Regulatory gene networks and the properties of the developmental process (original) (raw)
Abstract
Genomic instructions for development are encoded in arrays of regulatory DNA. These specify large networks of interactions among genes producing transcription factors and signaling components. The architecture of such networks both explains and predicts developmental phenomenology. Although network analysis is yet in its early stages, some fundamental commonalities are already emerging. Two such are the use of multigenic feedback loops to ensure the progressivity of developmental regulatory states and the prevalence of repressive regulatory interactions in spatial control processes. Gene regulatory networks make it possible to explain the process of development in causal terms and eventually will enable the redesign of developmental regulatory circuitry to achieve different outcomes.
The diverse developmental processes of Bilateria are generated with very similar “tool kits” of regulatory and signaling genes. It is the manner in which these genes are deployed that determines the specific features of development in any given animal (1). Developmental processes are, of course, the definitive, heritable character of each species in that body plans are the morphological outcomes of developmental processes. In physical terms, the control system for development consists of the genomic cis-regulatory sequences that control expression of the genes required to execute each developmental episode. Cis-regulatory elements provide the chemical code that specifies the interactions of transcription factors with their constituent sequences. Thereby is set in train the transcriptional gene expressions, and hence, the multitude of extranuclear events that build the cell types and create the morphological structures of the organism. But the genomic regulatory sequences that control developmental gene expression can be thought of in a more abstract way as well: they are the genomic source code for development. Like the DNA sequences specifying protein sequence, they are essentially a combinatorial digital code, consisting only of particular patches of As, Cs, Gs, and Ts.
The unique function of the cis-regulatory source code for development is to generate spatial patterns of gene expression, i.e., to specify at the right time the cellular domains in which given sets of genes are expressed. This function turns out to be very expensive in terms of cis-regulatory interactions. It is carried out by modular assemblages of clustered DNA target sites for diverse transcription factors. Specific regulatory modules execute control of expression at particular times or in specific spatial domains or execute other functions such as repression in specific places. As a generality, there are often four to eight diverse inputs into each developmental cis-regulatory module, and many of the target sites occur several times within the module. In a few well-studied cases the functional meaning of each species of target site has been examined (a thoroughly studied case is the endo16 gene of the sea urchin; for review of this and similar examples from several different animal species see ref. 1). With respect to the character of the genomic code for development, the most important conclusion is that cis-regulatory modules that control spatial and temporal gene expression behave as information processing elements. The transcription factors that each module sees, according to its primary DNA sequence, provide its inputs. The module acts as a conditional logic gate, the output of which depends on its various inputs. The genomic regulatory code in this way enables each gene required in development to respond to all of the ambient situations to be encountered throughout the life cycle and to be expressed specifically only in those cell types in which the appropriate combinations of transcription factors is presented.
A very important feature of the regulatory DNA code for animal development is that much of it is used to control expression of genes encoding regulatory proteins, in addition to the many genes encoding the structural proteins of which the animal will be built. In that each cis-regulatory element processes multiple inputs, it interacts with multiple transcription factors produced by multiple distinct genes. And each transcription factor interacts with multiple cis-regulatory elements. From these elemental facts the genomic regulatory code for development can be seen to generate a system of interactions that has the architecture of a network (rather than, say, a simple linear or branching pathway). Each node of a developmental gene regulatory network (GRN) thus consists of a gene encoding a transcription factor or a signaling component, together with the cis-regulatory module(s) controlling expression of that gene. As do transcription factors, signaling components may immediately affect the program of gene expression downstream of their own transcription. The architecture of the GRN can be considered to consist of the functional linkages among the nodes of the network. These linkages connect the output of the gene at each node to its target genes, where they serve as cis-regulatory inputs.
The progression of developmental states observed phenomonologically is controlled by the underlying progression of regulatory states, where “regulatory state” means the set of transcription factors present at given levels of activity in given cell nuclei. Regulatory state changes over time and defines the different fates of the cells composing the various spatial elements of the system. The regulatory state is the specific, overall output of the GRN running in each cell, qualitatively and quantitatively. Except for a fair idea of the quantitative boundaries of the fluctuations in transcription factor level and the overall kinetics of the developmental process, almost all knowledge of developmental GRNs is yet qualitative. That is, we are so far learning mainly about GRN architecture. But it is already evident that from the GRN architecture alone emerges an unprecedented explanatory and predictive power, in respect to understanding and even controlling developmental phenomenology.
