Gene Expression Profile of Glioblastoma Multiforme Invasive Phenotype Points to New Therapeutic Targets (original) (raw)
Abstract
The invasive phenotype of glioblastoma multiforme (GBM) is a hallmark of malignant process, yet molecular mechanisms that dictate this locally invasive behavior remain poorly understood. Gene expression profiles of human glioma cells were assessed from laser capture-microdissected GBM cells collected from paired patient tumor cores and white matter-invading cell populations. Changes in gene expression in invading GBM cells were validated by quantitative reverse transcription polymerase chain reaction (QRT-PCR) and immunohistochemistry in an independent sample set. QRT-PCR confirmed the differential expression in 19 of 21 genes tested. Immunohistochemical analyses of autotaxin (ATX), ephrin B3, B-cell lymphoma-w (BCLW), and protein tyrosine kinase 2 beta showed them to be expressed in invasive glioma cells. The known GBM markers, insulin-like growth factor binding protein 2 and vimentin, were robustly expressed in the tumor core. A glioma invasion tissue microarray confirmed the expression of ATX and BCLW in invasive cells of tumors of various grades. GBM phenotypic and genotypic heterogeneity is well documented. In this study, we show an additional layer of complexity: transcriptional differences between cells of tumor core and invasive cells located in the brain parenchyma. Gene products supporting invasion may be novel targets for manipulation of brain tumor behavior with consequences on treatment outcome.
Keywords: Glioma, invasion, cDNA microarray, expression profile, tissue microarray
Introduction
Glioblastoma multiforme (GBM) is the most common and most lethal primary malignant brain tumor. These nonmetastatic tumors are highly locally invasive [1], diffusely disseminating into the brain and placing cells outside the margin of therapeutic resection. Current therapies address the bulk of the tumor mass, whereas recurrence is most often within 3 cm of the resection margin [2] and accounts for the fatal outcome of the disease. The infiltrative path of GBM into the normal brain is not random; it often follows white matter tracts and extends along perivascular spaces, the glia limitans externa and the subependyma [3]. Little is known about the distinct biology of invasive glioblastoma cells in situ, but their diffuse infiltration suggests the activation of genetic and cellular programs that distinguish them from cells in the tumor core.
Microarray technology has proven to be very useful in the molecular classification of astrocytic tumor grades [4é9], generating evidence of a molecular evolution driving progressive stages of astrocytoma malignancy. Gene expression analysis enhances histopathologic diagnosis [7,8], specifically of nonclassic tumor histologies, providing a more accurate prognosis [4,10]. It is hoped that molecular characterization of tumor subtypes will lead to the application of therapy customized to a particular tumor's biology. This report illustrates the usage of cDNA microarrays to discern differential gene expression comparing glioma cells at a stationary, proliferative site within the tumor core to cells invading the surrounding brain exhibiting a diffuse, motile behavior. Patterns of gene expression by cells at the tumor core, such as the presence of insulin-like growth factor binding protein 2 (IGFBP2), were consistent with published cDNA microarray characterizations of GBM [5,6,9]. Genes upregulated in invasive cells depict a commitment to motility and invasion, such as the autocrine motility factor, autotaxin (ATX), and protein tyrosine kinase 2 beta (PYK2). Increased expression of the antiapoptotic BCLW and death-associated protein 3 (DAP3), a protein previously found to be transcriptionally upregulated in invasive glioma [11], points to potential interrelationships between motility and apoptosis resistance.
We propose that there is an invasion-specific gene expression profile—one that enables GBM cells to move through the brain parenchyma, creating two distinct subpopulations: the stationary, proliferative tumor core and the motile, invading tumor rim cells. Their distinct gene expression profiles suggest novel therapeutic targets that address dispersed, infiltrating tumor cells. This introduces the possibility of multiagent treatment modalities, specifically targeting invasive cells in conjunction with classic treatments aimed at the proliferating tumor core cells.
Materials and Methods
Clinical Samples and Histology
Human glioblastoma tumor samples were obtained from patients who underwent primary therapeutic subtotal or total tumor resection performed under image guidance. All specimens (13 in number) were collected and submitted to the study under institutional review board-approved protocols. None of the patients had been subjected to chemotherapy or radiotherapy prior to resection. The samples, which were obtained from the main tumor mass and the invasive edge, were immediately frozen on dry ice to be used in laser capture microdissection (LCM). Another portion was fixed in paraformaldehyde and paraffin-embedded for histologic evaluation. Histologic diagnosis was made by standard light microscopic evaluation of hematoxylin and eosin-stained sections. All tumor samples were classified as WHO grade IV GBM [12].
LCM
LCM was performed as described previously [13]. Briefly, 1000 to 2000 tumor core and invasive cells were dissected from 8-µm sections cut from four flash-frozen glioblastoma (WHO grade IV) tumors. Cells in the tumor core were identified and captured; tumor cells immediately adjacent to necrotic areas; cortical areas; cells with small, regular nuclei; and endothelial and blood cells were avoided. White matter-invading GBM cells were identified by means of their nuclear atypia and heteropyknotic staining, which was consistent with that of the cells within the tumor core. Reactive astrocytes were discriminated through their distinct stellate morphology with eosinophilic cytoplasm and large, acentric, round nuclei, and were avoided.
RNA Isolation and Amplification
Total RNA was isolated from 1000 to 2000 LCM cells using the PicoPure RNA Isolation Kit (Arcturus, Mountain View, CA), and quantified by real-time reverse transcription polymerase chain reaction (RT-PCR) performed with the LightCycler (Roche, Mannheim, Germany). This consisted of performing PCR with Histone 3B primers using a serial dilution of cDNA of known concentration as a standard. The remaining RNA (approximately 10 ng) was amplified in two rounds with the RiboAmp RNA Amplification kit (Arcturus), yielding between 30 and 60 mg of copy RNA. RNA from one sample of very diffusely invaded white matter was also isolated (after microscopic inspection) and amplified to address the possible contribution of genetic material admixed from normal brain surrounding the captured invading cells.
cDNA Microarray Analysis
Six micrograms of amplified RNA was labeled in a RT in the presence of dUTP Cy3 (Amersham, Piscataway, NJ) utilizing random hexamers as primers. Universal reference RNA (Stratagene, La Jolla, CA) was amplified for one round in the same manner and labeled with Cy5 dUTP (Amersham). Labeled cDNA was hybridized overnight to 5750 gene cDNA microarray slides (Arizona Cancer Center, Tucson, AZ). (The complete gene list can be found in Supplementary Figure 1.) Following hybridization, slides were washed, scanned, and quantitated with the Axon GenePix 4000 microarray reader (Axon Instruments, Foster City, CA). Gene expression results were analyzed using GeneSpring (Silicon Genetics, Redwood City, CA) software. The measured intensity of each gene was divided by its reference channel (signal from the universal reference RNA) in each sample. When the fluorescence intensity of the reference channel was below 10, the data point was considered uninformative. Intensity-dependent normalization was also applied, where the ratio was reduced to the residual Lowess fit of the intensity versus ratio curve. A fold change analysis was performed to identify differentially expressed genes. The ratios (sample over reference) for the three tumor core experiments and four invasive rim experiments were averaged and compared. Genes that were more than two-fold upregulated or downregulated were selected. Next, to address potential bias due to outliers, the gene lists were further screened by verifying that the same rim/core trend was present across samples that had matched core and rim populations. Genes following the trend in two of the three matched core/invasive rim sets were selected and tabulated. Complete lists of differentially expressed genes can be found in Supplementary Figure 2.
