Removal of Arginine 332 Allows Human TRIM5α To Bind Human Immunodeficiency Virus Capsids and To Restrict Infection (original) (raw)
Abstract
Human TRIM5α (TRIM5αhu) only modestly inhibits human immunodeficiency virus type 1 (HIV-1) and does not inhibit simian immunodeficiency virus (SIVmac). Alteration of arginine 332 in the TRIM5αhu B30.2 domain to proline, the residue found in rhesus monkey TRIM5α, has been shown to create a potent restricting factor for both HIV-1 and SIVmac. Here we demonstrate that the potentiation of HIV-1 inhibition results from the removal of a positively charged residue at position 332 of TRIM5αhu. The increase in restricting activity correlated with an increase in the ability of TRIM5αhu mutants lacking arginine 332 to bind HIV-1 capsid complexes. A change in the cyclophilin A-binding loop of the HIV-1 capsid decreased TRIM5αhu R332P binding and allowed escape from restriction. The ability of TRIM5αhu to restrict SIVmac could be disrupted by the presence of any charged residue at position 332. Thus, charged residues in the v1 region of the TRIM5αhu B30.2 domain can modulate capsid binding and restriction potency. Therapeutic strategies designed to neutralize arginine 332 of TRIM5αhu might potentiate the innate resistance of human cells to HIV-1 infection.
Primates express dominant restriction factors that block retrovirus infection soon after entry but prior to reverse transcription (1, 2, 5). Genetic studies of virus variants and restriction factor competition studies indicate that the viral capsid is the determinant of susceptibility to restriction (3, 3b, 6, 13, 14). Most early restriction in primates is mediated by TRIM5α (7, 10, 16, 23, 26). TRIM5α is a member of the tripartite motif family of proteins and contains RING, B-box 2, and coiled-coil (RBCC) domains (17). TRIM5α also contains a C-terminal B30.2/SPRY domain, which is required for retroviral restriction (23). Deletion of the B30.2 domain disrupts the ability of the TRIM5α protein to bind viral capsid complexes (20, 24). Differences in the B30.2 domains of TRIM5α proteins from distinct primate species account for patterns of retrovirus restriction. For example, the rhesus monkey TRIM5α (TRIM5αrh) potently blocks human immunodeficiency virus type 1 (HIV-1), which is only weakly inhibited by human TRIM5α (TRIM5αhu) (23). Neither TRIM5αrh nor TRIM5αhu efficiently restricts simian immunodeficiency virus (SIVmac) (23). Four variable regions (v1 to v4) are found in the B30.2 domains of TRIM5α proteins from different primates (19, 21). Differences in the v1 regions of TRIM5αrh and TRIM5αhu account for the differences in anti-HIV-1 potency of these TRIM5α variants (15, 19, 24, 27). Alteration of arginine 332 in the v1 region of TRIM5αhu to the proline residue found in TRIM5αrh results in a protein that can potently restrict HIV-1 and, surprisingly, SIVmac infection (24, 27). Here we investigate the specific v1 sequences in TRIM5αhu required for efficient antiviral activity against HIV-1 and SIVmac and provide a mechanistic explanation for the observed enhancement of restriction that results from changes in this region.
MATERIALS AND METHODS
Plasmids and mutants.
The pLPCX plasmids containing the TRIM5 cDNA from humans and rhesus monkeys have been previously described (23). Plasmids expressing TRIM5αhu with single-amino-acid changes were created by QuikChange mutagenesis (Stratagene). The nomenclature for these mutants is TRIM5αhu followed by the wild-type amino acid residue in single-letter code, amino acid position, and the amino acid residue to which the change was made. In the ΔR332 mutant of TRIM5αhu, arginine 332 is deleted.
Creation of cells stably expressing TRIM5α variants.
Retroviral vectors encoding the wild-type TRIM5αhu or TRIM5αrh proteins or the mutant TRIM5αhu proteins were created using the pLPCX plasmid (23). The LPCX vectors contain only the amino acid-coding sequence of the TRIM5α cDNA. In all constructs, the TRIM5α proteins possess C-terminal epitope tags derived from influenza virus hemagglutinin (HA). Recombinant viruses were produced in 293T cells by cotransfecting these pLPCX plasmids with the pVPack-GFP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. The resulting virus particles were used to transduce approximately 1 × 106 HeLa cells in the presence of 5 μg/ml Polybrene. Cells were selected in 1 μg/ml puromycin (Sigma).
