Hepatocyte nuclear factor 4α orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver (original) (raw)
Abstract
Epithelial formation is a central facet of organogenesis that relies on intercellular junction assembly to create functionally distinct apical and basal cell surfaces. How this process is regulated during embryonic development remains obscure. Previous studies using conditional knockout mice have shown that loss of hepatocyte nuclear factor 4α (HNF4α) blocks the epithelial transformation of the fetal liver, suggesting that HNF4α is a central regulator of epithelial morphogenesis. Although HNF4α-null hepatocytes do not express E-cadherin (also called CDH1), we show here that E-cadherin is dispensable for liver development, implying that HNF4α regulates additional aspects of epithelial formation. Microarray and molecular analyses reveal that HNF4α regulates the developmental expression of a myriad of proteins required for cell junction assembly and adhesion. Our findings define a fundamental mechanism through which generation of tissue epithelia during development is coordinated with the onset of organ function.
Keywords: cell junctions, organogenesis, transcription
The function of many organs, such as the liver, gastrointestinal tract, and kidney, depends inherently upon the generation of an epithelium. The transition from a set of loosely connected nonpolarized cells to organized sheets of closely associated polarized epithelial cells requires the assembly of specialized cell junctions. These junctions, which are linked to the cytoskeleton, separate the cell membrane into functionally distinct apical and basolateral regions (1). In addition, such junctions interact with molecules in the extracellular matrix, thereby providing a mechanism for cells to receive signals from their environment (2). In vertebrates, adherens junctions, tight junctions, and desmosomes are the three major types of junctions responsible for epithelial integrity. Studies of junction assembly in cultured cells revealed that the formation of cell junctions occurs in an orderly and defined manner and is initiated by the intercellular interaction of cadherin and nectin proteins (3, 4). This initial adhesion event is followed by maturation of the junctions, which is facilitated by rearrangements of the underlying cytoskeleton and recruitment of additional junction proteins in a process that is controlled in part by the Rho family of small GTPases (2). The number of proteins that contribute to junction formation and function is very large. Adherens junctions contain E-cadherin, nectins, and α- and β-catenin along with a host of cytoplasmic linker proteins (2). The primary tight junction components include the transmembrane proteins JAM-A (encoded by the F11r gene), claudins, and occludin (OCLN), which interact with the PAR6-PAR3-aPKC and CRB3-PALS-PATJ signaling complexes as well as with cytoplasmic linker proteins (3–6). Desmosomes contain desmocollins, desmogleins, plakoglobulin, desmoplakin, and plakophilins (6).
Although our knowledge of junction cell biology is now detailed, our understanding of how the expression of such an extensive array of proteins is coordinated during embryonic development remains rudimentary. This is an important question to address because epithelial formation is a potent driving force during tissue morphogenesis and organogenesis and, when reversed, results in uncontrolled cellular proliferation and tumorigenesis. Analyses of transcriptional regulatory elements have implicated the transcription factors CDX1, hepatocyte nuclear factor 1α (HNF1α), and β-catenin/T cell factor in regulating expression of claudin 2 (7). Regulatory regions important for expression of E-cadherin during embryogenesis have also been identified, and the transcriptional repressors Snail and Slug have been shown to down-regulate E-cadherin expression in cancer cells (8–13). Recent studies using conditional knockout mice have shown that the nuclear hormone transcription factor HNF4α is required for the epithelial transformation of the liver during development (14, 15). This finding identifies HNF4α as a potential key regulator of cell adhesion and junction gene expression. Here we use HNF4α conditional knockout mice to establish that HNF4α coordinates the developmental expression of an extensive array of proteins that are essential for diverse aspects of junction assembly and function during hepatogenesis. Our studies describe a molecular framework through which epithelial formation coincides with the onset of organ function during embryonic development.