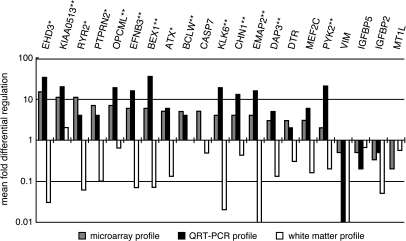
Figure 1.
QRT-PCR validation of gene candidates differentially expressed between invasive rim cells and tumor core cells in cDNA microarray analysis. Names of transcripts analyzed are on the x-axis and the mean fold differential regulation (difference in relative copy number, where 1 represents equal expression in both populations) is on the y-axis. Grey bars represent the mean gene expression levels of invasive tumor cells over tumor core cells seen in the cDNA microarray analysis. Black bars represent the mean gene expression levels of invasive tumor cells over tumor core cells as evaluated by QRT-PCR using seven matched tumor samples. White bars indicate the levels of gene expression (evaluated by QRT-PCR) in the diffusely invaded white matter from one of the samples used in the cDNA microarray analysis divided by the expression levels from either invasive cells (left side, where candidates genes are upregulated) or tumor core cells (right side, where candidate genes are downregulated) of the same sample. *Denotes genes with a significance of P ≤ .05. **Denotes genes with a differential expression that reaches a significance of P ≤ .025 as calculated using the Wilcoxon signed-rank test.
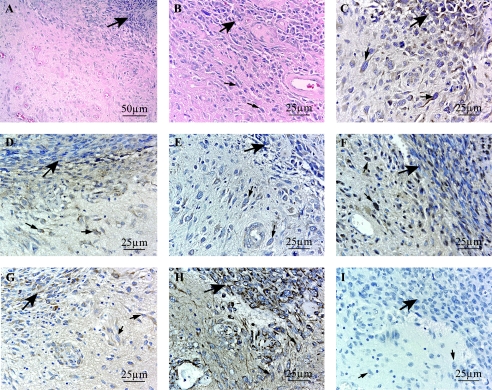
Figure 2.
Immunohistochemical analysis of six candidate genes ATX (C), BCLW (D), EFNB3 (E), PYK2 (F), IGFBP2 (G), and VIM (H); four overexpressed in the invasive rim cells and validated by QRT-PCR; and two underexpressed in the rim, respectively. (A and B) H&E stains of the glioblastoma invasion front. (I) Representative negative control. Large bold arrows point to the tumor core, and smaller arrows point at individually invasive glioblastoma cells. Original magnification, x400, except (A) at x200.
Quantitative RT-PCR (QRT-PCR) Validation
Total RNA was isolated from 500 to 1000 microdissected tumor core and invasive rim cells from 11 additional tumor samples as above. Seven samples were used to transcriptionally validate each gene candidate. Tumor RNA and RNA derived from the white matter adjacent to one of the tumor samples were reverse-transcribed with oligo dT primers using SuperScriptII (Invitrogen, Carlsbad, CA). The resulting cDNA was amplified by PCR with gene-specific primers (the list of primer sequences is available in online Supplementary Figure 3) (Operon/Qiagen, Alameda, CA) using the Light-Cycler and FastStart DNA Master SYBR Green I reagent (Roche). Log-linearity of the amplification curve for each primer set was confirmed down to the picogram range of cDNA. Specificity of PCR products was confirmed by melting curve analysis [14] and agarose gel electrophoresis. PCR protocols are disclosed in online Supplementary Figure 3. Quantification was done using Fit Points method of the LightCycler software version 3.5 [14]. The cDNA amount in each sample was normalized to the crossing point of the housekeeping gene Histone 3B. Relative mRNA fold upregulation in the invasive cells for each gene was calculated using the respective crossing points applied in the following formula: F = 2_(IH-IG)-(CH-CG)_ (adapted from Ref. [15]), where F = fold difference, C = core cells, I = invasive rim cells, G = gene of interest, and H = housekeeping (Histone 3B). Statistical significance of the differential gene regulation was assessed using the Wilcoxon signed-rank test.
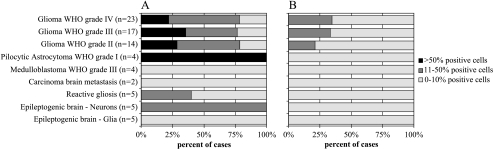
Figure 3.
Summary of immunohistochemical evaluation of ATX and BCLW in a glioma invasion tissue microarray. The tissue type examined and the number of samples in each category are listed on the y-axis. The percentage of cases with positively staining cells within each category is on the x-axis. The shading scale represents the percentage of positively stained cells within a sample. ATX TMA (A) evaluation showing a high degree of positive cells in gliomas of different grades, but also in some neurons and reactive astrocytes. The TMA stained for BCLW (B) reveals its presence in gliomas of all grades.
Immunohistochemical Confirmation of Gene Candidates
The protein products of six gene candidates validated by QRT-PCR were examined by immunohistochemistry on three GBM specimens. Briefly, 6-µm sections were heated for 2 hours at 65°C, deparaffinized in xylene and hydrated in a graded alcohol series, and subjected to antigen-specific epitope retrieval.
This was followed by quenching of endogenous peroxidase activity through incubation in 3% hydrogen peroxide in methanol. Slides were blocked with 10% normal serum in 0.1% Triton X-100 TBS and incubated overnight with the respective antibody at 4°C. Secondary antibodies appropriate to the primary antibody (Vectastain Kits; Vector Laboratories, Burlingame, CA) were added for 1 hour at room temperature, washed, and developed with DAB (Sigma, St. Louis, MO). The slides were counterstained with hematoxylin 2 (Richard-Allen Scientific, Kalamazoo, MI) prior to visualization. Antibody sources and epitope retrieval were as follows: for ATX, slides were microwaved in 10 mM sodium citrate, and antibody 100A (a generous gift from Dr. Tim Clair) was used at a 1:500 dilution. Slides stained for BCLW were digested for 30 minutes at 37°C in 0.5% pepsin in 0.01 N HCl, using a 1:25 dilution BCLW N-19 (Santa Cruz Biotechnology, Santa Cruz, CA). Treatment for EFNB3 included microwaving in 10 mM sodium citrate and a 1:100 concentration of EFNB3 antibody (R&D Systems, Minneapolis, MN). Slides for VIM were digested for 30 minutes at 37°C in 0.5% pepsin in 0.01 N HCl, followed by a 1:200 dilution of VIM 3B4 (DakoCytomation, Carpinteria, CA). Conditions for PYK2 pY402 (1:25; Biosource, Camarillo, CA) were as previously described [16], as were those for IGFBP2 sc-6001 (1:100; Santa Cruz Biotechnology) [17].