Brief Overview of the GRN for Specification of Endomesoderm in Sea Urchin Embryos
The sea urchin embryo is a particularly felicitous developmental system for GRN analysis: the form of development is simple, in evolutionary terms basal, and because of extensive modern experimental embryology, particularly well known (1–8). Specification of cell fate occurs within predictable lineage elements and to a large extent before any movement of the cells with respect to one another (2). Furthermore, it is easy to inject sea urchin eggs with regulatory DNA expression constructs and with micro- or macromolecules that efficiently perturb expression of given genes or sets of genes. A GRN for specification of endomesoderm in the sea urchin embryo has been published (9–11). The GRN covers the period from the time of the initial divisions that divide the eggs up into the first cells (cleavage), through the several hundred-cell stage when the embryo is a hollow cellular sphere (blastula), to the point when invagination of the gut is about to begin (gastrulation). Endomesoderm here means the cellular domains that will give rise to the gut, the skeletogenic mesenchyme, and various other mesodermal cell types (particularly pigment cells). These all arise from early cleavage lineages formed canonically at one end of the egg (i.e., the vegetal pole). The GRN model is essentially a provisional theory. Its object is to state the genomically encoded mechanism by which the cells of these lineages acquire their specific fates and by which further diversification of fate occurs within particular lineage elements. At present it includes ≈50 genes, mostly regulatory genes. This may be close to the real scale of the transcriptional regulatory apparatus specifically controlling this aspect of embryonic specification in the 0- to 30-h time frame. However, at present the GRN includes only a small, although possibly representative, fraction of the downstream structural genes that confer differentiated properties on endodermal and mesodermal cell types. Some global regulators are included, although most are not. The products encoded by regulatory genes expressed in all regions of the embryo may perform important mechanistic functions within the cis-regulatory elements included in the endomesoderm GRN (see, e.g., their functions in the endo16 gene, ref. 12). But specification cannot in the end depend on the transcriptional control apparatus of genes expressed globally; it must depend on regulatory genes expressed only in the endomesoderm. The GRN is continuously updated on the basis of new data, and the current, complete version can be found at www.its.caltech.edu/∼mirsky/endomes.htm (Endomes Gene Network Update). Here can also be seen those components of the GRN omitted from Fig. 1, where their existence is indicated by peripheral rectangles. The central portion of the GRN that is reproduced in Fig. 1 is constituted entirely of genes (and their cis-regulatory elements) that encode transcription factors, excepting only a few genes encoding signaling ligands or receptors. The interactions among these genes determine the mechanism of endomesodermal specification: the GRN portrays the regulatory “brains” behind the developmental phenomenon of endomesoderm formation.
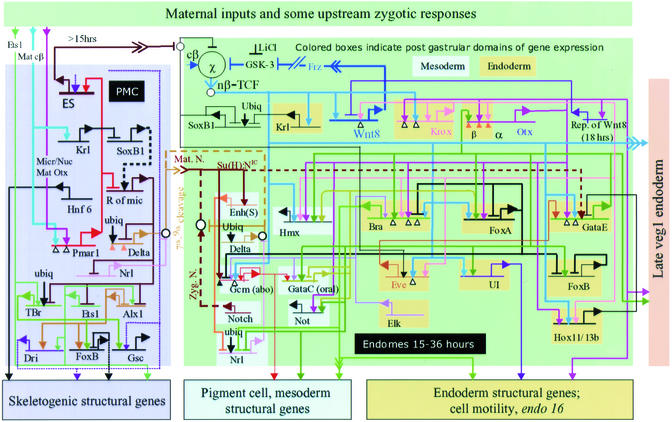
Figure 1.