Results and Discussion
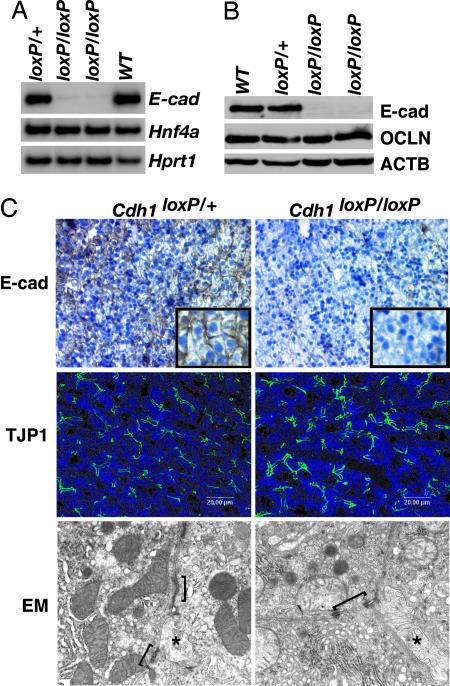
Several studies have implicated E-cadherin as a driving force in cell junction formation (2). In Madin–Darby canine kidney cells, inhibition of E-cadherin-mediated cell adhesion using blocking antibodies prevented the assembly not only of adherens junctions but also of tight junctions and desmosomes (16). Moreover, E-cadherin-null embryos die at 3.5–4.5 days postcoitum (dpc) because the trophoectoderm epithelium fails to form (17). Expression of E-cadherin is absent in HNF4α-null livers, and so we initially proposed that loss of E-cadherin in HNF4α-deficient embryonic livers is responsible for the failure of HNF4α-null hepatocytes to form an epithelium (15). To test this hypothesis, we used a conditional knockout approach that had previously been used to delineate the role of E-cadherin in development of the mouse mammary gland, epidermis, and peripheral nervous system (18–22). Using mice in which _lox_P elements flank exons 6–10 of the E-cadherin gene (Cdh1 tm2Kem, herein designated Cdh1 loxP; see ref. 19) and a transgenic mouse line in which expression of the Cre recombinase gene is controlled by the hepatoblast-specific albumin promoter and alphafetoprotein enhancer [(Tg(_Alb1-cre_)1Khk, herein designated _Alfp-Cre_; see ref. 14], we generated Cdh1 loxP/+;Alfp-Cre control and Cdh1 loxP/loxP;Alfp-Cre experimental animals. Surprisingly, Cdh1 loxP/loxP;Alfp-Cre mice were viable, fertile, and showed no signs of illness. Analyses of E-cadherin mRNA and protein levels confirmed it was lost in both adult (Fig. 1A and B) and fetal 18.5-dpc (Fig. 1C) Cdh1 loxP/loxP;Alfp-Cre livers. We also found no change in expression of the tight junction protein OCLN, and localization of tight junction protein 1 (TJP1, also known as ZO1) to the apical domain of the cell surface was normal in mutant hepatocytes (Fig. 1C). A comparison of E-cadherin-null and control adult livers by electron microscopy revealed no obvious differences in the formation of adherens junctions, tight junctions, or desmosomes (Fig. 1C); bile canaliculi were present in both control and mutant livers, suggesting that the E-cadherin-deficient hepatocytes were correctly polarized. Based on these data, we conclude that E-cadherin expression in hepatocytes is dispensable for establishing the hepatic epithelium during development.
Fig. 1.
Hepatocyte-specific loss of E-cadherin does not affect the formation of cell junctions in the liver. (A) RT-PCR showed loss of E-cadherin (E-cad) mRNA in livers of Cdh1 loxP/loxP;AlfpCre mice compared with control Cdh1 loxP/+;AlfpCre and WT littermates. Hnf4a levels were unchanged, and hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) confirmed equal loading. (B) Immunoblot analysis of liver extracts indicated that E-cadherin (E-cad) protein is undetectable in the Cdh1 loxP/loxP;AlfpCre livers compared with controls (Cdh1 loxP/+;AlfpCre and WT). Total protein levels of the tight junction protein OCLN were unchanged, and β-actin (ACTB) demonstrated equal loading. (C) Immunohistochemistry detected E-cadherin between hepatocytes in control livers (Top Left) but not between hepatocytes in Cdh1 loxP/loxP;AlfpCre livers (Top Right) (Inset is higher magnification). Confocal immunofluorescence microscopy was used to detect TJP1 (also known as ZO1) at the apical surface of the hepatocytes in both control (Cdh1 loxP/+;AlfpCre, Middle Left) and Cdh1 loxP/loxP;AlfpCre (Middle Right) livers. Junctional complexes (indicated by brackets) were identified in both control (Cdh1 loxP/+;AlfpCre, Bottom Left) and Cdh1 loxP/loxP;AlfpCre (Bottom Right) livers by transmission electron microscopy. Asterisks indicate bile canaliculi, which confirm that hepatocytes are polarized in the absence of E-cadherin. High-resolution electron microscopy images are provided in Fig. 5, which is published as supporting information on the PNAS web site.