Tissue Microarray (TMA) Assembly
A specific glioma invasion TMA was assembled by Dr. D. H. Friedrich using patient-consented cases selected from a database of histologic reports. Gliomas of WHO grades I to IV (n = 69) and control cases (n = 25) including other tumors, reactive gliosis, and “normal” brain specimens from epilepsy surgery were included. Briefly, five equidistant microsamples (600 µm cross section) were punched out of donor paraffin blocks along a histologically verified invasion gradient and arrayed into the TMA using an arraying device (Beecham Instruments, Sun Prairie, WI) as described elsewhere [18]. The TMA paraffin block was then cut in 5-µm slices, which were tape-transferred and subjected to the described staining methods.
Results
Microarray Analysis of Laser Capture-Microdissected Glioma Cells Reveals Two Transcriptional Profiles
Using LCM, we collected two distinct GBM subpopulations based on their pathologic and anatomic context. cDNA gene expression profiling, followed by fold change analysis, resulted in a list of differentially expressed genes that are well documented in glioma biology. Among the genes expressed in the tumor core whose expression is downregulated in invasive GBM cells were IGFBP-2 and IGFBP-5 (Table 1). Several transcriptional regulators involved in growth control were expressed in the core, including ZNF258, EYA2, EGR1, and JUNB. Genes whose products are involved in signal transduction cascades such as PSHL, PIM1, IQGAP, RDC1, and RGS16 were transcriptionally depressed in invasive cells, as were the cytoskeleton-related genes PFN1, MSN, PLEK, VIM, and CAPG. Angiogenesis-related VCAM1 and VEGF, two genes well known in glioma biology, reflect the high degree of neovascularization of GBM. Heightened cellular proliferation, another definitive characteristic of GBM, is represented by MCM2. Interestingly, genes involved in drug resistance such as ABCC3 (multidrug resistance protein 3, MRP3) and metallothioneins, which could play a role in intrinsic drug resistance in gliomas, are highly expressed in tumor core samples and downregulated in invasive cells. Finally, there was evidence of a three-fold lower level of proapoptotic PIG3 (p53-induced gene 3) message in motile cells.
Table 1.
Genes Downregulated in Invasive GBM.
| I/C | Accession Number | Number | Description |
|---|---|---|---|
| Extracellular | |||
| 0.16 | N91385 | MS4A1 | Membrane-spanning 4-domains, A1 |
| 0.2 | AA429895 | ABCC3 | ATP-binding cassette C (CFTR/MRP) |
| 0.25 | AA448569 | SRPX | Sushi repeat-containing protein, X chromosome |
| 0.25 | AA598653 | OSF2 | Osteoblast-specific factor 2 (fasciclin 1-like) |
| 0.33 | N76878 | DEPP | Decidual protein induced by progesterone |
| 0.33 | T77595 | TNC | Tenascin C (hexabrachion) |
| 0.33 | H79047 | IGFBP2 | Insulin-like growth factor binding protein 2 |
| 0.33 | R7563.5 | COLA1 | Collagen, type V, alpha 1 |
| 0.33 | T49159 | SERPIN | Serine (or cysteine) proteinase inhibitor, clade B 2 |
| 0.5 | R71440 | SERPINH2 | Serine (or cysteine) proteinase inhibitor, clade H2 (hsp47) |
| 0.5 | M65062 | IGFBP5 | Insulin-like growth factor binding protein 5 |
| Vascular involvement/angiogenesis | |||
| 0.25 | AA029842 | MTCP1 | Mature T-cell proliferation 1 |
| 0.25 | AA401693 | CD163 | CD163 |
| 0.33 | H16637 | VCAM1 | Vascular cell adhesion molecule 1 |
| 0.33 | AA421296 | CD68 | CD68 antigen |
| 0.33 | A4491191 | IF116 | Interferon gamma-inducible protein 16 |
| 0.5 | R19956 | VEGF | Vascular endothelial growth factor |
| Signal transduction | |||
| 0.09 | W05628 | PSHL | Phosphoserine phosphatase-like |
| 0.09 | W07300 | AP1G1 | Adaptor-related protein complex 1, gamma 1 |
| 0.25 | N63635 | P1M1 | Pim-1 protooncogene gene |
| 0.25 | AA598496 | IQGAP | IQ motif containing GTPase-activating protein 1 |
| 0.25 | AA019996 | PTGER4 | Prostaglandin E receptor 4 (subtype EP4) |
| 0.25 | AA397813 | CKS2 | CDC28 protein kinase regulatory subunit 2 |
| 0.25 | AA446290 | ST5 | Suppression of tumorigenicity 5 |
| 0.25 | AA489246 | ST14 | Suppression of tumorigenicity 14 |
| 0.25 | N53172 | RDCl | G protein-coupled receptor |
| 0.33 | AA443506 | ARHGAP1 | Rho GTPase-activating protein 1 |
| 0.33 | H62028 | DYRK3 | Dual-specificity tyrosine (Y) phosphorylation-regulated kinase 3 |
| 0.33 | AA453774 | RGS16 | Regulator of G-protein signalling 16 |
| 0.33 | AA487560 | CAV1 | Caveolin 1, caveolae protein, 22 kDa |
| 0.33 | AA478542 | AKAP12 | AKAP12 A kinase (PRKA) anchor protein (gravin) 12 |
| 0.5 | AA029737 | TK2 | Thymidine kinase 2, mitochondrial |
| 0.5 | AA496785 | ABL1 | Abelson murine leukemia viral oncogene homolog 1 |
| Cytoskeleton | |||
| 0.09 | AA521431 | PFN1 | Profilin 1 |
| 0.2 | R22977 | MSN | Moesin |
| 0.25 | AA490267 | PLEK | Plekstrin |
| 0.33 | AA486942 | CAPG | Capping protein (actin filament), gelsolin-like |
| 0.33 | AA411440 | VIL2 | Villin (ezrin) |
| 0.4 | AA069414 | GFAP | Glial fibrillary acidic protein |
| 0.5 | AA487812 | VIM | Vimentin |
| Apoptosis | |||
| 0.33 | AA668595 | PIG3 | p53-induced gene 3 |
| 0.33 | AA228130 | PSIP2 | PC4-and SFRSI-interacting protein 2 |
| 0.37 | H45000 | CASP4 | Caspase 4 |
| Transcription | |||
| 0.08 | AA280677 | ZNF258 | Zinc finger protein 258 |
| 0.33 | AA402207 | EYA2 | Eyes absent (Drosophila) homolog 2 |
| 0.33 | N94468 | JUNB | Jun B protooncogene |
| 0.33 | H26183 | CEBPB | CCAAT/enhancer-binding protein (C/EBP), beta |
| 0.5 | P18146 | EGR1 | Early growth response 1 |
| Proliferation | |||
| 0.33 | AA454572 | MCM2 | MCM2 minichromosome maintenance-deficient 2, mitotin |
| Unknown function | |||
| 0.045 | H93118 | H93118 | Hypothetical protein FLJ12592 |
| 0.04 | R76499 | R76499 | Hypothetical protein BCOO7384 |
| 0.11 | N29376 | MNDA | Myeloid cell nuclear differentiation antigen |
| 0.12 | R78516 | SELT | Selenoprotein T |
| 0.14 | AA481758 | DNAJBl | DnaJ (Hsp40) homolog, subfamily B, member 1 |
| 0.2 | AA490991 | HNRPF | Heterogeneous nuclear ribonucleoprotein F |
| 0.2 | N8Ol29 | MTIL | Metallothionein 1L |
| 0.2 | R64251 | DDX38 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 38 |
| 0.25 | AA486518 | CLIC1 | Chloride intracellular channel 1 |
| 0.33 | N49629 | UBD | Ubiquitin D |
| 0.33 | AA459318 | TPD52 | TPD52 tumor protein D52 |
The transcriptome of invasive GBM cells illustrates their biologic distinctivenes from their cognate tumor cores. Gene candidates found to be upregulated two-fold or greater in invasive cells suggests that they are functionally distinct cells (Table 2). There was a preponderance of genes involved in adhesion (OPCML and SPOCK), extracellular signal transduction (PTPRN2, DKK3, EFNB3, GRIN2A, FGFR3, EGFR, GPR19, and DTR), and cytoskeletal rearrangement (INA, EMAP2, CHN1, and PYK2). The serine proteinase KLK6 was the only extracellular matrix-degrading enzyme differentially expressed among the genes on the chip. We also observed genes involved in intracellular signal transduction (RGS7, EHD3, CS1, ITPK1, GRB2, and STK2), as well as a subset of genes linked to apoptosis (CASP7, BCLW, and DAP3). Some highly upregulated genes were difficult to classify, such as ATX, an extracellular protein involved in melanoma migration, and the intracellular calcium channel, RYR2. In conclusion, it was possible to transcriptionally differentiate cell populations from the same tumor that resides in different microenvironments.
