clinical Outcomes
Participants were enrolled in five longitudinal cohorts. The AIDS Link to the Intravenous Experience (ALIVE) is a community-based cohort of adult injection drug users in Baltimore.11 The racial distribution is 92.4% AA and 7.6% EA. The Hemophilia Growth and Development Study (HGDS) is a multicenter prospective study that enrolled children with hemophilia.12 The cohort consists of 126 HIV-1–uninfected and 207 HIV-1–infected children who were exposed to HIV-1 through blood products between 1982 and 1983. The racial distribution is 72% EA, 15% Hispanic, and 11% AA. The Multicenter Hemophiliac Cohort (MHCS) is a prospective study that enrolled persons with hemophilia.13 The racial distribution is 90% EA, 6% AA, and 3% Hispanic. The Multicenter AIDS Cohort Study (MACS) is a longitudinal study of men who have sex with men (MSM) from four U.S. cities: Chicago, Baltimore, Pittsburgh, and Los Angeles.14,15 The racial distribution is 83.3% EA, 10% AA, and 5% Hispanic. The San Francisco City Clinic Study (SFCC) is a cohort of MSM originally enrolled in a hepatitis B study in 1978–1980.16 The cohort consists of 211 individuals, 203 of whom are EA. The majority of subjects were enrolled into the cohorts during the following years: ALIVE, 1988–1989; MACS, 1984–1985; MHCS, 1982–1985; and SFCC, 1978–1980.
All individuals used for association analyses were (1) uninfected individuals who have undocumented levels of exposure but who belong to an HIV-1 risk group; (2) high-risk, exposed, uninfected (HREU) individuals with documented high risk for HIV-1 exposure; (3) HIV-1–positive seroconvertors, with seroconversion dates estimated as the midpoint between the last visit with seronegative test results and the first visit with seropositive test results; or (4) seroprevalent individuals who were infected with HIV-1 at study enrollment. The HREU participants were exposed to HIV-1 by receptive anal intercourse with multiple partners,15,16 by transfusion with nonheated units of factor VIII or IX between 1982 and 1983,17 or by injection drug use.11 A concise summary of these cohorts is available.18 Over 3,000 individuals were genotyped, and the data were used to estimate allele frequencies, but genotypes from only 1,326 individuals were used in disease association analyses: 449 AAs (159 seronegative individuals and 290 seroconvertors) and 877 EAs (216 seronegative individuals and 661 seroconvertors). Since antiretroviral treatment history was unavailable for a majority of subjects, participants were censored for highly active antiretroviral therapy (HAART), instead of the effects of HAART being adjusted for in the models.19 The censoring date was the earlier of two dates: the last recorded follow-up visit or December 31, 1995, for the MACS, MHCS, HGDS, and SFCC cohorts and the last recorded visit or July 31, 1997, for the ALIVE cohort. A later censoring date was used for the ALIVE cohort because administration of HAART was delayed.20 The median follow-up times were 5.1, 7.3, 10.7, and 13.4 years for the ALIVE, MACS, MHCS, and SFCC cohorts, respectively.
Three separate end points reflecting advancing AIDS pathogenesis were considered for seroconvertors: (1) the Centers for Disease Control and Prevention (CDC) definition of AIDS in 1993 (AIDS-93): HIV-1 infection plus a CD4+ T cell count of <200/μl or AIDS-defining conditions21; (2) the CDC definition of AIDS in 1987 (AIDS-87): HIV-1 infection plus AIDS-defining illness22; and (3) AIDS-related death during follow-up of an HIV-1–infected patient.
SNP Discovery and Genotyping
The CCL18, CCL4, and CCL3 genes are located within 40 kb of one another at chromosome 17q12 (fig. 1). Each gene contains three exons and two introns.24 SNPs were identified by using the SSCP technique, by resequencing the CCL3 and CCL4 genes, or by surveying the dbSNP database (build 110). For SSCP or resequencing, 12 different genomic regions covering >3,000 bp were analyzed on DNAs from 36 unrelated EAs and 36 unrelated AAs presenting with various HIV-1/AIDS phenotypes. A total of 21 SNPs were genotyped (table 1) by use of either PCR-RFLP or TaqMan allelic discrimination (Applied Biosystems) techniques (table 2).25
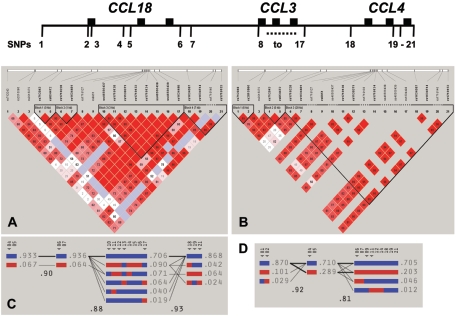
Figure 1. .
Molecular map of the CCL18, CCL3, and CCL4 genes. Each gene has three exons, depicted as blackened boxes, and SNPs are numbered 1–21. The distance between SNPs 1 and 21 is 47,637 bp. Below the map are pairwise _D_′ plots and haplotype blocks obtained from HAPLOVIEW for 420 AAs (A) and 972 EAs (B) (in panel B, seven tracks are blank because of MAFs <0.01). Dark-red squares signify high _D_′ values, light-blue squares indicate high _D_′ values with low LOD scores, and light-red and white squares indicate low _D_′ values. Block boundaries are based on the 95% CIs for _D_′.23 Panels C and D show the structure of haplotype blocks found in AAs and EAs, respectively. Haplotype-tagging SNPs identified by HAPLOVIEW are indicated by arrowheads. Numbers below the blocks are interblock, multiallelic _D_′ values.
Table 1. .