Central portion of the Strongylocentrotus purpuratus embryo endomesoderm GRN, from fertilization to just before gastrulation. The diagram is a recent version of that initially presented in refs. 9–11. Suspected interactions at the cis-regulatory elements represented by the horizontal lines are shown, irrespective of when in the 0- to 30-h period or where in the embryo they are expected to occur [a “view from the genome” GRN (24); for interactions occurring only in given domains and at given periods see ref. 10 and www.its.caltech.edu/∼mirsky/endomes.htm]. Transcriptional regulatory interactions are shown in the indicated spatial domains of the embryo: pmc domain, the skeletogenic micromere lineage; endomes domain, endomesoderm descendant from the sixth cleavage ring of eight “veg2” cells (2, 13, 24). Transcriptional inputs into the cis-regulatory elements of each named gene are indicated by arrows (activation, or permissive of activation) or bars (repression). Outputs from each gene (where known) are indicated by color-coded lines emanating from the bent arrows that symbolize transcription. For evidence see text, refs. 9–11, 15, 16, and 18, and www.its.caltech.edu/∼mirsky/endomes.htm. An arrowhead inserted in an arrow tail indicates an intercellular signaling interaction; small open circles indicate cytoplasmic interactions or specific events off the DNA, e.g., that by which the Soxb1 factor interferes with nuclearization of β-catenin (26). For further details see refs. 9 and 10 and www.its.caltech.edu/∼mirsky/endomes.htm.
The GRN is constructed on the basis of four kinds of information. First, is a large body of prior experimental embryological and molecular data. From this we know a great deal about how this embryo forms its endomesoderm. Now established are the fate choices available to cells of each lineage element, at least the major signaling interactions required to execute these choices, the maternal inputs that trigger endomesodermal specification at the vegetal pole of the embryos, and the behavior of the progeny of cells from various parts of the embryo if they are isolated in early cleavage or transplanted to ectopic locations (1–8, 13). In addition, many genes involved in endomesoderm specification were already known, and molecular markers of states of specification have been available for some years (for reviews see refs. 1–3). A large-scale program of differential array screening was carried out (e.g., refs. 14–16) to recover further genes expressed specifically in the context of endomesoderm specification, even if the level of expression is only 10–50 mRNA molecules per expressing cell (as is the case for most of the transcriptional regulators that drive this process). The second kind of data on which the GRN rests are new and detailed observations of just when and where each of the endomesoderm-specific genes is expressed. The third is experimental and computational cis-regulatory analysis (17, 18). The fourth, which speaks immediately to the architecture of the GRN, is an ongoing, large-scale perturbation analysis, in which expression of each gene in the GRN is blocked or otherwise altered and the effects on all other relevant genes in the GRN measured by quantitative PCR (QPCR; for the total current QPCR data set see www.its.caltech.edu/∼mirsky/endomes.htm). The major method of perturbation used here is blockade of mRNA translation by injection of morpholino-substituted antisense oligonucleotides or alternatively, the normal mRNA is made to be translated ectopically. In addition, some transcription factors have been converted into obligate repressors by fusing their DNA-binding domains with a sequence encoding the Engrailed repressor domain (9–11, 19). Other more general perturbations that result in specific, complete arrest of all endomesoderm specification (except skeletogenic), or arrest of all mesoderm but not endoderm specification, were also applied (refs. 9 and 10 and see below).
The GRN in Fig. 1 explains many observed aspects of endomesoderm specification in terms of the genomic regulatory code. A quick summary of some major functional aspects is important for what follows. Very briefly, in the left (lavender) block of Fig. 1 are shown regulatory interactions that purport to account for the fate and function of cells descendant from the four micromeres, which are born at the vegetal pole of the embryo at the 16-cell stage. The micromere lineage has three functions: it gives rise to exactly 32 skeletogenic (or “primary”) mesenchyme cells (pmcs) and these uniquely express skeletogenic genes; it expresses a signal, Delta (at seventh-ninth cleavage; refs. 5 and 6), which is received by the adjacent cells and sets in train the transcriptional specification of these cells as mesoderm; it also expresses an “early signal” (fourth-sixth cleavage; ref. 20), which is required for the founder cells of most of the endoderm and the mesoderm to be specified (the green domain of Fig. 1). The GRN shows that the micromere-specific expression of all three functions depends on a double transcriptional repression system. In such regulatory circuit elements a gene encoding a repressor that keeps other downstream genes off is repressed by another gene encoding a repressor of the first, so the downstream genes are allowed to run only where the second repressor gene is active. Here key micromere-specific genes are blocked elsewhere by expression of a globally active repressor encoded by the “_rep of mic_” gene, but this repressor is prevented from functioning in micromeres by the activation, only in micromeres, of a gene encoding a repressor of the rep of mic gene. This gene, pmar1, is expressed in micromeres immediately after they are born and for a few hours remains active in their progeny (11). Two of the spatially specific inputs into the pmar1 cis-regulatory system appear to be β-catenin/Tcf and Otx. The latter is a transcription factor localized in fourth-cleavage micromere nuclei, as is β-catenin (11, 21). At the bottom of the lavender domain of Fig. 1 are six transcriptional regulatory genes, which are all under the control of the pmar1 double repression system, directly or indirectly, and are themselves responsible for expression of skeletogenic structural genes.