Table 2.
Genes Upregulated in Invasive GBM Cells.
| I/C | Accession Number | Name | Description |
|---|---|---|---|
| Extracellular | |||
| 7 | AA436142 | SPOCK | Sparc/osteonectin, cwcv and kasal-like domains proteglycan (testican) |
| 6 | AA425947 | DKK3/RIG | Dickkopf (Xenopus laevis) homolog 3 |
| 6 | AA115876 | PI12 | Serine (or cystein) proteinase inhibitor, clade 1 (neuroserpin) 1 |
| 4 | R76614 | NTN14 | Netrin4 |
| Transmembrane proteins | |||
| 69 | H42679 | HLA-DRA | Major histocompatibility complex, class II DM alpha |
| 8 | N62620 | KCNK1 | Potassium channel, subfamily K, member 1 (TWIK-1) |
| 7 | R38201 | OPCML | Opioid-binding protein/cell adhesion molecule-like |
| 7 | AA464590 | PTPRN2 | Protein tyrosine phosphatase receptor type, N polypeptide 2 |
| 6 | AA485795 | EFNB3 | Ephrin B3 |
| 6 | H08933 | GRIN2A | Glutamate receptor, ionotropic, _N_-methyl D-aspartate 2A |
| 6 | R40790 | GABRG2 | Gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| 5 | T80232 | ATX | Autotaxin (ectonucleotide pyrophosphatase/phosphodiesterase 2) |
| 5 | AA417654 | FGFR3 | Fibroblast growth factor receptor 3 |
| 5 | AA393408 | PDE1A | Phosphodiesterase 1A, calmodulin-dependent |
| 4 | W48713 | EGFR | Epidermal growth factor receptor |
| 4 | AA454743 | KLK6 | Kallikrein 6 |
| 4 | N94270 | TPARL | TPA-regulated locus |
| 3 | R17717 | CDH13 | Cadherin 13 |
| 3 | H07878 | GPRl9 | G protein-coupled receptor 19 |
| 3 | R14663 | DTR | Diphtheria toxin receptor (heparin-binding EGF-like growth factor) |
| 3 | AA479243 | AMFR | Autocrine motility factor receptor |
| Intracellular signaling | |||
| 23 | H23046 | RGS7 | Regulator of G-protein signaling 7 |
| 15 | R22326 | EHD3 | EH domain containing 3 |
| 11 | R15791 | RYR2 | Ryanodine receptor 2 (cardiac) |
| 10 | AA064973 | CS-1 | Calcineurin-binding protein calsarcin-1 |
| 6 | R69354 | SAC2 | Sac domain containing inositol phosphatase 2 |
| 4 | AA464067 | ITPK1 | Inositol 1,3,4-triphosphate 5/6 kinase |
| 3 | AA449831 | GRB2 | Growth factor receptor-bound protein 2 |
| 3 | AA454947 | AKAP1 | A kinase (PRKA) anchor protein 1 |
| 3 | AA496013 | STK2 | Serine/threonine kinase 2 |
| Cytoskeleton rearrangement | |||
| 59 | AA448015 | INA | Internexin, neuronal intermediate filament |
| 4 | R27680 | EMAP2 | Microtubule-associated protein like echinoderm EMAP |
| 4 | AA598668 | CHN1 | Chimerin (chimaerin) 1 |
| 3 | H24688 | SMARCC2 | SWI/SNF-related, matrix-associated, actin dep reg of chromatin, C2 |
| 2 | R85257 | PYK2 | Protein tyrosine kinase 2 beta |
| 2 | AA457036 | P85SPR | PAK-interacting exchange factor beta (beta-pix) |
| Apoptosis | |||
| 5 | BCL2L2 | Bcl2-like 2 (Bcl-w) | |
| 5 | T50828 | CASP7 | Caspase 7, apoptosis-related cysteine protease |
| 3 | R43325 | DAP3 | Death-associated protein 3 |
| Transcription factors | |||
| 9 | AA459941 | PEG3 | Paternally expressed 3 |
| 5 | H60572 | TRABID | TRAF-binding protein domain |
| 4 | N99243 | TBX2 | T-box 2 |
| 4 | AA234897 | MEF2C | MADS box transcription enhancer factor 2C |
| 4 | W00959 | HLF | Hepatic leukemia factor |
| 3 | R42479 | ETS2 | V-ets E26 oncogene |
| Unknown function | |||
| 77 | R67147 | CRYM | Crystallin mu |
| 16 | H54364 | MAST3 | Microtubule associated serine/threonine kinase 3 |
| 11 | H24428 | KIAA0513 | KIAA0513 gene product |
| 8 | AA452725 | NUCB1 | Nucleobindin 1 |
| 7 | AA456008 | AF1Q | ALL1-fused gene from chromosome 1q |
| 7 | W48780 | NP25 | Neuronal protein |
| 6 | W60581 | BEX1 | Brain expressed, X-linked 1 |
| 5 | AA227594 | MAL | Mal, T-cell differentiation protein |
| 4 | H19439 | DSCR1L1 | Down syndrome critical region gene 1-like 1 |
| 4 | R59579 | PGDS2 | Prostaglandin D2 synthase (21 kDa, brain) |
| 4 | H66616 | GLG1 | Golgi apparatus protein 1 |
| 3 | H22481 | NPTX1 | Neuronal pentraxin I |
| 3 | H45376 | NELL2 | Nel (chicken)-like 2 |
| 3 | T84156 | LNX | Multi-PDZ-domain-containing protein |
Gene Candidate Validation by QRT-PCR
Gene candidates that discriminate invasive from tumor core cells were validated by real-time QRT-PCR using unamplified RNA (Figure 1). The genes chosen reflect various cellular processes that may be involved in the biology of the invasive phenotype, as well as some unknown, yet highly differentially expressed candidate genes. Candidates for validation were also chosen along the magnitude of the range of differential gene expression to validate the selection algorithm. Each gene's differential expression was assayed pairwise in corresponding tumor core and invasive rim from seven different tumor samples. To address possible contamination from surrounding normal brain tissues, RNA from a section of white matter surrounding one of the tumors was used to measure the expression of candidate genes in white matter. We found that 15 of 17 of the gene candidates were expressed 10-fold lower in the white matter compared to the invasive tumor. The Wilcoxon signed-rank test was used to analyze the statistical significance of the difference in gene expression between the tumor core and the invasive cell populations. Statistically significant changes in gene expression at the P ≤ .025 level were seen with KIAA0513, OPCML, EFNB3, BCLW, KLK6, CHN1, EMAP2, DAP3, and PYK2 transcripts. Significant change in gene expression at the .025 < P < .05 level was reached by an additional three transcripts, EHD3, PTPRN2, and ATX. CASP7 was not increased in six of seven tumor pairs examined, and thus did not verify the cDNA microarray analysis. We also examined four gene candidates decreased in the invading GBM cells. IGFBP2 and VIM transcript levels were greater than two-fold lower in the majority of tumors analyzed (four of seven and six of seven, respectively). IGFBP5 and MT1L were only decreased in less than half of the seven tumors analyzed. The directionality of the expression of these four tumor core gene candidates did not reach statistical significance using the Wilcoxon test. Differential transcription of invasion gene candidates derived from microarray analysis was validated by QRT-PCR and showed that the invasion transcriptome was not significantly influenced by admixture of genetic material from the white matter context in which they were located.