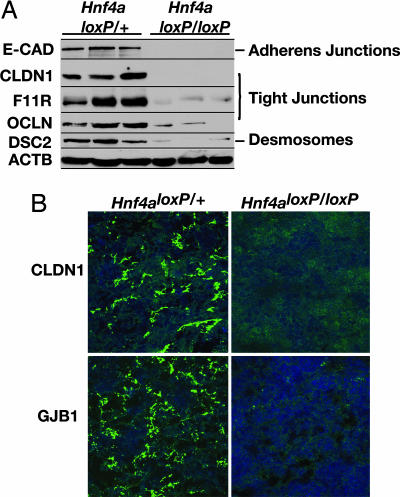
Because the loss of E-cadherin in hepatocytes does not explain the severe disruption to the formation of the hepatic epithelium seen in HNF4α-deficient livers, we hypothesized that the absence of HNF4α alters the expression of additional junction proteins that may compensate for loss of E-cadherin. Studies of mouse F9 embryonic carcinoma cells that overexpress HNF4α support this hypothesis. In these cells, HNF4α induces the expression of the tight junction proteins OCLN, claudin 6, claudin 7, and the F11 receptor (F11R, also known as JAM-A; see refs. 23 and 24). Therefore, to establish whether multiple aspects of cell adhesion and junction formation were affected in HNF4α-deficient livers, we examined the expression of proteins representing different types of junctions: E-cadherin represented adherens junctions; OCLN, F11R, and claudin 1 (CLDN1) represented tight junctions; and desmocollin 2 (DSC2) represented desmosomes. When we compared expression of these junction proteins in control and HNF4α-null 18.5-dpc livers by immunoblot analysis, we found that each was reduced or absent in mutant livers (Fig. 2A). We also examined the expression and localization of CLDN1 and gap junction protein β1 (GJB1, also known as connexin 32) in control and HNF4α-deficient livers by confocal fluorescence microscopy. We found that both CLDN1 and connexin 32 were expressed and appropriately localized in control livers but that these proteins were undetectable in HNF4α-null livers (Fig. 2B). From these data, we conclude that HNF4α is essential for the expression of genes encoding proteins involved in all major types of cell junctions, including tight junctions, adherens junctions, desmosomes, and gap junctions.
Fig. 2.
HNF4α is required for expression of cell junction and adhesion proteins in fetal mouse liver. (A) Immunoblots revealed a loss of or reduction in expression of proteins required for the formation of adherens junctions [E-cadherin (E-CAD)], tight junctions (CLDN1, F11R, and OCLN), and desmosomes (DSC2) in 18.5-dpc fetal livers lacking HNF4α (Hnf4a loxP/loxP;AlfpCre) compared with control livers (Hnf4a loxP/+;AlfpCre). β-Actin (ACTB) was used as a loading control. (B) Confocal microscopy demonstrated that the expression and localization of the tight junction protein CLDN1 and GJB1 (also known as connexin 32) were disrupted in 18.5-dpc fetal livers lacking HNF4α (Hnf4a loxP/loxP;AlfpCre) compared with control livers (Hnf4a loxP/+;AlfpCre).
The finding that junction formation in hepatocytes was extensively and generally diminished by loss of HNF4α implied that this single transcription factor could act as a coordinator of the expression of genes encoding junction and adhesion proteins. From a developmental perspective, this is appealing because it would suggest a mechanism through which epithelial morphogenesis of the liver could be efficiently orchestrated. We therefore used Affymetrix mouse oligonucleotide gene array analysis to determine whether loss of HNF4α in the developing liver comprehensively alters cell junction and adhesion gene expression. We compared the gene expression patterns between control Hnf4_a_loxP/+; Alfp-Cre and experimental Hnf4_a_loxP/loxP; Alfp-Cre livers harvested at 18.5 dpc. We used RT-PCR to verify that Hnf4a mRNA was absent in the Hnf4_a_loxP/loxP; Alfp-Cre livers (Fig. 6_A_, which is published as supporting information on the PNAS web site) and to confirm that Afp and Alb1 transcripts were present in the HNF4α-null livers because these markers of the hepatocyte population have been shown not to be affected by the loss of HNF4α (15). Using DNA-Chip (dchip) array analysis software (25), we identified 563 genes whose expression was down-regulated and 34 genes whose expression was up-regulated ≥2.5-fold (P ≤ 0.05) in the HNF4α-null embryonic livers compared with control livers (Table 1, which is published as supporting information on the PNAS web site). This large number of genes that depend on HNF4α for expression is consistent with the recent finding that HNF4α occupies sequences in >1,200 genes expressed in human liver cells (26). Of the down-regulated genes, 479 encode known proteins, and the remaining 84 encode ESTs. It is notable that expression of so few genes was up-regulated in mutant livers, suggesting that HNF4α is predominantly an activator of transcription in the liver. In agreement with our data is a study in which human hepatoma cells overexpressing HNF4α were analyzed by using gene arrays, and ≈10 times more genes were found to be up-regulated compared with those down-regulated (27). To validate the array data, we arbitrarily selected 40 genes whose expression was predicted to be reduced by array analysis and compared their steady-state mRNA levels in control and HNF4α-null embryonic livers using RT-PCR. In all cases, the changes in gene expression predicted by dchip software agreed with those determined by RT-PCR (Fig. 6_B_). We categorized the down-regulated genes by biological function using the Mouse Genomics Informatics GO_SLIM Chart tool (Fig. 6_C_). We found that every category queried by the GO_SLIM biological process tool was affected in HNF4α-null embryonic livers, demonstrating that HNF4α regulates diverse molecular pathways in the liver. Many genes down-regulated in HNF4α-null livers function in metabolic pathways, including amino acid metabolism, carbohydrate metabolism, cholesterol metabolism, lipid metabolism, and steroid metabolism. Additional pathways affected include lipid transport, ion transport, blood coagulation, immune function, regulation of proteolysis, and, as expected, cell adhesion.