In the mesodermal domain several regulatory genes [gray blue background (mesoderm) in Fig. 1] are activated in consequence of the Delta signal, via the Notch (N) signal transduction pathway. An important example is gcm. The gcm gene (probably a direct target of the N pathway) lies upstream of genes encoding several pigment synthesis enzymes. Interference with N signaling or expression of gcm or downstream genes produces albino embryos (refs. 5 and 16, and A. Ransick, C. Calestani, and E.H.D., unpublished data).
In the endodermal domain each of the many regulatory genes (Fig. 1, yellow background) has a unique biological function in that (where known) it has a unique set of downstream targets, and each answers to a unique set of cis-regulatory inputs. Expression of all is required for endoderm formation and/or gastrulation. The gatae gene is particularly important, as it appears to provide inputs into most of the other endodermal regulatory genes. For instance, gatae expression is required for activation of ui, a gene encoding a transcription factor that directly controls expression of the well-studied endo16 gene in the gut (12). The expression of the brachyury gene also depends on gatae. Expression of brachyury results in the activation of cell motility genes needed for gastrulation and invagination to occur (15, 22). Another example of a gatae target is the foxa gene (Fig. 1). The FoxA transcription factor is a repressor of other regulatory genes in the endoderm: it has multiple roles in the spatial control of gene expression patterns, as we see below. These examples, gatae, brachyury, and foxa, illustrate the point that each of the regulatory nodes of the GRN has a particular biological meaning, with respect to its downstream function(s).
The GRN in Fig. 1 is couched in terms of cis-regulatory interactions with the genomic DNA. As such, it is directly testable in detail by experimental cis-regulatory analysis. But as we now illustrate, it can also be tested in a precise way by experimentally manipulating the embryo.
From the GRN to Developmental Phenomenology
How closely related to the observable phenomena of development is the GRN? A great fund of experimental observations on single gene effects from developmental genetics, what we know about gene regulation molecular biology, and logic, say that the GRN should provide direct access to the causes of the developmental process (1). But this question has hardly been examined at the systems level of GRN architecture. We have begun to do this (11, 26) and here provide an illustration.
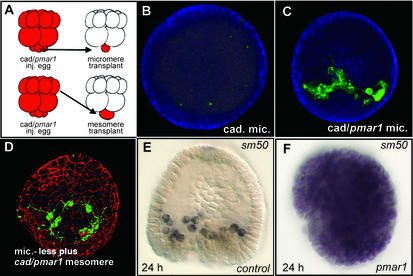
Tests of logic predictions from the “pmc” portion of the GRN are shown in Fig. 2. These have specifically to do with the role of the pmar1 gene, as explicitly given in the GRN (Fig. 1). The first test (26) was done by removing the micromeres of a normal embryo (at fourth cleavage, right after they are born), and replacing them with micromeres from another embryo in which the regulatory state had been altered experimentally (Fig. 2A Upper). The β-catenin/Tcf input (blue lines in Fig. 1) is required for all three micromere lineage functions, because they are all lost if this input is blocked (4). The GRN proposes that the sole required target that transduces this input in the micromeres, and is thereupon causally responsible for all three micromere lineage functions, is the cis-regulatory element of the pmar1 gene. The method used to block the β-catenin/Tcf input is injection of cadherin mRNA, which traps the β-catenin in the cytoplasm and results in a total absence of endomesoderm specification (4, 9), including skeletogenesis. Fig. 2B shows that if a micromere bearing cadherin mRNA is transplanted to the micromereless host embryo no skeletal matrix is formed (nor does normal specification of the endomesoderm occur; refs. 20 and 27). But if the micromere also contains pmar1 mRNA as in Fig. 2C its progeny execute normal skeletogenic functions. Thus, in fact, there are no other required β-catenin/Tcf targets downstream of the pmar1 gene, and upstream of the skeletogenic regulatory apparatus (i.e., the tbr, alx1, ets1, gsc, foxb, and dri genes; Fig. 1). An even more aggressive test is