Immunohistochemical Evaluation of Gene Candidates
Verification of protein product was undertaken in three GBM samples for IGFBP2, VIM, ATX, EFNB3, BCLW, and PYK2 (Figure 2). The invasion gene candidates ATX and EFNB3 (Figure 2, C and E) are transmembrane proteins that displayed predominantly cytoplasmic staining in the invading cells, but also in the tumor core. BCLW was present in invasive cells (Figure 2_D_), as well as in vascular endothelium and, to a lesser degree, in reactive astrocytes (data not shown). The biologically active, phosphorylated form of PYK2 showed perinuclear localization in invasive glioma cells as well as in some tumor core cells (Figure 2_F_). The two candidates from the tumor core that were transcriptionally downregulated in the infiltrating cell population were visualized immunohistochemically in three GBM samples. IGFBP2 (Figure 2_G_) showed distinct cytoplasmic staining in the tumor core and somewhat lighter cytoplasmic staining in invading cells. Cytoplasmic staining for IGFBP2 was also visible, to a lesser extent, in astrocytes and reactive astrocytes present in the invaded white matter surrounding the tumor. The intermediate filament VIM (Figure 2_H_) was strongly present in the GBM tumor core, vascular endothelium, astrocytes, and reactive astrocytes, but was markedly reduced in invasive glioma cells. These data indicate that the protein product of the selected genes is produced in glioma cells.
TMA Analysis of ATX and BCLW
We further examined ATX and BCLW expression on an invasion TMA assembled to reflect the dispersion of infiltrative glioma of various grades and cellular origins (Figure 3). ATX is strongly expressed in glioblastoma cells and is also clearly expressed by WHO grade II and grade III gliomas. Interestingly, it is highly expressed in the four pilocytic astrocytomas (WHO grade I) examined. Positive staining is evident in normal vascular endothelium and, to a lesser extent, in tumor vasculature. Weaker expression is observed in reactive astrocytes and Nissl bodies of some neurons. It is weakly or not expressed by carcinoma metastasis and not expressed by normal astrocytes and oligodendrocytes (Figure 3_A_). Further analysis of 10 GBM cases revealed that of the invasive cells, 51% were ATX-positive, whereas only 30% of tumor core cells had ATX immunopositivity (data not shown). One such representative tumor is illustrated in Supplementary Figure 4. BCLW immunopositivity is weaker, but it is nonetheless detectable in glioma cells. Its expression mildly increases, with progressive malignancy grades reaching its peak in GBM (Figure 3_B_). As with ATX, no expression was seen in normal astrocytes and oligodendrocytes, nor was it present in metastatic adenocarcinoma or medulloblastoma. These data show that ATX and BCLW, two proteins not previously associated with glioma biology, are expressed in invasive glioma cells.
Discussion
Glioblastomas display a notoriously heterogeneous phenotypic presentation [19], yet there are key genetic changes that define these tumors [20]. Epidermal growth factor receptor (EGFR) overexpression/amplification occurs in primary GBM, which constitutes roughly 50% of gliomas. GBM that progresses from lower grades (secondary GBM) does not overexpress EGFR, but exhibits a loss of p53 [21]. These molecular subtypes of glioblastoma have distinct transcriptional profiles [22], which will be useful in targeting new therapies to a potentially more responsive subset of tumors. Previous gene expression profiles of glial tumors show that GBM can be differentiated from lower grades of astrocytic tumors through a characteristic group of upregulated genes [6]. Our studies reflect the expression of such GBM hallmark genes, which include IGFBP2, IGFBP5, VEGF, VCAM1, EGFR, MCM2, and TNC [4–6] in both tumor core and invasive cells. Interestingly, most of these genes are downregulated in the invasive cell population relative to the tumor core. Expression of these genes in conjunction with the histopathologic diagnosis confirms the identity of the infiltrating cells as GBM cells. Analysis of the gene expression profile from the white matter surrounding one of the tumors indicates that the gene expression profile of invasive glioma cells is not attributable to contaminating mRNA from the white matter that they invade (Figure 1). Furthermore, glial fibrillary acidic protein (GFAP) is moderately transcriptionally downregulated in invasive glioma cells as compared to the tumor core population (Table 1). Immunohistochemical staining for GFAP further corroborates this finding (Supplemental Figure 5) and reveals that GFAP levels are much higher in normal and reactive astrocytes than in invasive tumor cells. Because GFAP levels are reduced with increasing astrocytic malignancy [23], it follows that the invasive transcriptome is not influenced by contribution of genetic material stemming from reactive astrocytes.
The importance of the tumor's microenvironment as a contributing factor to gene expression changes should not be overlooked [24]. Cells at the tumor core are densely packed, proliferative, and may experience considerable hypoxia leading to extensive areas of necrosis. Individually infiltrating cells interact with the extracellular matrix and diverse cells residing in the brain parenchyma, incorporating signals as they invade. Interactions with such diverse microenvironments likely contribute significantly to the initiation and maintenance of these discrete transcription profiles.