In experiments examining the antiretroviral activity of ΔR332 TRIM5αhu, LPCX retroviral vectors expressing this mutant or the wild-type TRIM5αhu and TRIM5αrh proteins were used to transduce Cf2Th canine thymocytes. These cells, as well as cells transduced with the empty LPCX vector, were selected in 1 μg/ml puromycin (Sigma).
Immunoblotting.
HA-tagged proteins were expressed in HeLa or Cf2Th cells by transduction with LPCX vectors as described above. The HA-tagged TRIM5α proteins were detected in whole-cell lysates in 100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 7.5), 10% glycerol, and 1% Nonidet P40 by Western blotting with horseradish peroxidase (HRP)-conjugated 3F10 antibody (Roche). As a control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected with an HRP-conjugated anti-GAPDH antibody (Abcam). HIV-1 p24 capsid protein was detected using an HRP-conjugated murine anti-p24 monoclonal antibody (ImmunoDiagnostics).
Infection with viruses expressing GFP.
Recombinant HIV-1 and SIVmac vectors expressing green fluorescent protein (GFP) (HIV-1-GFP and SIV-GFP, respectively) were prepared as described previously (8, 13, 14, 23). The Moloney murine leukemia virus vector expressing GFP (MLV-GFP) was prepared by cotransfecting 293T cells with 15 μg of pFB-hrGFP, 15 μg of pVPack-GFP, and 4 μg of pVPack-VSV-G (all from Stratagene). HIV-1 stocks were quantified by measuring reverse transcriptase activity as described previously (18). MLV reverse transcriptase activity was determined by the same procedure except that 20 mM MnCl2 was used instead of MgCl2. For infections, 3 × 104 HeLa or Cf2Th cells seeded in 24-well plates were incubated in the presence of virus for 24 h. Cells were washed and returned to culture for 48 h and then subjected to fluorescence-activated cell sorting (FACS) analysis with a FACScan (Becton Dickinson).
Capsid-binding assay.
Purification of recombinantly expressed HIV-1 capsid-nucleocapsid (CA-NC) protein from Escherichia coli was carried out as previously described (4). High-molecular-weight HIV-1 capsid complexes were assembled using 300 μM CA-NC protein and 60 μM (TG)50 DNA oligonucleotide in a volume of 100 μl of 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl. The reaction was allowed to proceed overnight at 4°C as previously described (4). The assembled CA-NC complexes were stored at 4°C until needed.
For a source of TRIM5 protein, 106 293T cells seeded in a six-well dish were transfected with 1 μg of the appropriate pLPCX-TRIM5 plasmid, using Lipofectamine 2000 according to the manufacturer's protocol. Forty-eight hours later, the cells were harvested in phosphate-buffered saline containing 5 mM EDTA and resuspended in 250 μl of hypotonic lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM EDTA) and placed on ice for 15 min. The cells were lysed by using a 2-ml Dounce homogenizer (pestle B, 15 strokes), and the cell debris was removed by centrifugation at 4°C for 10 min at maximum speed (14,000 × g) in an Eppendorf microcentrifuge. Fifty microliters of lysate was saved for assessment of the input amount of TRIM5 in the assay. Two hundred microliters of the cleared cell lysate was combined with 5 μl of HIV-1 CA-NC complexes from the assembly reaction mixture, and the concentration of NaCl was adjusted to 150 mM. The mixture was incubated for 1 hour at room temperature with gentle mixing. After incubation, the mixture was layered onto a 3.5-ml 70% sucrose cushion (prepared in 1 × phosphate-buffered saline) and centrifuged at 110,000 × g for 1 hour at 4°C in a Beckman SW55Ti rotor. The pellet was resuspended in 50 μl of 1 × sodium dodecyl sulfate (SDS) sample buffer and subjected to SDS-polyacrylamide gel electrophoresis and Western blotting for TRIM5α and HIV-1 p24 capsid protein as described above.
RESULTS AND DISCUSSION
Impairment of anti-HIV-1 activity by a basic residue at position 332 of TRIM5αhu.