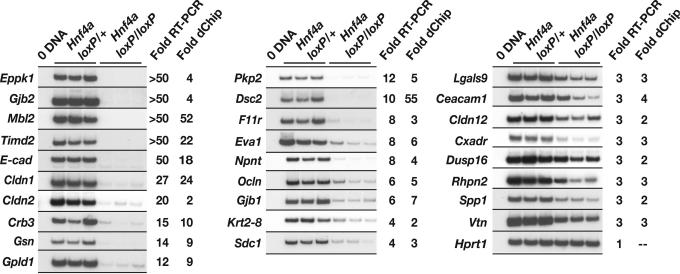
Because we were interested in determining the extent to which loss of HNF4α in the developing liver affects expression of cell junction and adhesion genes, we expanded our search for such genes by relaxing our screening stringency to genes predicted to be down-regulated ≥2.0-fold (P ≤ 0.05) by dchip software. Using the GO_SLIM tool and literature searches to classify down-regulated genes into the cell junction and adhesion category combined with RT-PCR to validate the fold changes predicted by dchip software, we identified 27 genes with either defined or predicted roles in cell junctions and adhesions (Fig. 3). These genes encode proteins contributing to tight junctions (Cldn1, Cldn2, Cldn12, Ocln, F11r, Cxadr, and Crb3), adherens junctions (E-cadherin), desmosomes (Dsc2, Pkp2, and Krt2–8), gap junctions (Gjb1 and Gjb2), and other cell–cell and cell–extracellular matrix adhesions (Ceacam1, Eva1, Gpld1, Lgals9, Mbl2, Npnt, Sdc1, Spp1, Timd2, and Vtn). Genes encoding cytoskeletal regulatory proteins (Eppk1 and Gsn) as well as downstream effectors of the mitogen-activated protein kinase and RHO signaling pathways (Dusp16 and Rhpn2, respectively) were also found to be down-regulated.
Fig. 3.
Loss of HNF4α in hepatocytes disrupts diverse pathways in the developing liver, including those associated with cell adhesion and junction formation. RT-PCR analysis of control Hnf4a loxP/+; AlfpCre and mutant Hnf4a loxP/loxP; AlfpCre 18.5-dpc livers confirmed that the mRNA levels of multiple cell junction and adhesion genes were down-regulated in mutant livers. Genes with decreased expression by RT-PCR of ≥2.5-fold in HNF4α-null livers compared with control livers are shown. Fold changes predicted from the array analysis using dchip software are listed. All genes tested, with the exception of Gjb2 and Cldn2, were predicted by dchip to have fold changes ≥2.0-fold; P ≤ 0.05. The P value of the fold changes predicted for Gjb2 and Cldn2 exceeded 0.05. Hprt1 was used as a standard to normalize loading.
To identify direct targets of HNF4α within this set of 27 down-regulated cell adhesion protein-encoding genes, we analyzed the complete genomic sequence of each, including −10 kb relative to the transcriptional start site (+1). Our first level of analysis consisted of comparing these genomic sequences with a dataset of known HNF4α binding sites using a Knuth–Morris–Pratt exact match algorithm (28). We identified 14 sites in nine genes matching previously identified HNF4α binding sequences (Table 2, which is published as supporting information on the PNAS web site). These 14 sites were located both upstream and downstream of the transcriptional start site, and each sequence was confirmed to bind HNF4α by EMSA (data not shown). To identify previously undescribed HNF4α binding sites, we used a newly developed permutated Markov model to search the same genomic sequences queried with the exact match algorithm (29). We found that 25 of the 27 cell adhesion genes contained at least one putative HNF4α binding site (Table 2). These sites were also located both upstream and downstream of the transcriptional start site. Because of the large number of sites predicted by the Markov model (97 sites), we decided to focus on those located upstream of the transcriptional start site, presumably in promoter regions (25 sites). To determine whether HNF4α interacts with any of these sequences, we tested each for its ability to compete with a well characterized HNF4α binding site from the human apolipoprotein C-III (APOC3) promoter for binding to exogenous HNF4α protein (30) by EMSA (Fig. 7, which is published as supporting information on the PNAS web site). Of the 25 sequences analyzed, 15 inhibited binding of HNF4α to the radiolabeled APOC3 probe by ≥90%. Together, our prediction methods combined with verification by EMSA yielded 29 confirmed HNF4α binding sites in 18 cell junction and adhesion genes (Fig. 8, which is published as supporting information on the PNAS web site).