MAFs and Gene Positions for 21 SNPs in the CCL18, CCL3, and CCL4 Chemokine Genes
| MAFa | ||||||
|---|---|---|---|---|---|---|
| SNP | dbSNP | AA | EA | Location | Position | Inter-SNP Distanceb(bp) |
| 1 | rs712040 | .497 | .108 | CCL18 promoter | −6111 C/T | … |
| 2 | rs2015086 | .384 | .13 | CCL18 promoter | −86 C/T | 6,025 |
| 3 | rs2015070 | .039 | .087 | CCL18 intron 1 | +81 A/G | 166 |
| 4 | rs712043 | .058 | .302 | CCL18 intron 1 | +4965 C/T | 4,884 |
| 5 | rs854472 | .062 | .304 | CCL18 intron 1 | +5906 A/G | 941 |
| 6 | rs1719220 | .055 | .277 | CCL18 3′ UTR | +12575 A/G | 6,669 |
| 7 | rs1634481 | .055 | .271 | CCL3 3′ UTR | +11730 A/G | 1,398 |
| 8 | rs1719127 | .121 | .004 | CCL3 3′ UTR | +1829 A/G | 9,901 |
| 9 | rs8951 | .081 | .218 | CCL3 exon 3 | +1685 C/T | 144 |
| 10 | ss46566437 | .083 | .004 | CCL3 exon 3 | +1342 G/T (E79D) | 343 |
| 11 | rs1719130 | .194 | .24 | CCL3 intron 2 | +1159 C/T | 183 |
| 12 | rs1130371 | .154 | .234 | CCL3 exon 2 | +868 C/T (P60P) | 291 |
| 13 | rs1719133 | .065 | .005 | CCL3 intron 1 | +740 A/G | 128 |
| 14 | rs1719134 | .151 | .24 | CCL3 intron 1 | +459 A/G | 281 |
| 15 | ss46566438 | .083 | .005 | CCL3 intron 1 | +113 C/T | 346 |
| 16 | ss46566439 | .08 | .004 | CCL3 promoter | −891 T/C | 1,002 |
| 17 | rs1634498 | .077 | .003 | CCL3 promoter | −2021 C/T | 1,131 |
| 18 | rs1634507 | .113 | .237 | CCL4 promoter | −5725 A/C | 6,149 |
| 19 | rs1719144 | .054 | .222 | CCL4 intron 1 | +104 A/T | 5,829 |
| 20 | rs1719146 | .032 | 0 | CCL4 exon 2 | +663 C/T (T39T) | 558 |
| 21 | rs1719153 | .077 | .233 | CCL4 3′ UTR | +1931 A/T | 1,268 |
Table 2. .
Primers and Probes and the Restriction Enzymes Used in Genotyping Assays
| SNP | dbSNP | PCR Primer(s) | TaqMan Probes or Restriction Enzyme |
|---|---|---|---|
| 1 | rs712040 | ABI Assays-on-Demand, hCV2555159 | ABI Assays-on-Demand |
| 2 | rs2015086 | ABI Assays-on-Demand, hCV2555168 | ABI Assays-on-Demand |
| 3 | rs2015070 | ABI Assays-on-Demand, hCV7449055 | ABI Assays-on-Demand |
| 4 | rs712043 | ABI Assays-on-Demand, hCV2262137 | ABI Assays-on-Demand |
| 5 | rs854472 | ABI Assays-on-Demand, hCV7449039 | ABI Assays-on-Demand |
| 6 | rs1719220 | ABI Assays-on-Demand, hCV2555178 | ABI Assays-on-Demand |
| 7 | rs1634481 | ABI Assays-on-Demand, hCV2555183 | ABI Assays-on-Demand |
| 8 | rs1719127 | 5′-GGGAAATAATAAAGATGCTCTTTTAAAA-3′;5′-TGCCTATGATTCCTCTTAACC-3′ | _Bst_NI |
| 9 | rs8951 | 5′-CCCTTCCCTCACACCGC-3′;5′-AACTCAATACTGGTTTACCTTTTAAAAGAG-3′ | 5′-CCTACACAGGCTGATGACAGCCACT-3′;5′-CCTACACAGGCCGATGACAGCC-3′ |
| 10 | ss46566437 | 5′-ACAGCTTCCTAACCAAGCGAA-3′;5′-AGCTCCAGGTCGCTGACA-3′ | _Bsl_I |
| 11 | rs1719130 | 5′-GGCCACCCCTACTGAGTCAC-3′;5′-TCAGGGCTTGCTCCTCTTTC-3′ | 5′-AAGCTCTCTAGACAGAGATAGGCAGGG-3′;5′-AGCTCTCTAGACGGAGATAGGCAGG-3′ |
| 12 | rs1130371 | 5′-AATTTCATAGCTGACTACTTTGAGACGA-3′;5′-GGCCTGTCTCTGCCCCA-3′ | 5′-TGCTCCAAGCCCGGTGTCATGTA-3′;5′-CAGTGCTCCAAGCCTGGTGTCATGT-3′ |
| 13 | rs1719133 | 5′-AAGCCTGCCTTCCTCAACTG-3′;5′-AGCTATGAAATTCTGTGGAATCTGC-3′ | _Ban_II |
| 14 | rs1719134 | 5′-CACTCTAGGTCTCCCAGGAGCT-3′;5′-GACTGTTCTCTTATCTCAGTTCTCTTCAG-3′ | 5′-AGAGATGTCCAAGGCTTCTCTTGGGTT-3′;5′-AGAGATGTCCAAGGTTTCTCTTGGGTTG-3′ |
| 15 | ss46566438 | 5′-CCTCCTCTGCACCATGGCT-3′;5′-GGGCTTTTAGGCCACAAGAAA-3′ | 5′-ACCATGGCCAGAGAGTGGT-3′;5′-ACCATGACCAGAGAGTGGT-3′ |
| 16 | ss46566439 | 5′-TCAGACTTTGTAGAATTTGTATAATGTCG-3′;5′-AGGGAATGATAAATTATCCACTAGATCA-3′ | _Mnl_I |
| 17 | rs1634498 | 5′-CAGACACTTAGAAAGGACAGAATTCC-3′;5′-TGATAAAGCTAAATTGGGACCAAAC-3′ | _Hin_cII |
| 18 | rs1634507 | 5′-TCTTGCTGGAGTATTCCCTATGA-3′;5′-GCCACAACCTGCTCTTGC-3′ | _Hph_I |
| 19 | rs1634514 | 5′-GCCTTCTGCTCTCCAGCG-3′;5′-TTCTTAAGAAAATAGGAACTTGAATGTTAT-3′ | _Hin_fI |
| 20 | rs1719146 | 5′-CATGTTAGGTGGGAATGGATATTT-3′;5′-GGTGGATGGGATCCTTCCT-3′ | _Bst_UI |
| 21 | rs1719153 | 5′-CAAGGGTTTTAACACCCTTATGAAC-3′;5′-CCAAGCAGGCCTACAAGCTT-3′ | 5′-TTTCCTTAACTGTGAAACT-3′;5′-TTTCCTTAACAGTGAAACT-3′ |
Statistical Analyses
Allele frequencies and genotype frequencies were calculated and tests for Hardy-Weinberg equilibrium were performed using SAS Genetics software (SAS Institute). Unphased diploid genotype data were partially phased by use of PHASE,26 and the output from PHASE was input into HAPLOVIEW27 for pairwise _D_′ and _r_2 calculation, haplotype estimation, and haplotype-block construction. Comparisons of allele, genotype, and haplotype frequencies between the seronegative HREU individuals and the HIV-1–infected seroconvertors were done with Fisher’s exact test or logistic regression by use of SAS. Cox proportional hazards regression and Kaplan-Meier survival statistics assessed rates of disease progression among seroconvertors. Regression and survival analyses were performed twice—once by using each SNP or haplotype alone and once by including three SNPs from CCL5 (RANTES) as covariates: RANTES-3′ 222, RANTES-In1.1 (rs2280789), and RANTES-(−403) (rs2107538).28,29 Participants were stratified by sex (results are shown for males only) and age at HIV-1 seroconversion: <20 years, 20–40 years, and >40 years. The results stratified by age were included in the model and appear in table 3.