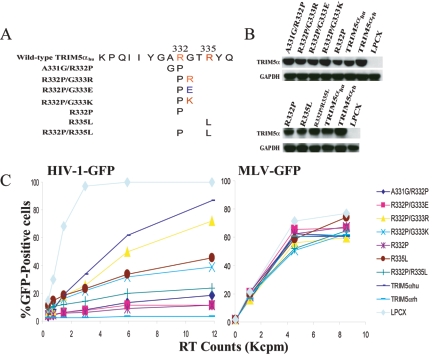
A TRIM5αhu mutant, TRIM5αhu R332P, exhibits significantly greater inhibitory activity against HIV-1 than wild-type TRIM5αhu does (24, 27). To investigate whether the proline substitution is required for potentiating the anti-HIV-1 ability of TRIM5αhu, mutant proteins in which arginine 332 is replaced by different amino acid residues were created (Fig. 1A). HeLa cells were transduced by LPCX retroviral vectors expressing the wild-type and mutant TRIM5αhu proteins as described previously (23). All of the TRIM5αhu proteins have a carboxy-terminal HA tag, allowing documentation of expression levels in the transduced cells. Figure 1B shows the similar steady-state expression levels of the different mutant TRIM5αhu proteins. Compared with HeLa cells transduced with the empty LPCX vector, HeLa cells expressing TRIM5αhu R332P were strongly resistant to infection with HIV-1-GFP (Fig. 1C). A modest decrease in HIV-1-GFP infection was observed in cells expressing wild-type TRIM5αhu. HeLa cells expressing every TRIM5αhu mutant tested, except TRIM5αhu R332K, were almost as resistant to infection by HIV-1-GFP as cells expressing TRIM5αhu R332P. An intermediate level of HIV-1-GFP inhibition was observed in the cells expressing the TRIM5αhu R332K protein. Infection of the cells expressing the various TRIM5α variants by Moloney MLV-GFP was similar to that of the control cells transduced with the empty pLPCX vector. We conclude that removal of a positively charged side chain in residue 332 can significantly potentiate the anti-HIV-1 activity of TRIM5αhu. Acquisition of a proline residue at this position is not essential for this potentiation.
FIG. 1.
Impairment of TRIM5αhu anti-HIV-1 activity by a positively charged residue at position 332. (A) The TRIM5αhu protein is depicted, with the domains shown (B2, B-box 2). Residue numbers are indicated, as are the locations of the v1 to v3 variable regions of the B30.2 domain. The primary amino acid sequence of a segment of v1 is shown, with the changes in residue 332 for each of the mutants indicated. The analogous region of TRIM5αrh is shown below for comparison. (B) The expression of the TRIM5α variants was measured by Western blotting HeLa cell lysates with an antibody directed against the HA epitope tag. As a control, the lysates were Western blotted with an anti-GAPDH antibody. (C) HeLa cells expressing the wild-type and mutant TRIM5α proteins or control HeLa cells transduced with the empty LPCX vector were incubated with various amounts of HIV-1-GFP or Moloney MLV (MLV-GFP). Infected GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. RT, reverse transcriptase.
To investigate further the effect of v1 region charge on HIV-1 restriction, additional TRIM5αhu R332P mutants were created in which basic and acidic amino acid residues were introduced at positions adjacent to residue 332 (Fig. 2A). After verifying that approximately equivalent levels of the mutant proteins were made in transduced HeLa cells (Fig. 2B), the susceptibility of the cells to HIV-1-GFP infection was tested. The mutants that contain amino acid residues with an uncharged side chain (A331G/R332P) or negatively charged side chain (R332P/G333E) adjacent to proline 332 strongly inhibited HIV-1 infection (Fig. 2C and Table 1). In contrast, the inhibition observed for the R332P/G333R and R332P/G333K mutants, both of which have basic residues adjacent to proline 332, was much less than that seen for the R332P protein. We conclude that positively charged amino acids adjacent to residue 332 diminish the restriction potency of R332P against HIV-1.
FIG. 2.