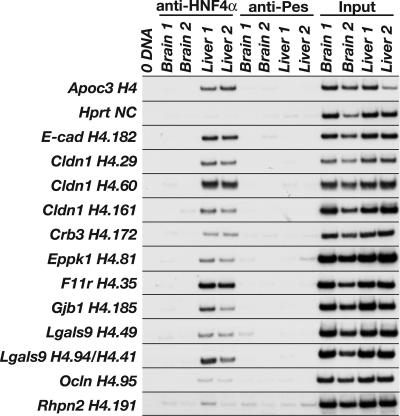
Identifying a particular DNA–protein complex in vitro using EMSA, however, does not demonstrate that such an interaction occurs in vivo. We therefore performed chromatin immunoprecipitation (ChIP) to determine whether HNF4α occupies genomic sequences containing these binding sites in the context of fetal liver. Chromatin isolated from either WT mouse embryonic livers or brains at 18.5 dpc was immunoprecipitated with either an HNF4α-specific antibody, to isolate the fraction of DNA associated with HNF4α, or a nonrelated antibody (anti-Pescadillo), to control for nonspecific immunoprecipitation of DNA. Because HNF4α is not expressed in brain (30), immunoprecipitation of chromatin isolated from embryonic brains served as a control for specificity of the anti-HNF4α antibody. We identified sequences enriched in the pool of anti-HNF4α-precipitated liver chromatin compared with the negative controls using PCR with oligonucleotides flanking the HNF4α binding sites. As expected, we detected HNF4α bound to a previously identified HNF4α binding site in the promoter of the mouse Apoc3 gene (31) and failed to detect HNF4α associated with chromatin from the coding region of exon 9 of the mouse Hprt1 gene, which lacks HNF4α binding sites (Fig. 4). We detected HNF4α occupying its binding sites in 8 of 18 cell junction genes assayed: E-cadherin, Cldn1, Crb3, Eppk1, F11r, Gjb1, Lgals9, and Ocln (Fig. 4). Not all sites that bound HNF4α by EMSA were occupied in vivo; for example, a predicted binding site in the Rhpn2 gene was not precipitated by anti-HNF4α (Fig. 4). Four of the sites that we identified as in vivo binding sequences for HNF4α in fetal livers are located upstream of the transcriptional start site in the presumptive promoter region: E-cadherin (H4.182), Crb3 (H4.172), Gjb1 (H4.185), and Lgals9 (H4.49); the remaining 11 sites are located downstream of the transcriptional start site (Fig. 8). We found HNF4α associated with sites in intron 1 of both the F11r gene (H4.35) and the Ocln gene (H4.95). Two of three positive sites in the Cldn1 gene are also in intron 1 (H4.60 and H4.161); the third site (H4.29) is located in exon 4 in the 3′-UTR. In addition to the site found in the Lgals9 promoter, we detected HNF4α bound to two sites within the gene sequence. Because of their proximity to each other, these sites cannot be distinguished by ChIP. One site (H4.94) is located within exon 3, and the other (H4.41) is located in intron 3. The final site found to bind to HNF4α is located in the unusually large exon 2 of the Eppk1 gene (H4.81) (32). A recent study mapping the location of transcription factor binding sites on human chromosomes 21 and 22 found a greater percentage of sites located within or 3′ to a known gene (36%) compared with those located within or 5′ to the gene (22%), suggesting that regulation of transcription through cis-regulatory elements positioned downstream of transcriptional start sites is a general phenomenon (33). In addition, global genomic analyses of HNF4α binding sites in HepG2 cells revealed that HNF4α was commonly bound to distal sites located both upstream and downstream of the transcriptional start site (34). Because we limited our analysis of novel HNF4α binding sites to sequences located upstream of the transcriptional start site, we cannot exclude the possibility that additional sequences predicted to be within these genes function as bona fide HNF4α binding sites. Therefore, the number of genes encoding cell adhesion proteins directly regulated by HNF4α is likely underrepresented by this analysis.
Fig. 4.
HNF4α occupies sites in several cell junction and adhesion genes requiring HNF4α for their expression. ChIP showed that HNF4α occupies sites in 8 of the 18 genes assayed. We performed ChIP using chromatin isolated from independent 18.5-dpc WT mouse brains and livers and antibodies that immunoprecipitate either HNF4α (anti-HNF4α) or a nonrelated protein, PES1 (anti-Pescadillo). Input samples confirmed that equivalent amounts of chromatin were used in each ChIP reaction. ChIP of a known HNF4α binding site from the Apoc3 gene is shown as a positive control, and ChIP of a sequence lacking an HNF4α binding site from the Hprt1 gene is shown as a negative control. Two sites in the Lgals9 gene, H4.94 and H4.41, were treated as a single site because their proximity to each other prevents them from being discerned by ChIP. With the exception of site H4.191 in the Rhpn2 gene, which provides an example of a predicted HNF4α site we scored as negative by ChIP, only sites found to be occupied by HNF4α are shown.