shown in Fig. 2D. This experiment (Fig. 2A Lower) challenges the statement of the GRN that the pmar1 double repression system is all that is required to elicit skeletogenic function, i.e., that there are no other micromere-specific components required for installation of the skeletogenic regulatory state. Skeletogenesis in the embryo in Fig. 2D is again shown being executed by the descendants of a cell transplanted to a micromereless host embryo, but here the cell is a prospective ectoderm cell (“mesomere”) expressing injected cadherin plus pmar1 mRNAs, rather than a micromere. Fig. 2 D and E (11) makes this same point differently. This is another test of the logical implication of the GRN that pmar1 expression is what causally distinguishes micromeres from any other normally nonskeletogenic cells of the embryo. We see that pmar1 mRNA expression suffices by itself to convert all embryonic cells into skeletogenic mesenchyme. Here the readout is expression of a structural gene encoding a skeletogenic matrix protein, Sm50, normally expressed only in pmcs (Fig. 2E). When pmar1 is expressed globally, the sm50 gene is also expressed in all cells (Fig. 2F), and the embryo can be seen to be falling apart into a pile of mesenchyme-like cells, in place of the compact, hollow, epithelial sphere described by the normal embryo. In other words, the whole embryo is now expressing the functions normally executed only by cells arising from the micromere lineage.
Figure 2.
Experimental tests of GRN predictions regarding the pmar1 gene. (A) Diagrams of cell transplantation experiments. (Upper) Transplantation of micromere. (Lower) Transplantation of ectoderm precursor cell, or mesomere. Sixteen cell-stage embryos are shown. The red color symbolizes injected constituents, here cadherin mRNA or cadherin mRNA plus pmar1 mRNA, originally introduced into the egg from which the donor fourth-cleavage embryo develops. The micromeres were removed by microsurgery from the recipient embryo and replaced with a micromere or mesomere from the donor embryo, and the embryos were then cultured for 48 h (B_–_D). (B) Replacement with a micromere expressing cadherin mRNA. (C) Replacement with a micromere expressing cadherin plus pmar1 mRNAs. (D) Replacement with a mesomere expressing cadherin plus pmar1 mRNAs as in C. The green stain identifies Msp130, a skeletogenic cell-specific protein, by fluorescence immunocytology. The red stain in D similarly identifies β-catenin to mark the cell junctions. A_–_D are adapted from ref. 26. (E) Normal localization of sm50 gene expression in skeletogenic pmcs by whole-mount in situ hybridization. (F) Global expression of sm50 in embryo grown from an egg into which pmar1 mRNA had been injected. E and F are adapted from ref. 11.
The relationships in the pmc domain of the GRN also provide a direct interpretation of a remarkable fact of experimental sea urchin embryology first observed in the 1920s (13). This is that transplantation of micromeres to an ectodermal position opposite their normal location induces formation of an ectopic second gut from the adjacent cells. Modern studies (20, 27) show that indeed the whole endomesoderm specification program is thus induced. A key aspect of the mechanism was discovered by the Angerers and colleagues (25): this is that a maternally encoded factor, Soxb1, must be cleared from prospective endomesoderm territory or else no endomesoderm specification takes place. The reason is that Soxb1 interferes with β-catenin/Tcf nuclearization (25, 28), which as the blue lines in Fig. 1 show, is needed for regulatory gene expression throughout the endomesoderm GRN. But as the GRN indicates, control of Soxb1 function in turn depends on the “early signal” (ES) from the micromeres to the adjacent endomesodermal founder cells, and expression of the ES is also under pmar1 control. For example, if a prospective ectodermal cell or mesomere expressing pmar1 mRNA is transplanted to the top of a host embryo it acts just like a transplanted micromere, emits the ES, and thus produces an ectopic patch of cleared Soxb1 in the adjacent cells similar to the endogenous patch in the normal endomesodermal domain at the bottom of the embryo (26).