Effects of amino acid substitutions adjacent to TRIM5αhu residue 332 on HIV-1 infection. (A) A diagram of amino acid substitutions in the v1 region of the TRIM5αhu B30.2 domain is shown. (B) The expression of the TRIM5α variants was measured by Western blotting HeLa cell lysates with an antibody directed against the HA epitope tag. As a control, the lysates were Western blotted with an anti-GAPDH antibody. (C) HeLa cells expressing the wild-type and mutant TRIM5α proteins or control HeLa cells transduced with the empty LPCX vector were incubated with various amounts of HIV-1-GFP or MLV-GFP. Infected GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. Reverse transcriptase (RT) activity (in counts per minute [in thousands] [Kcpm]) is shown.
TABLE 1.
Antiviral activities of human TRIM5α variants
| TRIM5αhu variant | Amino acid sequencea | Charge on residue 332 | Charge on residues adjacent to residue 332 | Restrictionbof: |
|---|---|---|---|---|
| HIV-1 | SIVmac | |||
| Wild-type | GARGTRYQ | Positive | + | + |
| R332A | A | +++ | ++ | |
| R332G | G | +++ | ++ | |
| R332P | P | +++ | +++ | |
| R332K | K | Positive | + | + |
| R332D | D | Negative | +++ | + |
| R332E | E | Negative | +++ | + |
| R332H | H | +++ | ++ | |
| R332Q | Q | +++ | +++ | |
| R332S | S | +++ | ++ | |
| ΔR332 | - | ++ | +++ | |
| R335L | L | ++ | + | |
| A331G/R332P | GP | +++ | +++ | |
| R332P/G333E | PE | Negative | +++ | +++ |
| R332P/G333R | PR | Positive | + | + |
| R332P/G333K | PK | Positive | ++ | + |
| R332P/R335L | PL | ++/+++ | + |
To examine whether the arginine residue at position 335 influences HIV-1 restriction, we altered this residue in TRIM5αhu and TRIM5αhu R332P. The TRIM5αhu variant R335L, in which the arginine residue at position 335 is substituted by a corresponding residue in TRIM5αrh, exhibited only a modest level of HIV-1-GFP inhibition (Fig. 2C and Table 1). The double mutant R332P/R335L exhibited no more inhibitory effect against HIV-1 than TRIM5αhu R332P did. Thus, the positively charged residue at position 335 interferes much less with the anti-HIV-1 activity of TRIM5αhu than the positive charge at position 332.
Sequence requirements in the TRIM5αhu B30.2 v1 region for SIVmac restriction.
The TRIM5αhu R332P mutant has been shown to inhibit SIVmac infection potently, a property possessed by neither TRIM5αhu nor TRIM5αrh (24, 27). The abilities of the TRIM5αhu mutants to restrict SIVmac infection were studied. Most of the TRIM5αhu mutants inhibited SIVmac infection similarly to their ability to restrict HIV-1 infection (Fig. 3 and Table 1). Two variants, TRIM5αhu R332D and TRIM5αhu R332E, with acidic residues at position 332, were exceptions. Both mutants, which exhibited potent restriction against HIV-1 (Fig. 1C), only minimally inhibited SIV-GFP infection (Fig. 3 and Table 1). We conclude that the influence of amino acid variation in the TRIM5αhu B30.2 v1 region on SIVmac inhibitory activity is different than the effect on HIV-1 inhibition. In particular, SIVmac inhibition is sensitive to the presence of any charged residue at position 332 of TRIM5αhu, whereas HIV-1 inhibition is sensitive only to positively charged residues at this position. Also, the R335L change in TRIM5αhu R332P is more detrimental to SIVmac inhibition than to HIV-1 inhibition.
FIG. 3.
Effects of the expression of TRIM5αhu mutants on SIVmac infection. (A and B) HeLa cells expressing the wild-type and mutant TRIM5α proteins or control HeLa cells transduced with the empty LPCX vector were incubated with various amounts of SIV-GFP. Infected GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. RT, reverse transcriptase.
Effects of changes in the TRIM5αhu B30.2 v1 region on specific binding to the HIV-1 capsid.