In summary, we conclude that HNF4α is essential for the expression of a multitude of genes encoding cell junction and adhesion proteins during embryonic development of the mouse liver. These genes encode proteins involved in all aspects of cell adhesion, including the formation of adherens junctions, tight junctions, desmosomes, and gap junctions, as well as proteins involved in epithelial polarization, cytoskeletal organization, and signal transduction. Our data demonstrate that HNF4α is bound to regulatory elements within many of these genes in vivo. Thus, HNF4α appears to directly activate the expression of genes encoding many cell adhesion molecules. However, the phenotype associated with loss of HNF4α is complex, and it is likely that indirect mechanisms also participate. For example, we know from other studies that HNF4α regulates the expression of other liver transcription factors, including HNF1α (35, 36). Moreover, disruption of the signaling cascades regulated by the junctions themselves is also likely to affect the phenotype of HNF4α-null livers. Although our data demonstrate that expression of several key adhesion and junction proteins is lost from HNF4α-null livers, it does not necessarily mean that these factors alone are sufficient to form a hepatic epithelium. Nevertheless, our finding that HNF4α is responsible for expression of such a large and diverse repertoire of cell adhesion and cell junction proteins strongly supports the proposal that HNF4α acts as an orchestrator of epithelial morphogenesis in the developing liver primarily by coordinating the formation of cell and junction adhesions. As such, HNF4α has a critical role in the development of a functional hepatic epithelium, which in turn is a prerequisite for the correct physiological activity of the liver.
Materials and Methods
Production of Mice.
Derivation of Hnf4_a_loxP/+, Hnf4_a_loxP/loxP, Cdh1 loxP/+, Cdh1 loxP/loxP, and AlfpCre mice has been described (14, 15, 19). Embryonic mice were generated by timed matings, considering noon on the day we found a vaginal plug as 0.5 dpc. Genotypes were determined by using PCR analysis of tail or ear punch DNA following standard protocols. PCR primers used were: Cre, gttcgcaagaacctgatggaca, ctagagcctgttttgcacgttc; Cdh1 loxP, gtgacaggaaaggcatatcagcaacaagat, gtgagctggtacccatggaggacactga; and Hnf4a loxP, ccgaagatagggccatgttgga, ccgaggtggatttccaacaga. The Medical College of Wisconsin's Animal Care Committee approved all animal procedures used in this study.
RT-PCR.
We carried out RT-PCR as described (37) using total RNA isolated from either adult (5-wk) or embryonic (18.5-dpc) livers [Qiagen (Valencia, CA) RNeasy kit]. To quantify fold changes in gene expression, a PhosphorImager (Molecular Dynamics) scanner was used, and samples were normalized to the level of Hprt1 expression. Primer sequences are provided in Table 2.
Immunoblotting.
Protein was extracted from livers of 18.5-dpc or 5-wk-old mice, and 75 or 100 μg was used for immunoblotting. Proteins separated by SDS/PAGE were transferred to Immun-Blot poly(vinylidene difluoride) membrane (Bio-Rad) by wet transfer by using buffer containing 25 mM Tris, 192 mM glycine, and 10% methanol. Antibodies against the following proteins were used: E-cadherin (mouse monoclonal; BD Biosciences; 1:2,500), OCLN (rabbit polyclonal; Zymed; 1:500), β-actin (mouse monoclonal; Sigma; 1:10,000), CLDN1 (rabbit polyclonal; Zymed; 1:500), F11r (rabbit polyclonal; Zymed; 1:250), and DSC2 (7G6, mouse monoclonal, a gift of M. Wheelock, University of Nebraska Medical Center, Omaha, NE; 1:5). Secondary antibodies used were goat anti-rabbit–horseradish peroxidase (Bio-Rad; 1:6,000) or goat anti-mouse–horseradish peroxidase (Bio-Rad; 1:10,000). SuperSignal West Pico chemiluminescent substrate (Pierce) was used according to the manufacturer's instructions.
Immunohistochemistry, Immunofluorescence, and Confocal Microscopy.
For immunohistochemistry, paraffin sections (5 μm) of 18.5-dpc mouse livers were stained by using an antibody against E-cadherin (mouse monoclonal, BD Biosciences; 1:16,000) as described (15). For immunofluorescence and confocal microscopy, frozen sections (10 μm) of livers from control and experimental 18.5-dpc embryos or 5-wk-old mice were stained as described (38). Antibodies against the following proteins were used: claudin-1 (rabbit polyclonal; Zymed; 1:100), connexin 32 (rabbit polyclonal; Zymed; 1:200), ZO1 (rabbit polyclonal; Zymed; 1:200), and Alexa-Fluor 488 goat anti-rabbit (Invitrogen; 1:500). TO-PRO-3 iodide (Invitrogen) was used to stain nuclei. Confocal images (×100) were acquired by using a Leica (Deerfield, IL) TCS SP2 laser-scanning confocal microscopic imaging system.
Electron Microscopy.
Adult (5-wk) livers were fixed in 2.5% gluteraldehyde in 0.1 M cacodylate buffer and embedded in EPON 812 epoxy resin. Sections of 60-nm thickness were contrasted with uranyl acetate and lead citrate and examined by using a Hitachi (Tokyo) 600 transmission electron microscope.
Oligonucleotide Array Analysis.