Table 3. .
Survival Analyses of Progression to Three AIDS End Points for CCL18, CCL3, and CCL4 SNPs and One Haplotype[Note]
| AIDS-93 | AIDS-87 | Death | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Group and SNP or Haplotype | NE | RH (95% CI) | P | NE | RH (95% CI) | P | NE | RH (95% CI) | P |
| AAs in all cohorts (_n_=290): | … | … | … | … | … | … | … | … | … |
| rs712040 | 124 | 1.01 (.60–1.52) | .96 | 57 | .86 (.49–1.52) | .6 | 33 | 1.37 (.61–.50) | .45 |
| rs2015086 | 125 | 1.09 (.75–1.57) | .66 | 59 | .95 (.56–1.61) | .84 | 32 | 1.12 (.55–2.28) | .76 |
| rs2015070 | 125 | 1.11 (.55–2.22) | .77 | 59 | .79 (.28–2.23) | .65 | 34 | .71 (.17–3.06) | .65 |
| rs712043 | 125 | 1.18 (.72–1.94) | .5 | 59 | 1.09 (.53–2.25) | .81 | 34 | .75 (.26–2.15) | .59 |
| rs854472 | 128 | 1.11 (.68–1.82) | .68 | 61 | 1.03 (.50–2.12) | .93 | 35 | .73 (.25–2.07) | .55 |
| rs1719220 | 130 | 1.26 (.75–2.11) | .39 | 63 | 1.18 (.55–2.53) | .68 | 36 | .98 (.34–2.81) | .97 |
| rs1634481 | 129 | .06 (.70–2.01) | .52 | 63 | 1.10 (.51–2.38) | .8 | 36 | .92 (.32–2.65) | .88 |
| rs1719127 | 131 | .96 (.62–1.51) | .87 | 62 | .84 (.43–1.62) | .6 | 35 | 1.18 (.53–2.64) | .68 |
| rs8951 | 131 | 1.34 (.85–2.11) | .2 | 61 | 1.26 (.66–2.43) | .48 | 35 | 1.69 (.75–3.81) | .21 |
| ss46566437 | 132 | .97 (.58–1.61) | .9 | 62 | .77 (.36–1.65) | .5 | 35 | .77 (.27–2.19) | .62 |
| rs1719130 | 125 | 1.3 (.89–1.94) | .18 | 57 | 1.27 (.73–2.20) | .4 | 32 | 1.29 (.62–2.68) | .49 |
| rs1130371 | 131 | 1.34 (.91–1.94) | .14 | 62 | 1.21 (.69–2.11) | .5 | 35 | .90 (.41–1.95) | .78 |
| rs1719133 | 132 | .91 (.52–1.59) | .74 | 62 | 1.09 (.51–2.32) | .82 | 35 | 1.16 (.45–3.02) | .76 |
| rs1719134 | 125 | 1.25 (.83–1.88) | .29 | 56 | .96 (.51–1.82) | .91 | 32 | .79 (.32–1.96) | .62 |
| ss46566438 | 129 | .90 (.53–1.53) | .69 | 60 | .85 (.40–1.81) | .67 | 35 | .80 (.28–2.28) | .68 |
| ss46566439 | 130 | .92 (.54–1.56) | .75 | 60 | .65 (.28–1.52) | .32 | 35 | .91 (.32–2.59) | .86 |
| rs1634498 | 129 | 1.02 (.62–1.69) | .93 | 60 | 1.13 (.56–2.26) | .74 | 34 | 1.29 (.53–3.16) | .58 |
| rs1634507 | 130 | 1.53 (1.01–2.34) | .05* | 60 | 1.33 (.71–2.50) | .37 | 35 | .64 (.24–1.69) | .36 |
| rs1719144 | 129 | 1.52 (.91–2.52) | .11 | 60 | 1.34 (.61–2.95) | .47 | 34 | .66 (.19–2.31) | .52 |
| rs1719146 | 126 | 2.37 (1.11–5.06) | .03* | 59 | 2.72 (.99–7.49) | .05* | 34 | 1.39 (.30–6.37) | .67 |
| rs1719153 | 129 | 1.55 (.96–2.48) | .07 | 62 | 1.60 (.81–3.13 | .17 | 35 | .71 (.24–2.13) | .54 |
| Haplotype [block 3-.071] | 123 | 1.37 (.77–2.34) | .21 | 60 | 1.19 (.55–2.47) | .38 | 33 | .84 (.33–2.26) | .51 |
| rs1719146 | 104 | 2.38 (1.12–5.04) | .02* | 46 | 2.72 (.97–7.56) | .06 | 22 | 2.17 (.47–9.97) | .32 |
| EAs in all cohorts (_n_=661): | … | … | … | … | … | … | … | … | … |
| rs712040 | 406 | .96 (.75–1.23) | .75 | 293 | .95 (.71–1.28) | .74 | 250 | .93 (.66–1.29) | .65 |
| rs2015086 | 413 | .96 (.76–1.21) | .72 | 299 | .98 (.75–1.29) | .89 | 256 | .97 (.72–1.31) | .85 |
| rs2015070 | 406 | .90 (.69–1.17) | .43 | 293 | .80 (.58–1.12) | .19 | 250 | .82 (.57–1.17) | .27 |
| rs712043 | 411 | 1.09 (.89–1.32) | .4 | 298 | 1.16 (.92–1.46) | .22 | 255 | 1.08 (.84–1.39) | .55 |
| rs854472 | 408 | 1.11 (.91–1.35) | .3 | 294 | 1.21 (.96–1.53) | .11 | 250 | 1.12 (.87–1.44) | .38 |