The expression profile of invasive glioma provides new insight into the interplay of the concerted molecular phenomena activated during invasion. Two such apparently linked mechanisms are motility and apoptosis resistance. Various types of cancer, such as glioma [11,25], gastric cancer [26], Kaposi's sarcoma [27], and pancreatic cancer [28], show evidence for this relationship. Recent evidence suggests that this occurs at the level of gene expression in breast cancer [29] and glioma [30], corroborating our findings that the invasion transcriptome shows a concomitant upregulation of genes involved in motility and apoptosis resistance.
Genes Involved in Motility-Related Pathways Are Differentially Regulated in Invasive Cells
Motility is dependent on cytoskeletal rearrangement and the extension of filopodia and then lamellipodia at the leading edge; these phenomena are modulated by Cdc42, Rac, and Rho [31]. The tight regulation of actin polymerization also emerges from this molecular portrait. Heightened expression of capping protein and profilin in the tumor core may inhibit filamentous actin polymerization and elongation, as overexpression of profilin in breast cancer reduces migration and invasion [32]. ERM (ezrin, radixin, and moesin) proteins, two of which are overexpressed in the core, act as linkers between the plasma membrane and the actin cytoskeleton, impairing migration-associated processes, such as cell spreading, attachment, and motility [33,34]. Involvement of small G-protein signaling and motility-related cytoskeleton rearrangement are illustrated by the differential expression of chimaerin alpha 1 (CHN1), a GTPase-activating protein for Cdc42 and Rac1 [35], which induces the formation of lamellipodia and filopodia in neuroblastoma cells [36]—key hallmarks of Cdc42 and Rac1 activation. Microinjection of full-length CHN1 colocalizes with filamentous actin microspikes as well as with membrane ruffles, and is involved in the redistribution of focal adhesion protein vinculin. PYK2 is a member of the focal adhesion kinase family of nonreceptor tyrosine kinases; it is closely involved with src-induced increased actin polymerization at the fibroblastic cell periphery [37]. Its role in glioma migration/invasion is becoming clearer, as overexpression of PYK2 induced glioblastoma cell migration in culture [16]. Levels of activated PYK2 positively correlated with the migration phenotype in four glioblastoma cell lines (SF767, G112, T98G, and U118) tested in a two-dimensional migration assay [16]. Our analysis of activated PYK2 in GBM invasion in situ revealed strong staining in infiltrating GBM cells.
Tumor mitogens such as the cytokine ATX, which is an autocrine motility factor in melanoma [38] and breast cancer [39], as well as an autocrine motility factor receptor [40] and Netrin 4 [41], are also involved in promoting cell movement. A growing body of literature documents ATX's role in cancer invasion; we therefore chose to examine its expression in a glioma invasion-specific TMA. Evaluation of ATX staining revealed that it is expressed in all grades of glioma, but not in normal astrocytes. It also appears that almost twice the number of invasive tumor cells expresses ATX when compared to its expression in the tumor core. These findings suggest that the role of ATX in glioma invasion should be examined further.
Invasive Cells May Pre-empt Apoptosis
There is a positive correlation between apoptosis index (AI) and progressive grades of astrocytoma malignancy [42,43]. However, within GBM, there is a direct correlation between AI and patient survival, indicating that the most malignant GBM (measured by a shorter progression-free survival) has a lower rate of apoptosis [44]. It is of interest that the most malignant tumors are highly invasive [45]. Higher expression of antiapoptotic bcl-2 family proteins in recurrent GBM, even in patients who did not receive adjuvant chemotherapy or radiotherapy, points to the intrinsic resistance to apoptosis of these tumors [46]. We report BCLW (a member of this family), which is transcriptionally upregulated and expressed in invasive glioma cells. This finding reflects the previously observed expression of BCLW in infiltrative morphotypes of gastric cancer [47]. The mechanism by which BCLW acts in glioma cells is not known, but this family of apoptosis suppressors has been implicated in coordinating Ca2+ balance between the endoplasmic reticulum (ER) and mitochondria [48]. Recently, Ca2+ homeostasis has been linked to apoptosis [49], showing that Ca2+ release from the ER protects cells from apoptosis; interestingly, Ca2+ release is modulated, in part, by ryanodine receptors [50] and we found RYR2 to be consistently upregulated in invasive glioma cells.
A direct correlation between invasion and apoptosis resistance can also be effected by modulation of apoptotic signaling. Such may be the case with DAP3, originally described as a proapoptotic protein that transduces tumor necrosis factor ligand-dependent signals from death receptor DR4 through FADD (Fas-associated by death domain) [51] in fibrosarcoma cells. DAP3, however, was also described as an antiapoptotic factor in migrating glioma cells [11]. We propose that the equilibrium between proapoptotic and antiapoptotic proteins may be regulated, in part, by transcriptional activation of apoptosis modulators (such as DAP3) and antiapoptotic genes (such as BCLW) during activation of the invasive phenotype.
Current treatment for GBM includes surgical resection, chemotherapy, and radiotherapy, but despite continuous improvements in these approaches, patients' median survival remains at 1 year. New treatment modalities such as targeted therapy with monoclonal antibodies and immunotoxin-conjugated antibodies [52] are aimed at tyrosine kinase receptors such as EGFR and PDGFR, which are frequently overexpressed in glioblastomas. Gene therapy for GBM has met with some success in clinical trial [53], but is still in the early stages of development. Signaling pathways such as those involving EGFR, PDGFR, PI3K/AKT, and RAS can be targeted with small molecule inhibitors. However, most of these approaches predominantly address key pathways involved in cell proliferation, whereas recurrent tumors regrow from the cells that have invaded the brain and may be temporally less proliferative [54]. This preliminary study of glioma invasion-related gene regulation suggests targets that are potentially upregulated in gliomas regardless of their molecular etiology. Further transcriptional profiling of invasive GBM cells in more tumors with known EGFR and p53 status should clarify if this profile can be subcategorized according to current molecular classifications. An expanded approach including transcriptional profiling of diffusely infiltrating gliomas of lower grades may lead to insight into general biochemical mechanisms necessary for invasion.
In conclusion, we propose that the gene expression profile of invading glioma reveals a pattern unique to this discrete population of cells. These transcriptional differences point to reasons why invasive GBM cells are unlikely to respond to conventional therapies aimed at a proliferative and stationary tumor mass that has been the reference tissue for the molecular genetic analysis of this disease. Understanding the genetic basis of the invasive behavior may lead to novel combination therapies that not only address the tumor core, but also this distinct subpopulation of cells that have proven refractory to treatment. We anticipate that this will suggest novel intervention strategies through combined modification of apoptotic cascades, or potential use of small compounds targeting extracellular receptors expressed by invading glioma cells.