Total RNA (15 μg) isolated from three independent control (Hnf4_a_loxP/+; Alfp-Cre) and experimental (Hnf4_a_loxP/loxP; Alfp-Cre) 18.5-dpc livers by using the Qiagen (Valencia, CA) RNeasy kit was used to prepare biotinylated cRNA following the protocol described in the Affymetrix Expression Analysis Technical Manual. We hybridized a total of six GeneChip Mouse Genome 430 2.0 arrays (Affymetrix), three for control samples and three for experimental samples, with 15 μg of fragmented cRNA. Images were acquired by using a GeneChip Scanner 3000 (Affymetrix). genechip Operating Software (gcos) from Affymetrix and dchip Ver. 1.3 software (25) were used in combination to analyze the data. For genes to be considered down-regulated in experimental livers, we required that the gene's expression be called present in all control samples using presence and absence calls generated by gcos software. Conversely, for genes to be considered up-regulated, we required that the signal be called present in all experimental samples. Mean values for gene expression along with P values were determined by using dchip software. Genes were classified by biological process using the Mouse Genome Informatics GO_SLIM Chart tool (www.spatial.maine.edu/∼mdolan/MGI_GO_Slim_Chart.html).
EMSA.
Radiolabeled APOC3 probe was incubated with 150-fold molar excess of each cold putative HNF4α binding site and nuclear extract from HNF4α-transfected COS-7 cells, as described with minor modifications (39). The amount of APOC3 probe shifted in the presence of each competitor was quantified by using a PhosphorImager scanner. The amount of shifted probe present in two independent competition experiments was averaged, and those sequences able to inhibit binding of APOC3 to exogenous HNF4α by ≥90% were analyzed by ChIP. Binding site sequences tested by EMSA are provided in Table 2.
ChIP.
Livers and brains harvested from 18.5-dpc CD1 embryos were fixed in 1% formaldehyde for 15 min at room temperature. Cells were isolated by homogenizing tissue using a Medimachine (BD Biosciences). ChIPs were performed by using the Upstate Biotechnology (Lake Placid, NY) ChIP Assay Kit following the manufacturer's instructions with minor modifications. Each sample used for ChIP contained chromatin pooled from two independent livers or brains. Precleared liver or brain chromatin was immunoprecipitated with 1 μg of either anti-HNF4α (H-171; Santa Cruz Biotechnology) or anti-Pescadillo (nonspecific antibody control; see ref. 40) overnight at 4°C. After treatment of eluted complexes with proteinase K and RNase A, chromatin was recovered by phenol–chloroform extraction and ethanol precipitation. Immunoprecipitated chromatin was detected by PCR amplification using primers flanking each predicted HNF4α binding site and [α-_32_P]dATP. Primer sequences are provided in Table 2. Amplicons were separated in 4% polyacrylamide gels and visualized by using autoradiography. A fraction of diluted precleared chromatin was treated identically to immunoprecipitates to yield the input fraction. Input chromatin was further diluted 100-fold before PCR amplification.
Supplementary Material
Supporting Information
Acknowledgments
We thank Klaus Kaestner (University of Pennsylvania, Philadelphia) and B. Knowles (The Jackson Laboratory) for providing Alfp-Cre and Cdh1 loxP/loxP mice, respectively; M. Wheelock (University of Nebraska Medical Center, Omaha, NE) for providing the antibody against DSC2; M. Dolan (University of Maine, Orono) for assistance in using the Mouse Genome Informatics GO_SLIM Chart tool; and C. Wells (Medical College of Wisconsin) for performing electron microscopy. We also thank J. Besharse and P. Traktman for critically reading the manuscript. Funding for this project was provided by grants from the National Institutes of Health (to S.A.D., F.M.S., M.A.B., and G.K.).
Abbreviations
HNF4α
hepatocyte nuclear factor 4α
dpc
days postcoitum
ChIP
chromatin immunoprecipitation.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE3126).