| rs1719220 | 410 | 1.04 (.85–1.27) | .71 | 298 | 1.09 (.86–1.37) | .48 | 253 | .97 (.75–1.24) | .79 |
| rs1634481 | 396 | 1.06 (.87–1.30) | .55 | 286 | 1.14 (.90–1.44) | .29 | 240 | .99 (.77–1.29) | .96 |
| rs1719127a | … | … | … | … | … | … | … | … | … |
| rs8951 | 409 | 1.15 (.95–1.40) | .15 | 298 | 1.30 (1.03–1.63) | .03* | 262 | 1.06 (.83–1.36) | .64 |
| ss46566437 | … | … | … | … | … | … | … | … | … |
| rs1719130 | 400 | 1.24 (1.01–1.51) | .04* | 289 | 1.39 (1.10–1.76) | .01* | 250 | 1.25 (.97–1.60) | .09 |
| rs1130371 | 414 | 1.17 (.97–1.43) | .11 | 302 | 1.33 (1.06–1.67) | .01* | 263 | 1.15 (.90–1.47) | .26 |
| rs1719133 | … | … | … | … | … | … | … | … | … |
| rs1719134 | 418 | 1.18 (.98–1.44) | .09 | 304 | 1.34 (1.07–1.68) | .01* | 264 | 1.17 (.92–1.49) | .21 |
| ss46566438 | … | … | … | … | … | … | … | … | … |
| ss46566439 | … | … | … | … | … | … | … | … | … |
| rs1634498 | … | … | … | … | … | … | … | … | … |
| rs1634507 | 405 | 1.14 (.93–1.38) | .21 | 297 | 1.23 (.98–1.54) | .08 | 257 | 1.07 (.83–1.37) | .61 |
| rs1719144 | 392 | 1.20 (.98–1.47) | .07 | 287 | 1.32 (1.05–1.67) | .02* | 252 | 1.09 (.85–1.41) | .49 |
| rs1719146 | … | … | … | … | … | … | … | … | … |
| rs1719153 | 409 | 1.19 (.98–1.45) | .08 | 297 | 1.33 (1.06–1.68) | .01* | 253 | 1.20 (.94–1.54) | .15 |
| EAs in MACS cohort (_n_=403): | … | … | … | … | … | … | … | … | … |
| rs712043 | 248 | 1.14 (.89–1.47) | .3 | 183 | 1.38 (1.02–1.85) | .04* | 159 | 1.18 (.86–1.62) | .31 |
| rs854472 | 248 | 1.15 (.89–1.48) | .28 | 181 | 1.41 (1.05–1.91) | .02* | 156 | 1.20 (.87–1.65) | .27 |
| rs1634481 | 246 | 1.08 (.83–1.39) | .57 | 179 | 1.27 (.94–1.71) | .12 | 153 | 1.01 (.73–1.39) | .95 |
| rs8951 | 243 | 1.21 (.94–1.57) | .14 | 182 | 1.43 (1.06–1.91) | .02* | 162 | 1.12 (.82–1.53) | .46 |
| rs1719130 | 241 | 1.19 (.92–1.54) | .18 | 179 | 1.40 (1.04–1.88) | .02* | 158 | 1.22 (.89–1.68) | .21 |
| rs1130371 | 245 | 1.22 (.95–1.57) | .13 | 184 | 1.41 (1.06–1.89) | .02* | 162 | 1.15 (.85–1.57) | .37 |
| rs1719134 | 250 | 1.22 (.95–1.57) | .11 | 187 | 1.42 (1.07–1.90) | .02* | 164 | 1.19 (.87–1.62) | .28 |
| rs1634507 | 238 | 1.19 (.92–1.54) | .18 | 180 | 1.32 (.99–1.78) | .06 | 157 | 1.12 (.82–1.54) | .47 |
| rs1719144 | 228 | 1.19 (.92–1.55) | .19 | 172 | 1.42 (1.05–1.91) | .02* | 153 | 1.13 (.82–1.56) | .44 |
| rs1719153 | 247 | 1.22 (.95–1.57) | .12 | 183 | 1.45 (1.08–1.94) | .01* | 158 | 1.26 (.92–1.73) | .15 |
Results
Twenty-one SNPs covering a 47,637-bp region containing CCL18, CCL3, and CCL4 (fig. 1) were genotyped in up to 3,158 participants. Three SNPs occurred within coding regions as follows: (1) ss46566437 at +1342G→T changes amino acid position 79 (E79D) in exon 3 of the CCL3 gene, (2) rs1130371 at +868C→T is a silent substitution (P60P) in exon 2 of the CCL3 gene, and (3) rs1719146 at +663C→T is a silent substitution (T39T) in exon 2 of the CCL4 gene. All other SNPs were noncoding and were found in promoter regions, introns, and intergenic areas (fig. 1 and table 1). Of the 21 SNPs, 7 had minor-allele frequencies (MAFs) <0.01 in EAs and were excluded in all subsequent analyses of EAs, whereas all 21 SNPs had MAFs >0.03 in AAs and were retained in analyses of AAs (table 1).