Acknowledgements
We thank the following people for their contribution to the success of this project: Alf Giese, MD, Wendy Elder, MD, and Kris Smith, MD, for contributing the samples that made this study possible; Nikolai G. Rainov, MD, DSc, Mitsutoshi Nakada, MD, PhD, and Tim D. Demuth, MD, for expert review and helpful discussion of the manuscript; David H. Friedrich, MD, for the assembly of the invasion TMA; Galen Hostetter, MD, for informative pathology discussions; Jeanne Razevich and Kathy Goehring for expert technical histologic assistance; and the staff at Arcturus for excellent technical assistance.
Abbreviations
GBM
glioblastoma multiforme
LCM
laser capture microdissection
TMA
tissue microarray
Footnotes
1
This work was supported by NIH-NS 42262.
References
- 1.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 2.Burger PC, Dubois PJ, Schold SC, Jr, Smith KR, Jr, Odom GL, Crafts DC, Giangaspero F. Computerized tomographic and pathologic studies of the untreated, quiescent, and recurrent glioblastoma multiforme. J Neurosurg. 1983;58:159–169. doi: 10.3171/jns.1983.58.2.0159. [DOI] [PubMed] [Google Scholar]
- 3.Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, Cavenee WK. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. (discussion, 226–219) [DOI] [PubMed] [Google Scholar]
- 4.Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000;60:6617–6622. [PubMed] [Google Scholar]
- 5.Ljubimova JY, Khazenzon NM, Chen Z, Neyman YI, Turner L, Riedinger MS, Black KL. Gene expression abnormalities in human glial tumors identified by gene array. Int J Oncol. 2001;18:287–295. doi: 10.3892/ijo.18.2.287. [DOI] [PubMed] [Google Scholar]
- 6.Rickman DS, Bobek MP, Misek DE, Kuick R, Blaivas M, Kurnit DM, Taylor J, Hanash SM. Distinctive molecular profiles of highgrade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001;61:6885–6891. [PubMed] [Google Scholar]
- 7.Fuller GN, Hess KR, Rhee CH, Yung WK, Sawaya RA, Bruner JM, Zhang W. Molecular classification of human diffuse gliomas by multidimensional scaling analysis of gene expression profiles parallels morphology-based classification, correlates with survival, and reveals clinically-relevant novel glioma subsets. Brain Pathol. 2002;12:108–116. doi: 10.1111/j.1750-3639.2002.tb00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Dougherty ER, Shmulevich L, Hess KR, Hamilton SR, Trent JM, Fuller GN, Zhang W. Identification of combination gene sets for glioma classification. Mol Cancer Ther. 2002;1:1229–1236. [PubMed] [Google Scholar]
- 9.Fuller GN, Rhee CH, Hess KR, Caskey LS, Wang R, Bruner JM, Yung WK, Zhang W. Reactivation of insulin-like growth factor binding protein 2 expression in glioblastoma multiforme: a revelation by parallel gene expression profiling. Cancer Res. 1999;59:4228–4232. [PubMed] [Google Scholar]
- 10.Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 11.Mariani L, Beaudry C, McDonough WS, Hoelzinger DB, Kaczmarek E, Ponce F, Coons SW, Giese A, Seiler RW, Berens ME. Death-associated protein 3 (Dap-3) is overexpressed in invasive glioblastoma cells in vivo and in glioma cell lines with induced motility phenotype in vitro. Clin Cancer Res. 2001;7:2480–2489. [PubMed] [Google Scholar]
- 12.Kleihues P, Webster KC. Tumors of the Nervous System. Lyon, France: IARC Press; 2000. World Health Organization Classification of Tumors. Pathology and Genetics. [Google Scholar]
- 13.Mariani L, McDonough WS, Hoelzinger DB, Beaudry C, Kaczmarek E, Coons SW, Giese A, Moghaddam M, Seiler RW, Berens ME. Identification and validation of P311 as a glioblastoma invasion gene using laser capture microdissection. Cancer Res. 2001;61:4190–4196. [PubMed] [Google Scholar]
- 14.Roche Molecular Biochemicals ME, author. LightCycler Operator's Manual. Version 3.5. Mannheim D-68298, Germany: Roche Diagnostics GmbH; 2000. [Google Scholar]
- 15.Ivanov AI, Pero RS, Scheck AC, Romanovsky AA. Prostaglandin E(2)-synthesizing enzymes in fever: differential transcriptional regulation. Am J Physiol Regul Integr Comp Physiol. 2002;283:R1104–R1117. doi: 10.1152/ajpregu.00347.2002. [DOI] [PubMed] [Google Scholar]
- 16.Lipinski CA, Tran NL, Bay C, Kloss J, McDonough WS, Beaudry C, Berens ME, Loftus JC. Differential role of proline-rich tyrosine kinase 2 and focal adhesion kinase in determining glioblastoma migration and proliferation. Mol Cancer Res. 2003;1:323–332. [PubMed] [Google Scholar]
- 17.Zhang W, Wang H, Song SW, Fuller GN. Insulin-like growth factor binding protein 2: gene expression microarrays and the hypothesis- generation paradigm. Brain Pathol. 2002;12:87–94. doi: 10.1111/j.1750-3639.2002.tb00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 19.Paulus W, Peiffer J. Intratumoral histologic heterogeneity of gliomas. A quantitative study. Cancer. 1989;64:442–447. doi: 10.1002/1097-0142(19890715)64:2<442::aid-cncr2820640217>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Louis DN, Holland EC, Cairncross JG. Glioma classification: a molecular reappraisal. Am J Pathol. 2001;159:779–786. doi: 10.1016/S0002-9440(10)61750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 22.Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 23.Rutka JT, Murakami M, Dirks PB, Hubbard SL, Becker LE, Fukuyama K, Jung S, Tsugu A, Matsuzawa K. Role of glial filaments in cells and tumors of glial origin: a review. J Neurosurg. 1997;87:420–430. doi: 10.3171/jns.1997.87.3.0420. [DOI] [PubMed] [Google Scholar]
- 24.Gladson CL. The extracellular matrix of gliomas: modulation of cell function. J Neuropathol Exp Neurol. 1999;58:1029–1040. doi: 10.1097/00005072-199910000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Joy AM, Beaudry CE, Tran NL, Ponce FA, Holz DR, Demuth T, Berens ME. Migrating glioma cells activate the PI3-K pathway and display decreased susceptibility to apoptosis. J Cell Sci. 2003;116:4409–4417. doi: 10.1242/jcs.00712. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi H, Tanaka F, Sadanaga N, Ohta M, Inoue H, Mori M. Stimulation of CD40 inhibits Fas-or chemotherapy-mediated apoptosis and increases cell motility in human gastric carcinoma cells. Int J Oncol. 2003;23:1697–1702. [PubMed] [Google Scholar]