References
- 1.Gumbiner B. M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Moreno M., Jamora C., Fuchs E. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 3.Matter K., Balda M. S. Nat. Rev. Mol. Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 4.Schneeberger E. E., Lynch R. D. Am. J. Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 5.Wodarz A. Nat. Cell Biol. 2002;4:E39–E44. doi: 10.1038/ncb0202-e39. [DOI] [PubMed] [Google Scholar]
- 6.Getsios S., Huen A. C., Green K. J. Nat. Rev. Mol. Cell Biol. 2004;5:271–281. doi: 10.1038/nrm1356. [DOI] [PubMed] [Google Scholar]
- 7.Sakaguchi T., Gu X., Golden H. M., Suh E., Rhoads D. B., Reinecker H. C. J. Biol. Chem. 2002;277:21361–21370. doi: 10.1074/jbc.M110261200. [DOI] [PubMed] [Google Scholar]
- 8.Stemmler M. P., Hecht A., Kemler R. Development (Cambridge, U.K.) 2005;132:965–976. doi: 10.1242/dev.01662. [DOI] [PubMed] [Google Scholar]
- 9.Stemmler M. P., Hecht A., Kinzel B., Kemler R. Dev. Dyn. 2003;227:238–245. doi: 10.1002/dvdy.10301. [DOI] [PubMed] [Google Scholar]
- 10.Bolos V., Peinado H., Perez-Moreno M. A., Fraga M. F., Esteller M., Cano A. J. Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 11.Batlle E., Sancho E., Franci C., Dominguez D., Monfar M., Baulida J., Garcia De Herreros A. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 12.Cano A., Perez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 13.Hajra K. M., Chen D. Y., Fearon E. R. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 14.Parviz F., Li J., Kaestner K. H., Duncan S. A. Genesis. 2002;32:130–133. doi: 10.1002/gene.10058. [DOI] [PubMed] [Google Scholar]
- 15.Parviz F., Matullo C., Garrison W. D., Savatski L., Adamson J. W., Ning G., Kaestner K. H., Rossi J. M., Zaret K. S., Duncan S. A. Nat. Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 16.Gumbiner B., Stevenson B., Grimaldi A. J. Cell Biol. 1988;107:1575–1587. doi: 10.1083/jcb.107.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larue L., Ohsugi M., Hirchenhain J., Kemler R. Proc. Natl. Acad. Sci. USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young P., Boussadia O., Halfter H., Grose R., Berger P., Leone D. P., Robenek H., Charnay P., Kemler R., Suter U. EMBO J. 2003;22:5723–5733. doi: 10.1093/emboj/cdg560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boussadia O., Kutsch S., Hierholzer A., Delmas V., Kemler R. Mech. Dev. 2002;115:53–62. doi: 10.1016/s0925-4773(02)00090-4. [DOI] [PubMed] [Google Scholar]
- 20.Tunggal J. A., Helfrich I., Schmitz A., Schwarz H., Gunzel D., Fromm M., Kemler R., Krieg T., Niessen C. M. EMBO J. 2005;24:1146–1156. doi: 10.1038/sj.emboj.7600605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tinkle C. L., Lechler T., Pasolli H. A., Fuchs E. Proc. Natl. Acad. Sci. USA. 2004;101:552–557. doi: 10.1073/pnas.0307437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young P., Boussadia O., Berger P., Leone D. P., Charnay P., Kemler R., Suter U. Mol. Cell. Neurosci. 2002;21:341–351. doi: 10.1006/mcne.2002.1177. [DOI] [PubMed] [Google Scholar]
- 23.Chiba H., Gotoh T., Kojima T., Satohisa S., Kikuchi K., Osanai M., Sawada N. Exp. Cell Res. 2003;286:288–297. doi: 10.1016/s0014-4827(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 24.Satohisa S., Chiba H., Osanai M., Ohno S., Kojima T., Saito T., Sawada N. Exp. Cell Res. 2005;310:66–78. doi: 10.1016/j.yexcr.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 25.Li C., Wong W. H. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., et al. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naiki T., Nagaki M., Shidoji Y., Kojima H., Imose M., Kato T., Ohishi N., Yagi K., Moriwaki H. J. Biol. Chem. 2002;277:14011–14019. doi: 10.1074/jbc.M105403200. [DOI] [PubMed] [Google Scholar]
- 28.Knuth D. E., Morris J. J. H., Pratt V. R. SIAM J. Comput. 1977;6:323–350. [Google Scholar]
- 29.Ellrott K., Yang C., Sladek F. M., Jiang T. Bioinformatics. 2002;18(Suppl. 2):S100–S109. doi: 10.1093/bioinformatics/18.suppl_2.s100. [DOI] [PubMed] [Google Scholar]
- 30.Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 31.del Castillo-Olivares A., Campos J. A., Pandak W. M., Gil G. J. Biol. Chem. 2004;279:16813–16821. doi: 10.1074/jbc.M400646200. [DOI] [PubMed] [Google Scholar]
- 32.Spazierer D., Fuchs P., Proll V., Janda L., Oehler S., Fischer I., Hauptmann R., Wiche G. J. Biol. Chem. 2003;278:31657–31666. doi: 10.1074/jbc.M303055200. [DOI] [PubMed] [Google Scholar]
- 33.Cawley S., Bekiranov S., Ng H. H., Kapranov P., Sekinger E. A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A. J., et al. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 34.Rada-Iglesias A., Wallerman O., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O. M., Ellis P. D., et al. Hum. Mol. Genet. 2005;14:3435–3447. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- 35.Kuo C. J., Conley P. B., Chen L., Sladek F. M., Darnell J. E., Jr., Crabtree G. R. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 36.Tian J. M., Schibler U. Genes Dev. 1991;5:2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- 37.Li J., Ning G., Duncan S. A. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 38.Itoh M., Yonemura S., Nagafuchi A., Tsukita S. J. Cell Biol. 1991;115:1449–1462. doi: 10.1083/jcb.115.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang G., Nepomuceno L., Hopkins K., Sladek F. M. Mol. Cell. Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lerch-Gaggl A., Haque J., Li J., Ning G., Traktman P., Duncan S. A. J. Biol. Chem. 2002;277:45347–45355. doi: 10.1074/jbc.M208338200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information