Linkage Disequilibrium (LD) and Haplotype Analyses
Pairwise _D_′ values for AAs and EAs are shown (fig. 1A and 1B). In AAs, the three SNPs at both (5′ and 3′) ends of the region had much lower _D_′ values than did the 15 SNPs in the middle. Similarly, in EAs, the three SNPs at the 5′ end had lower _D_′ values than the remaining 11 SNPs. In addition, 16 (7.6%) of 211 pairwise _r_2 values exceeded 0.70 in AAs, whereas 30 (33%) of 91 pairwise _r_2 values exceeded 0.70 in EAs. These higher _r_2 values in EAs are reflected by the limited range in MAFs (0.218–0.304) for all but the three SNPs at the 5′ end, whereas MAFs in AAs showed a greater range across the entire region (0.032–0.497) (table 1).
Haplotype-block analysis identified four blocks in AAs, which were <1 kb, 1 kb, 3 kb, and 7 kb in size, and three blocks in EAs, which were 6 kb, <1 kb, and 28 kb in size (fig. 1_C_ and 1_D_). Diversity within each block was limited; only block 3 in AAs had more than two common haplotypes with frequencies exceeding 5%. In addition, multiallelic _D_′ values between blocks ranged from 0.81 to 0.93, indicating extensive disequilibrium between blocks in both AAs and EAs. Although there are recombinant haplotypes in AA blocks 3 and 4 and in EA block 3, only one common haplotype (frequency >0.05) contains information that is not available from analyzing the individual SNPs. This haplotype is found in AA block 3 at a frequency of 0.071 and is named “AA haplotype [block 3-0.071].” This haplotype is a recombinant containing only a portion of the minor alleles from rs1719130, rs1130371, and rs1719134 (fig. 1C). On the other hand, all other common haplotypes contain all minor alleles for at least one SNP and are thus redundant with that SNP. For example, AA haplotype [block 3-0.090] contains a portion of alleles from three SNPs but contains all alleles from SNPs ss46566437, ss46566438, and ss46566439 (fig. 1C).
Disease Associations
The influence of each SNP and the single unique haplotype—AA haplotype _[block 3-0.071]_—on HIV-1 transmission was examined by comparing HREU individuals with HIV-1–infected seroconvertors after stratifying by race (tables 4 and 5). Among AAs from all cohorts combined, ss46566439, occurring in CCRL3, had significantly elevated genotype frequency in the 79 HREU individuals (frequency 0.241) compared with the 296 HIV-1–positive seroconvertors (frequency 0.143) (P_=.04). To minimize inherent variability resulting from the combining of cohorts, the predominant AA cohort (the injection-drug-using ALIVE cohort) was considered separately. Among AAs in the ALIVE cohort, three CCL3 SNPs—_ss46566437, ss46566438, and _ss46566439_—were significantly elevated in frequency in the 73 HREU individuals compared with the 238 HIV-1–infected seroconvertors (_P_=.02–.03). These three SNPs are found together on one AA haplotype, [block 3-0.090], which spans 2,231 bp (fig. 1 and table 1). Among EAs from all cohorts combined, no SNPs were significant when the 128 HREU individuals were compared with the 661 HIV-1–positive seroconvertors. When the EAs were stratified by cohort, the dominant rs2015086 genotype had a frequency of 0.40 in the 25 hemophilic HREU individuals in the MHCS cohort and a frequency of 0.18 in the 170 seroconvertors in the MHCS cohort (_P_=.02) (tables 4 and 5).
Table 4. .
A Comparison of CCL18, CCL3, and CCL4 Dominant Genotype Frequencies between Seroconvertors (HIV+) and HREU Participants (HIV-1 Seronegative)[Note]
| AAs in All Cohorts | EAs in All Cohortsa | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | HIV+(_n_=290) | HIV-1Seronegative(_n_=79) | OR (95% CI) | P | HIV+(_n_=661) | HIV-1Seronegative(_n_=128) | OR (95% CI) | P |
| rs712040 | .714 | .708 | 1.03 (.58–1.82) | 1 | .189 | .183 | 1.04 (.64–1.70) | 1 |
| rs2015086 | .599 | .569 | 1.13 (.67–1.90) | .69 | .236 | .227 | 1.05 (.67–1.66) | .91 |
| rs2015070 | .076 | .118 | .61 (.27–1.39) | .25 | .165 | .164 | 1.00 (.60–1.69) | 1 |
| rs712043 | .129 | .149 | .84 (.39–1.80) | .69 | .498 | .461 | 1.15 (.79–1.69) | .51 |
| rs854472 | .135 | .143 | .93 (.44–1.98) | .85 | .505 | .461 | 1.19 (.82–1.75) | .38 |
| rs1719220 | .124 | .127 | .98 (.46–2.07) | 1 | .459 | .448 | 1.05 (.71–1.54) | .85 |
| rs1634481 | .125 | .115 | 1.10 (.51–2.39) | 1 | .453 | .437 | 1.06 (.73–1.57) | .77 |
| rs1719127 | .207 | .278 | .68 (.38–1.19) | .22 | … | … | … | … |
| rs8951 | .176 | .127 | 1.47 (.71–3.04) | .39 | .396 | .353 | 1.20 (.82–1.77) | .38 |
| ss46566437 | .153 | .241 | .57 (.31–1.04) | .09 | … | … | … | … |
| rs1719130 | .345 | .329 | 1.08 (.62–1.86) | .89 | .398 | .423 | .90 (.61–1.32) | .62 |
| rs1130371 | .295 | .304 | .96 (.56–1.65) | .89 | .417 | .409 | 1.03 (.71–1.50) | .92 |
| rs1719133 | .115 | .179 | .60 (.30–1.18) | .13 | … | … | … | … |
| rs1719134 | .263 | .316 | .77 (.45–1.33) | .39 | .414 | .411 | 1.01 (.70–1.46) | 1 |
| ss46566438 | .151 | .244 | .55 (.30–1.01) | .06 | … | … | … | … |
| ss46566439 | .143 | .241 | .53 (.29–.97) | .04* | … | … | … | … |
| rs1634498 | .154 | .208 | .69 (.37–1.31) | .3 | … | … | … | … |
Table 5. .