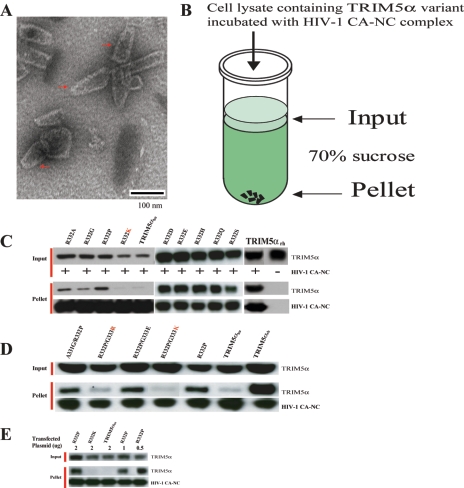
Next we investigated the mechanism whereby the above amino acid substitutions altered the anti-HIV-1 activity of TRIM5αhu. The HIV-1 capsid protein has been shown to be the major determinant of TRIM5α-mediated restriction (3, 3b, 6, 13, 14, 23). The ability of TRIM5α relatives from different species to associate with the assembled HIV-1 capsid complex has recently been shown to correlate with the ability to restrict HIV-1 infection (22). For example, TRIM5αrh, which potently restricts HIV-1 infection, strongly associated with the HIV-1 capsid complexes; TRIM5αhu, which only moderately restricts HIV-1 infection, only weakly associated with the HIV-1 capsid complexes. Therefore, we tested the hypothesis that the amino acid changes that potentiate the anti-HIV-1 activity of TRIM5αhu also enhance its ability to associate specifically with the HIV-1 capsid. HIV-1 capsid complexes were assembled from purified recombinant capsid-nucleocapsid (CA-NC) protein as described previously (4, 12). In the presence of nucleic acid and high salt, the CA-NC protein assembles into highly stable cylindrical and conical structures, which recapitulate the authentic surface lattice structure of native HIV-1 capsids (4, 12). We verified by electron microscopy that our CA-NC preparations contained capsid-like structures (Fig. 4A). After incubation of the HIV-1 CA-NC complexes with TRIM5α-containing cell lysates, the mixtures were layered onto 70% sucrose cushions (Fig. 4B). The TRIM5αrh protein did not sediment through 70% sucrose in the absence of added CA-NC complexes (Fig. 4C, TRIM5αrh lane). Consistent with our previous results (22), the TRIM5αrh protein efficiently cosedimented with the HIV-1 CA-NC complexes, whereas TRIM5αhu only weakly associated with the HIV-1 CA-NC complexes (Fig. 4C to E). The TRIM5αhu R332P mutant exhibited substantially greater binding to the HIV-1 CA-NC complexes than the parental TRIM5αhu protein did (Fig. 4C and D). Every TRIM5αhu variant with a basic residue at or adjacent to position 332 (i.e., TRIM5αhu R332K, TRIM5αhu R332P/G333R,TRIM5αhu R332P/G333K, and the parental TRIM5αhu) cosedimented with the HIV-1 CA-NC complexes much less efficiently than TRIM5αhu R332P did (Fig. 4C to E). In contrast, TRIM5αhu mutants lacking a positively charged residue at or near position 332 exhibited strong binding to the HIV-1 CA-NC complexes. We conclude that a positively charged amino acid at or adjacent to position 332 of TRIM5αhu reduces its binding to the HIV-1 capsid. Our results support the hypothesis that an increase in the affinity for the HIV-1 capsid contributes to the enhanced HIV-1-restricting ability of TRIM5αhu variants lacking a basic residue in the vicinity of position 332.
FIG. 4.
Contribution of HIV-1 capsid binding to the HIV-1-restricting activity of TRIM5αhu mutants. (A) Transmission electron microscopic image of negatively stained CA-NC complexes formed by the assembly of the HIV-1 CA-NC protein and DNA oligonucleotides. Conical structures are marked by the red arrows. (B) To determine the abilities of TRIM5α variants to bind the HIV-1 capsid complexes, in vitro-assembled HIV-1 CA-NC complexes were mixed with 293T cell lysates containing the TRIM5α proteins and layered onto 70% sucrose cushions before centrifugation. Immediately prior to mixing, aliquots of the cell lysates were removed and blotted with anti-HA antibodies to determine the steady-state expression levels of the different TRIM5α variants (Input). After centrifugation, the pellets were resuspended in SDS sample buffer and analyzed by Western blotting using an anti-HA antibody (to detect TRIM5α) or an anti-p24 antibody (to detect the HIV-1 CA-NC protein). (C, D, and E) The input cell lysates and pellets from the binding assays were analyzed for the presence of TRIM5α and the HIV-1 CA-NC protein by Western blotting. In panel E, the amount (in micrograms) of plasmid transfected into the 293T cells was varied as indicated to allow comparison of the HIV-1 capsid-binding ability of the wild-type and mutant TRIM5αhu proteins.