- 27.Buttiglieri S, Deregibus MC, Bravo S, Cassoni P, Chiarle R, Bussolati B, Camussi G. Role of PAX2 on apoptosis resistance and pro-invasive phenotype of Kaposi's sarcoma cells. J Biol Chem. 2004;279:4136–4143. doi: 10.1074/jbc.M306824200. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa S, Egami H, Kurizaki T, Akagi J, Tamori Y, Yoshida N, Tan X, Hayashi N, Ogawa M. Identification of genes related to invasion and metastasis in pancreatic cancer by cDNA representational difference analysis. J Exp Clin Cancer Res. 2003;22:299–306. [PubMed] [Google Scholar]
- 29.Wang W, Wyckoff JB, Frohlich VC, Oleynikov Y, Huttelmaier S, Zavadil J, Cermak L, Bottinger EP, Singer RH, White JG, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–6288. [PubMed] [Google Scholar]
- 30.Giese A. Glioma invasion—pattern of dissemination by mechanisms of invasion and surgical intervention, pattern of gene expression and its regulatory control by tumor-suppressor p53 and proto-oncogene ETS-1. Acta Neurochir Suppl. 2003;88:153–162. doi: 10.1007/978-3-7091-6090-9_21. [DOI] [PubMed] [Google Scholar]
- 31.Maidment SL. The cytoskeleton and brain tumour cell migration. Anticancer Res. 1997;17:4145–4149. [PubMed] [Google Scholar]
- 32.Roy P, Jacobson K. Overexpression of profilin reduces the migration of invasive breast cancer cells. Cell Motil Cytoskelet. 2004;57:84–95. doi: 10.1002/cm.10160. [DOI] [PubMed] [Google Scholar]
- 33.Speck O, Hughes SC, Noren NK, Kulikauskas RM, Fehon RG. Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature. 2003;421:83–87. doi: 10.1038/nature01295. [DOI] [PubMed] [Google Scholar]
- 34.Wick W, Grimmel C, Wild-Bode C, Platten M, Arpin M, Weller M. Ezrin-dependent promotion of glioma cell clonogenicity, motility, and invasion mediated by BCL-2 and transforming growth factor-beta2. J Neurosci. 2001;21:3360–3368. doi: 10.1523/JNEUROSCI.21-10-03360.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed S, Lee J, Kozma R, Best A, Monfries C, Lim L. A novel functional target for tumor-promoting phorbol esters and lysophosphatidic acid. The p21rac-GTPase activating protein-chimaerin. J Biol Chem. 1993;268:10709–10712. [PubMed] [Google Scholar]
- 36.Kozma R, Ahmed S, Best A, Lim L. The GTPase-activating protein n-chimaerin cooperates with Rac1 and Cdc42Hs to induce the formation of lamellipodia and filopodia. Mol Cell Biol. 1996;16:5069–5080. doi: 10.1128/mcb.16.9.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Xie Y, Du QS, Wu XJ, Feng X, Mei L, McDonald JM, Xiong WC. Regulation of the formation of osteoclastic actin rings by proline-rich tyrosine kinase 2 interacting with gelsolin. J Cell Biol. 2003;160:565–575. doi: 10.1083/jcb.200207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stracke ML, Krutzsch HC, Unsworth EJ, Arestad A, Cioce V, Schiffmann E, Liotta LA. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J Biol Chem. 1992;267:2524–2529. [PubMed] [Google Scholar]
- 39.Yang SY, Lee J, Park CG, Kim S, Hong S, Chung HC, Min SK, Han JW, Lee HW, Lee HY. Expression of autotaxin (NPP-2) is closely linked to invasiveness of breast cancer cells. Clin Exp Metastasis. 2002;19:603–608. doi: 10.1023/a:1020950420196. [DOI] [PubMed] [Google Scholar]
- 40.Timar J, Raso E, Dome B, Ladanyi A, Banfalvi T, Gilde K, Raz A. Expression and function of the AMF receptor by human melanoma in experimental and clinical systems. Clin Exp Metastasis. 2002;19:225–232. doi: 10.1023/a:1015595708241. [DOI] [PubMed] [Google Scholar]
- 41.Kennedy TE. Cellular mechanisms of netrin function: long-range and short-range actions. Biochem Cell Biol. 2000;78:569–575. [PubMed] [Google Scholar]
- 42.Carroll RS, Zhang J, Chauncey BW, Chantziara K, Frosch MP, Black PM. Apoptosis in astrocytic neoplasms. Acta Neurochir (Wien) 1997;139:845–850. doi: 10.1007/BF01411402. [DOI] [PubMed] [Google Scholar]
- 43.Bogler O, Weller M. Apoptosis in gliomas, and its role in their current and future treatment. Front Biosci. 2002;7:e339–e353. doi: 10.2741/a928. [DOI] [PubMed] [Google Scholar]
- 44.Kuriyama H, Lamborn KR, O'Fallon JR, Iturria N, Sebo T, Schaefer PL, Scheithauer BW, Buckner JC, Kuriyama N, Jenkins RB, et al. Prognostic significance of an apoptotic index and apoptosis/proliferation ratio for patients with high-grade astrocytomas. Neuro-Oncology. 2002;4:179–186. doi: 10.1093/neuonc/4.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chicoine MR, Silbergeld DL. The in vitro motility of human gliomas increases with increasing grade of malignancy. Cancer. 1995;75:2904–2909. doi: 10.1002/1097-0142(19950615)75:12<2904::aid-cncr2820751218>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Strik H, Deininger M, Streffer J, Grote E, Wickboldt J, Dichgans J, Weller M, Meyermann R. BCL-2 family protein expression in initial and recurrent glioblastomas: modulation by radiochemotherapy. J Neurol Neurosurg Psychiatry. 1999;67:763–768. doi: 10.1136/jnnp.67.6.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HW, Lee SS, Lee SJ, Um HD. Bcl-w is expressed in a majority of infiltrative gastric adenocarcinomas and suppresses the cancer cell death by blocking stress-activated protein kinase/c-Jun NH2-terminal kinase activation. Cancer Res. 2003;63:1093–1100. [PubMed] [Google Scholar]
- 48.Pinton P, Ferrari D, Rapizzi E, Di Virgilio F, Pozzan T, Rizzuto R. A role for calcium in Bcl-2 action? Biochimie. 2002;84:195–201. doi: 10.1016/s0300-9084(02)01373-1. [DOI] [PubMed] [Google Scholar]
- 49.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 50.Fill M, Copello JA. Ryanodine receptor calcium release channels. Physiol Rev. 2002;82:893–922. doi: 10.1152/physrev.00013.2002. [DOI] [PubMed] [Google Scholar]
- 51.Miyazaki T, Reed JC. A GTP-binding adapter protein couples TRAIL receptors to apoptosis-inducing proteins. Nat Immunol. 2001;2:493–500. doi: 10.1038/88684. [DOI] [PubMed] [Google Scholar]
- 52.Mischel PS, Cloughesy TF. Targeted molecular therapy of GBM. Brain Pathol. 2003;13:52–61. doi: 10.1111/j.1750-3639.2003.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rainov NG, Ren H. Gene therapy for human malignant brain tumors. Cancer J. 2003;9:180–188. doi: 10.1097/00130404-200305000-00006. [DOI] [PubMed] [Google Scholar]
- 54.Giese A, Loo MA, Tran N, Haskett D, Coons SW, Berens ME. Dichotomy of astrocytoma migration and proliferation. Int J Cancer. 1996;67:275–282. doi: 10.1002/(SICI)1097-0215(19960717)67:2<275::AID-IJC20>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]