A Comparison of CCL18, CCL3, and CCL4 Dominant Genotype and Haplotype Frequencies between Seroconvertors (HIV+) and HREU Participants (HIV-1 Seronegative)[Note]
| AAs in ALIVE Cohorta | AAs in All Cohorts | EAs in MHCS Cohort | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP orHaplotype | HIV+ | HIV-1Seronegative | OR (95% CI) | P | HIV+(_n_=296) | HIV-1Seronegative(_n_=79) | OR (95% CI) | P | HIV+(_n_=170) | HIV-1Seronegative(_n_=25) | OR (95% CI) | P |
| ss46566437 | .143 | .26 | .47 (.25–.90) | .03* | … | … | … | … | … | … | … | … |
| ss46566438 | .145 | .264 | .48 (.25–.89) | .03* | … | … | … | … | … | … | … | … |
| ss46566439 | .135 | .26 | .44 (.23–.84) | .02* | … | … | … | … | … | … | … | … |
| Haplotype [block 3-.071] | .126 | .096 | 1.31 (.49–3.52) | .61 | .126 | .096 | 1.35 (.57–3.21) | .49 | … | … | … | … |
| rs2105086 | … | … | … | … | … | … | … | … | .176 | .4 | .32 (.13–.78) | .02* |
The influence of genetic variation in each SNP on disease progression was assessed. Cox proportional hazards regression, including CCL5 covariates, showed SNPs rs1634507 and rs1719146 to be significantly associated with more-rapid disease progression (to AIDS-93 or AIDS-87) when 290 AAs from all cohorts were combined (_P_=.03–.05) (table 3). Among all AAs, rs1719146 was significantly associated (relative hazard [RH] of 2.97; _P_=.03) when the CCL5 covariates were not included (data not shown). When the 238 members of ALIVE were examined separately, only rs1719146 was significantly associated (_P_=.02). Cox regression also found six SNPs to be significantly associated with more-rapid progression to AIDS-87 and one to AIDS-93, when 661 EAs from all cohorts combined were examined with CCL5 covariates included (_P_=.01–.04) (table 3). Five of the six SNPs associated with AIDS-87 are together on a single EA haplotype, [block 3-0.203], that spans 28,952 bp. The same six SNPs were significant (_RH_=1.29–1.39; _P_=.005–.03) among all EAs when the CCL5 covariates were not included (data not shown). Inspection of the three major EA cohorts indicated that these disease-accelerating effects are largely attributable to the MACS cohort rather than the MHCS, HGDS, or SFCC cohort (table 3). Eight SNPs were significantly associated (_P_=.01–.04) with AIDS-87 in the MACS cohort, whereas none was associated in the MHCS, HGDS, or SFCC cohort.
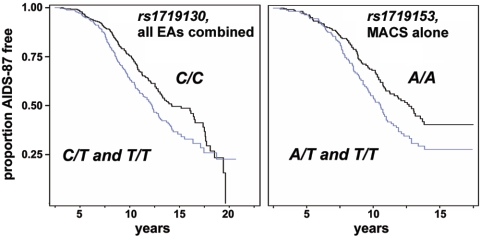
The results presented in table 3 were confirmed by Kaplan-Meier survival analyses without CCL5 covariates, in which 10 SNPs were associated with more-rapid disease progression in all EAs, including 9 SNPs in the MACS cohort (results not shown). As illustrated by the graphs in figure 2, the rate of AIDS-87 disease progression is increased among all EAs carrying one (C/T) or two (T/T) copies of the variant rs1719130 allele compared with those carrying the wild-type (C/C) genotype (fig. 2A) and is increased among individuals in the MACS cohort carrying one (A/T) or two (T/T) copies of the variant rs1719153 allele compared with those homozygous (A/A) for the wild-type allele (fig. 2B).
Figure 2. .
Kaplan-Meier survival curves showing the dependence of AIDS-87 progression (the proportion surviving who did not reach the disease end point) on the dominant rs1719130 genotype in all EAs (A) and the dominant rs1719153 genotype in the MACS cohort (B). A, The black curve (C/C) represents all individuals homozygous for the wild-type allele, and the blue curve (C/T and T/T) includes individuals with one or two copies of the minor allele. The total number of individuals was 636; 291 developed AIDS, and 345 were censored. _RH_=1.50; log-rank _P_=.0065; Wilcoxon _P_=.0023; −2log(likelihood ratio) _P_=.009. B, The black curve (A/A) represents individuals carrying two copies of the wildtype allele, and the blue curve (A/T and T/T) represents individuals with one or two copies of the variant allele. The total number of individuals was 402; 184 developed AIDS, and 218 were censored. _RH_=1.51; log-rank _P_=.0072; Wilcoxon _P_=.0159; −2log(likelihood ratio) _P_=.0149. RH values were obtained from Cox proportional hazards regression, and the P values were obtained from the Kaplan-Meier survival analyses.
Discussion
This study evaluated the role of genetic variation in 21 SNPs in the CCL18, CCL3, and CCL4 chemokine genes in HIV-1 infection and AIDS disease progression in five U.S. natural-history AIDS cohorts. There was extensive LD in the 47,637 bp surveyed in both EAs and AAs but particularly in EAs, among whom limited allelic and haplotype diversity was found. Four statistically significant associations with HIV-1 transmission or AIDS progression were identified, two of which replicate results of an earlier study. Importantly, replication has recently been reported to be the “gold standard” in genetic association studies.31 We further discuss the two replicated associations.
The first was a protective genetic influence on HIV-1 infection for three correlated CCL3 SNPs (ss46566437, ss46566439, and ss46566438, located on one haplotype covering 2,231 bp) in AAs from the ALIVE cohort. Although ss46566437 is a nonsynonymous (E79D) substitution, we have no evidence indicating that it is a functional mutation. These results confirm and extend those of Gonzalez et al.,10 who reported a significantly elevated frequency of the _ss46566438_-containing haplotype in HIV-1–negative AA subjects compared with HIV-1–positive AA subjects. The protective effect found in our study was slightly stronger for the ALIVE cohort alone (odds ratio [OR] 0.47) than for all AAs combined (OR 0.57). This may be a result of mode of infection, sampling artifacts, or population stratification. ALIVE is an injection-drug-user cohort from Baltimore. The second largest population of AA participants consists of 45 seroconvertors from the MACS, a cohort of homosexual men from four American cities.18 It is well established that AAs are admixed and that different populations may have different ancestral African and European genetic contributions.32 It is thus expected that different genetic backgrounds will reveal varying susceptibility to infectious agents.