Effects of deletion of TRIM5αhu arginine 332 on capsid binding and virus inhibition.
The above results suggest that arginine 332 inhibits the ability of TRIM5αhu to bind HIV-1 capsids and to inhibit viral infection. To test this model, the ΔR332 mutant of TRIM5αhu, in which arginine 332 is deleted, was made and tested. The ΔR332 mutant was expressed stably in Cf2Th canine thymocytes at levels comparable to those of the wild-type TRIM5αhu and TRIM5αrh proteins (Fig. 5A). The ΔR332 mutant inhibited HIV-1 infection more effectively than TRIM5αhu did but not as efficiently as TRIM5αrh did (Fig. 5B). Cells expressing the ΔR332 protein were very resistant to SIVmac infection (Fig. 5C). The ΔR332 mutant bound HIV-1 CA-NC complexes more efficiently than the wild-type TRIM5αhu protein did (Fig. 5D). Thus, removal of residue 332 in TRIM5αhu results in an improvement in HIV-1 capsid binding and in the restriction of infection by both HIV-1 and SIVmac.
FIG. 5.
Effects of deletion of TRIM5αhu residue 332 on viral inhibition and capsid binding. (A) The expression of the TRIM5α variants in Cf2Th cells was measured by Western blotting cell lysates with an antibody directed against the HA epitope tag. As a control, the lysates were Western blotted with an anti-GAPDH antibody. (B and C) Cf2Th cells expressing the indicated TRIM5α variants or control Cf2Th cells transduced with the empty LPCX vector were incubated with various amounts of HIV-1-GFP (B) or SIV-GFP (C). Infected GFP-positive cells were counted by FACS. Reverse transcriptase (RT) activity (in counts per minute [in thousands] [Kcpm]) is shown. (D) HIV-1 CA-NC complexes were mixed with 293T cell lysates containing the indicated TRIM5α proteins and layered onto 70% sucrose cushions before centrifugation. The input and pelleted proteins were analyzed by Western blotting as described in the legend to Fig. 4.
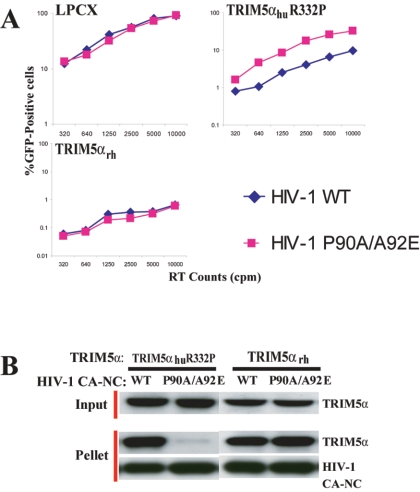
TRIM5αhu R332P binding and restriction of an HIV-1 capsid mutant.
Some HIV-1 mutants with alterations in the cyclophilin A-binding loop of the N-terminal capsid domain are less susceptible to restriction in monkey cells and, as virus-like particles, compete for the antiviral restricting factor(s) less effectively than wild-type HIV-1 does (11, 13). In the course of our studies, we identified an HIV-1 capsid mutant, P90A/A92E, that exhibited different susceptibility to TRIM5αrh and TRIM5αhu R332P. The changes in the cyclophilin A-binding loop of this capsid mutant also decrease cyclophilin A binding (3a, 6, 25). Figure 5A shows the results of infecting control HeLa cells or HeLa cells expressing TRIM5αrh or TRIM5αhu R332P with either wild-type HIV-1-GFP or HIV-1(P90A/A92E)-GFP. Both viruses infected the control HeLa cells transduced with the empty LPCX vector efficiently. Both viruses were blocked equally by TRIM5αrh. However, the HIV-1(P90A/A92E)-GFP was less sensitive to restriction by TRIM5αhu R332P than the wild-type HIV-1-GFP was (FIG. 6A). To test whether TRIM5α-capsid binding could explain these observations, we constructed and assembled HIV-1 CA-NC complexes containing the P90A/A92E changes. The association of TRIM5αrh and TRIM5αhu R332P with the assembled wild-type and P90A/A92E HIV-1 CA-NC complexes was studied as described above. The binding of TRIM5αhu R332P to the P90A/A92E HIV-1 capsid complexes was significantly less efficient than binding to the wild-type HIV-1 capsid complex (Fig. 6B). In contrast, TRIM5αrh associated equally with the wild-type and P90A/A92E HIV-1 CA-NC complexes. These results suggest that changes in the cyclophilin A-binding loop of the HIV-1 capsid protein can affect the efficiency of TRIM5α interaction with the assembled capsid. The results support a model in which the diminished sensitivity of the P90A/A92E HIV-1 mutant to TRIM5αhu R332P-mediated restriction is due to a specific decrease in the association of this TRIM5αhu variant with the altered capsid.