Second, we found that each of seven highly correlated SNPs was associated with more-rapid disease progression in EAs. These SNPs span 36,562 bp and contain all three genes. Given the extensive disequilibrium in the region, it is not possible to identify a particular SNP or even a single gene as causative. Although CCL18 has not yet been implicated in HIV-1/AIDS pathogenesis and its receptor is not known,33 the present genetic analysis cannot rule out this gene as a candidate for modulating HIV-1–related pathogenesis. Furthermore, pairwise protein alignment scores obtained using CLUSTALW34 were 54.4 for CCL3 and CCL4, 59.6 for CCL3 and CCL18, and 40.5 for CCL4 and CCL18. The high relative score for alignment between CCL3 and CCL18 suggests that there could be some redundancy in receptor binding between these two ligands. Importantly, this disease-accelerating association supports an earlier study in which borderline statistical significance (_P_=.03–.09) in Kaplan-Meier survival analysis of an EA population was reported for rs1719134.10 Interestingly, the level of LD in the present study is comparable to what was reported for the CCL2-CCL7-CCL11 chemokine gene cluster, which is 1.8 Mb upstream of the CCL18-CCL3-CCL4 cluster.25 In that region, a 31-kb haplotype containing all three genes was associated with decreased susceptibility to infection among EAs. Similar to the findings here, a single gene or SNP could not be singled out as being responsible for the association, given the observed extent of LD.
Although significant Cox regression P values were observed for EAs in the MACS cohort but not in the other cohorts, the RH values for these cohorts overlapped each other. Thus, the smaller P values for the MACS EAs may be influenced by the larger sample size. The MACS and SFCC cohorts consist of men who became infected with HIV-1 through unprotected anal intercourse, whereas the MHCS participants have hemophilia and were exposed to HIV-1–contaminated clotting factor; thus, route of infection does not seem to explain differences between cohorts in the association results. On the other hand, the DNA samples used in this study from the MHCS and the SFCC cohorts are enriched for long-term nonprogressors and thus represent a nonrandom segment of the population surviving with HIV-1 infection, whereas the MACS samples are a broader representation of the infected population.18 In other words, if people developing AIDS early after infection had been recorded by the MHCS and SFCC cohorts, then perhaps their disease-progression profiles would be more similar to that of the MACS cohort.
CCR5 binding ligands such as CCL3, CCL4, and CCL5 can limit HIV-1 infection via at least three different mechanisms. After a ligand binds to its receptor, the viral envelope protein is blocked from attaching to the cell. A second possibility is that the chemokines induce endocytosis of the receptor, thus limiting availability of the receptor to virus.35,36 It has also been suggested that chemokines can induce receptor dimerization that subsequently prevents viral proteins from interacting with the cellular membrane.37 Further, it has been shown that the ability of CCL5 to limit HIV-1 infection can vary between cell types.38 In addition, gene-by-gene and gene-by-environment interactions certainly act on HIV-1/AIDS pathology, as do various social and environmental variables.39 The idea that certain genotypes act only under specific environmental conditions or that a given genotype may produce different phenotypes in different environments is a distinct possibility.31 These interactions are more complex than was initially anticipated, reinforcing the need for further study and emphasizing the importance of replication among studies with adequate power.
Acknowledgments
Michael Sanford, Julie Sawitzke, Maidar Jamba, and Elizabeth Binns-Roemer provided laboratory expertise. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (contract NO1-CO-12400).
References
- 1.DeVico AL, Gallo RC (2004) Control of HIV-1 infection by soluble factors of the immune response. Nat Rev Microbiol 2:401–413 10.1038/nrmicro878 [DOI] [PubMed] [Google Scholar]
- 2.Winkler C, An P, O’Brien SJ (2004) Patterns of ethnic diversity among the genes that influence AIDS. Hum Mol Genet Spec No 1 13:R9–R19 10.1093/hmg/ddh075 [DOI] [PubMed] [Google Scholar]
- 3.Walker CM, Moody DJ, Stites DP, Levy JA (1986) CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science 234:1563–1566 [DOI] [PubMed] [Google Scholar]
- 4.Mackewicz C, Levy JA (1992) CD8+ cell anti-HIV activity: nonlytic suppression of virus replication. AIDS Res Hum Retroviruses 8:1039–1050 [DOI] [PubMed] [Google Scholar]
- 5.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P (1995) Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815 [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini M, Dewald B, Moser B (1994) Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv Immunol 55:97–179 [PubMed] [Google Scholar]
- 7.Luster AD (1998) Chemokines—chemotactic cytokines that mediate inflammation. N Engl J Med 338:436–445 10.1056/NEJM199802123380706 [DOI] [PubMed] [Google Scholar]
- 8.Modi WS (2004) CCL3L1 and CCL4L1 chemokine genes are located in a segmental duplication at chromosome 17q12. Genomics 83:735–738 10.1016/j.ygeno.2003.09.019 [DOI] [PubMed] [Google Scholar]
- 9.Xin X, Nakamura K, Liu H, Nakayama EE, Goto M, Nagai Y, Kitamura Y, Shioda T, Iwamoto A (2001) Novel polymorphisms in human macrophage inflammatory protein-1 alpha (MIP-1α) gene. Genes Immun 2:156–158 10.1038/sj.gene.6363759 [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, Anderson SA, Walter EA, Stephan KT, Hammer MF, Mangano A, Sen L, Clark RA, Ahuja SS, Dolan MJ, Ahuja SK (2001) Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci USA 98:5199–5204 10.1073/pnas.091056898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlahov D, Graham N, Hoover D, Flynn C, Bartlett JG, Margolick JB, Lyles CM, Nelson KE, Smith D, Holmberg S, Farzadegan H (1998) Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA 279:35–40 10.1001/jama.279.1.35 [DOI] [PubMed] [Google Scholar]