FIG. 6.
Sensitivity of TRIM5αhu R332P binding and restriction to a change in the HIV-1 capsid. (A) HeLa cells transduced with the empty LPCX vector or expressing TRIM5αrh or TRIM5αhu R332P were incubated with various amounts of wild-type (WT) HIV-1-GFP or HIV-1(P90A/A92E)-GFP. Infected GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. RT, reverse transcriptase. (B) Binding of TRIM5αrh and TRIM5αhu R332P to wild-type (WT) or P90A/A92E HIV-1 CA-NC complexes was measured using the assay described in the legend to Fig. 4B and Materials and Methods. Proteins in the input and pellet were detected by Western blotting.
The potentiation of the anti-HIV-1 activity of TRIM5αhu by removal of arginine 332 raises the possibility that pharmacologic means of neutralizing the positive charge on this residue might be discovered. As TRIM5αhu is constitutively expressed in most human cells (17), such therapies could enhance innate intracellular resistance to HIV-1 and prevent provirus formation. Recent studies of polymorphism in TRIM5αhu suggest that even small differences in the moderate degree of HIV-1 restriction afforded by this protein may influence the susceptibility of humans to HIV-1 infection (9). The clear contribution of capsid-binding ability to HIV-1-restricting activity supports attempts to understand the TRIM5α-capsid interaction in detail.
Acknowledgments
We thank Yvette McLaughlin for manuscript preparation.
This study was supported in part by the National Institutes of Health (grant AI063987 and a Center for AIDS Research Award [AI60354]), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, the William A. Haseltine Foundation for the Arts and Sciences, and the late William F. McCarty-Cooper. M.S. was supported by a National Defense Science and Engineering Fellowship and is a Fellow of the Ryan Foundation.
REFERENCES
- 1.Besnier, C., L. Ylinen, B. Strange, A. Lister, Y. Takeuchi, S. P. Goff, and G. J. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77**:**13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11**:**286-291. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., M. Bock, G. Towers, and J. P. Stoye. 2001. Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J. Virol. 75**:**5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70**:**5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3b.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99**:**11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283**:**80-83. [DOI] [PubMed] [Google Scholar]
- 5.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 38**:**61-85. [DOI] [PubMed] [Google Scholar]
- 6.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78**:**6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 101**:**10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73**:**10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javanbakht, H., A. Ping, B. Gold, D. C. Petersen, C. O'hUigin, S. J. O'Brien, G. D. Kirk, R. Detels, J. Goedert, S. Buchbinder, S. Donfield, S. Shulenin, B. Song, M. J. Perron, M. Stremlau, J. Sodroski, M. Dean, and C. Winkler. Effects of human TRIM5α polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Submitted for publication. [DOI] [PubMed]
- 10.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101**:**10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100**:**1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407**:**409-413. [DOI] [PubMed] [Google Scholar]
- 13.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78**:**5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77**:**726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79**:**8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101**:**11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20**:**2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rho, H. M., B. Poiesz, F. W. Ruscetti, and R. C. Gallo. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112**:**355-360. [DOI] [PubMed] [Google Scholar]
- 19.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102**:**2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sebastian, S., and J. Luban. 2005. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2**:**40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song, B., B. Gold, C. O'hUigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79**:**6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5a restriction factor. Proc. Natl. Acad. Sci. USA 103**:**5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427**:**848-853. [DOI] [PubMed] [Google Scholar]
- 24.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79**:**3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9**:**1138-1143. [DOI] [PubMed] [Google Scholar]
- 26.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101**:**10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15**:**73-78. [DOI] [PubMed] [Google Scholar]