- 12.Hilgartner MW, Donfield SM, Willoughby A, Contant CF Jr, Evatt BL, Gomperts ED, Hoots WK, Jason J, Loveland KA, McKinlay SM, Stehbens JA (1993) Hemophilia Growth and Development Study: design, methods, and entry data. Am J Pediatr Hematol Oncol 15:208–218 [DOI] [PubMed] [Google Scholar]
- 13.Goedert JJ, Kessler CM, Aledort LM, Biggar RJ, Andes WA, White GC II, Drummond JE, Vaidya K, Mann DL, Eyster ME, Ragni MV, Lederman MM, Cohen AR, Bray GL, Rosenberg PS, Friedman RM, Hilgartner MW, Blattner WA, Kroner B, Gail MH (1989) A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med 321:1141–1148 [DOI] [PubMed] [Google Scholar]
- 14.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR Jr (1987) The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol 126:310–318 [DOI] [PubMed] [Google Scholar]
- 15.Detels R, Liu Z, Hennessey K, Kan J, Visscher BR, Taylor JMG, Hoover DR, Rinaldo CR Jr, Phair JP, Saah AJ, Giorgi JV, for the Multicenter AIDS Cohort Study (1994) Resistance to HIV-1 infection. J Acquir Immune Defic Syndr 7:1263–1269 [PubMed] [Google Scholar]
- 16.Rutherford GW, Lifson AR, Hessol NA, Darrow WW, O’Malley PM, Buchbinder SP, Barnhart JL, Bodecker TW, Cannon L, Doll LS, Holmberg SD, Harrison JS, Rogers MF, Werdegar D, Jaffe HW (1990) Course of HIV-I infection in a cohort of homosexual and bisexual men: an 11 year follow up study. BMJ 301:1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroner BL, Rosenberg PS, Aledort LM, Alvord WG, Goedert JJ (1994) HIV-1 infection incidence among persons with hemophilia in the United States and western Europe, 1978–1990. Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr 7:279–286 [PubMed] [Google Scholar]
- 18.Modi WS, Scott K, Goedert JJ, Vlahov D, Buchbinder S, Detels R, Donfield S, O’Brien SJ, Winkler C (2005) Haplotype analysis of the SDF-1 (CXCL12) gene in a longitudinal HIV-1/AIDS cohort study. Genes Immun 6:691–698 [DOI] [PubMed] [Google Scholar]
- 19.Detels R, Munoz A, McFarlane G, Kingsley LA, Margolick JB, Giorgi J, Schrager LK, Phair JP, for the Multicenter AIDS Cohort Study Investigators (1998) Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. JAMA 280:1497–1503 10.1001/jama.280.17.1497 [DOI] [PubMed] [Google Scholar]
- 20.Celentano DD, Galai N, Sethi AK, Shah NG, Strathdee SA, Vlahov D, Gallant JE (2001) Time to initiating highly active antiretroviral therapy among HIV-infected injection drug users. AIDS 15:1707–1715 10.1097/00002030-200109070-00015 [DOI] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention (1992) 1993 Revised classification system for HIV infection and an expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm Rep 41:1–19 [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention (1987) Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. MMWR Morb Mortal Wkly Rep Suppl 1 36:1S–15S [PubMed] [Google Scholar]
- 23.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- 24.Nakao M, Nomiyama H, Shimada K (1990) Structures of human genes coding for cytokine LD78 and their expression. Mol Cell Biol 10:3646–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modi WS, Goedert JJ, Strathdee S, Buchbinder S, Detels R, Donfield S, O’Brien SJ, Winkler C (2003) MCP-1-MCP-3–Eotaxin gene cluster influences HIV-1 transmission. AIDS 17:2357–2365 10.1097/00002030-200311070-00011 [DOI] [PubMed] [Google Scholar]
- 26.Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 28.McDermott DH, Beecroft MJ, Kleeberger CA, Al-Sharif FM, Ollier WE, Zimmerman PA, Boatin BA, Leitman SF, Detels R, Hajeer AH, Murphy PM (2000) Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS 14:2671–2678 10.1097/00002030-200012010-00006 [DOI] [PubMed] [Google Scholar]
- 29.An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, Vlahov D, Buchbinder S, Farrar WL, Modi W, O’Brien SJ, Winkler CA (2002) Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci USA 99:10002–10007 10.1073/pnas.142313799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien SJ (1996) Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856–1862 [DOI] [PubMed] [Google Scholar]
- 31.Ober C (2005) Perspectives on the past decade of asthma genetics. J Allergy Clin Immunol 116:274–278 10.1016/j.jaci.2005.04.039 [DOI] [PubMed] [Google Scholar]
- 32.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunther C, Bello-Fernandez C, Kopp T, Kund J, Carballido-Perrig N, Hinteregger S, Fassl S, Schwarzler C, Lametschwandtner G, Stingl G, Biedermann T, Carballido JM (2005) CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. J Immunol 174:1723–1728 [DOI] [PubMed] [Google Scholar]
- 34.Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266:383–402 [DOI] [PubMed] [Google Scholar]
- 35.Amara A, Gall SL, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier JL, Arenzana-Seisdedos F (1997) HIV coreceptor downregulation as antiviral principle: SDF-1α-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med 186:139–146 10.1084/jem.186.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Signoret N, Rosenkilde MM, Klasse PJ, Schwartz TW, Malim MH, Hoxie JA, Marsh M (1998) Differential regulation of CXCR4 and CCR5 endocytosis. J Cell Sci 111:2819–2830 [DOI] [PubMed] [Google Scholar]
- 37.Vila-Coro AJ, Mellado M, Martin de Ana A, Lucas P, del Real G, Martinez AC, Rodriguez-Frade JM (2000) HIV-1 infection through the CCR5 receptor is blocked by receptor dimerization. Proc Natl Acad Sci USA 97:3388–3393 10.1073/pnas.050457797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross E, Amella CA, Pompucci L, Franchin G, Sherry B, Schmidtmayerova H (2003) Macrophages and lymphocytes differentially modulate the ability of RANTES to inhibit HIV-1 infection. J Leukoc Biol 74:781–790 10.1189/jlb.0403187 [DOI] [PubMed] [Google Scholar]
- 39.Weiss RA, McMichael AJ (2004) Social and environmental risk factors in the emergence of infectious diseases. Nat Med 10:S70–S76 10.1038/nm1150 [DOI] [PMC free article] [PubMed] [Google Scholar]