These are heuristic illustrations. They show commutative predictability: as in Fig. 2, specific aspects of GRN architecture predict specific phenomenological results at the level of experimental embryology and vice versa. The GRN is a powerful and direct tool for scientific understanding of development. Nothing less is, of course, to be expected, if the GRN in fact properly represents the genomic source code for development. Were it the case that the basic developmental source code lies elsewhere (just to take an example, say, in alternative splicing or spatially differential protein turnover rates) the causal, experimentally demonstrable relations between the transcriptional GRN and the observed events of development would be far less straightforward. With understanding comes the ability to reengineer the mechanism. The total change in developmental fates imposed in the experiment of Fig. 2F, and the light shed on the respecification of a whole new vegetal plate in the micromere transplantation experiment, are opening indications of the developmental transformations that will become possible at will when the genomic control program becomes understood.
Recurrent Properties of Developmental GRNs
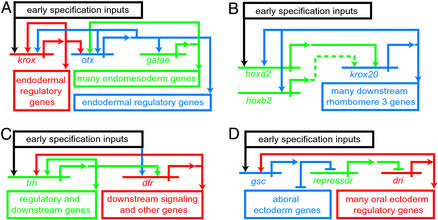
Knowledge of developmental GRNs is yet scarce, and it is too early for a systematic comparative effort. Even so, certain unique features of GRN architecture can already be seen to recur in distinct developmental contexts. Among the principles that emerge are the forward genomic programming of development and the use of genomically encoded spatial repression. The developmental process is moved forward by the use of intergenic feedback loops that serve to stabilize, or lock down, a newly set up-regulatory state. This feature was noted earlier in the endomesoderm GRN (9). A prominent example is the three gene loop including the krox, otx, and gatae genes (Fig. 3A, seen in context in the upper right of Fig. 1). The krox gene is activated early in development in endomesoderm lineages. After the middle of the blastula stage some cis-regulatory elements of the otx gene that respond to Krox input become active, but the output of the otx gene also feeds back on the krox gene. This loop now serves to drive gatae gene expression via an Otx input to the gatae gene. Remarkably, gatae then feeds back on the otx gene. The result is to ensure expression of gatae, a major contributor to the endodermal regulatory state, as we saw earlier. It is a fascinating fact that the same krox-otx-gatae feedback relations exist in a starfish (V. Hinman and E.H.D., unpublished data), considering that the last common ancestor shared by starfish and sea urchins lived no less than 500 million years ago. This is a testament to the value of an effective element of regulatory circuitry. Another example from the endomesoderm GRN, that includes signaling among its cells, is the reinforcing loop within which resides the wnt8 gene. This gene is both a target of the same β-catenin/Tcf input as is required by many other genes, and indirectly, a driver of β-catenin nuclearization (upper center of Fig. 1).
Figure 3.
Common GRN architectural feature: intergenic reinforcing loops that drive developmental state forward. (A) Relation among krox, otx, and gatae genes from the sea urchin endomesoderm gene network (Fig. 1). (B) Relation between hoxa2 and krox20 in mouse rhombomere specification (from ref. 1, after refs. 29–31). (C) Relations between trachealess (trh) and drifter (dfr) regulatory genes (from ref. 1, after ref. 32). (D) Relation between goosecoid (gsc) gene and deadringer (dri) gene in sea urchin oral ectoderm GRN (G. Amore and E.H.D., unpublished data).
Intergenic loops installed immediately downstream of initial specification events are found in many other GRNs as well. For example, in the hox gene network that controls rhombomere specification in the mouse hindbrain, the hoxa2 gene activates the krox20 gene in the region that will become rhombomere 3 (Fig. 3B). The krox20 gene in turn positively regulates the hoxa2 (and hoxb2) genes (1, 29–31). The significance is to stabilize the expression in rhombomere 3 of krox20, which is an activator of other genes downstream. Fig. 3C shows an example from the Drosophila tracheal placode. Two key regulatory genes required for placode specification are drifter and trachealess, and a GRN for the early events of placode specification shows that these two genes both autoregulate and positively cross-regulate one another. Thereupon they activate various downstream target genes (1, 32). A fourth case, excerpted from a different GRN of the sea urchin embryo, is shown in Fig. 3D. This derives from an initial exploration of the GRN underlying oral ectoderm specification (G. Amore and E.H.D., unpublished data). This study again revealed an essential, three-gene loop, consisting of a gene encoding a transcriptional repressor, goosecoid, which in the oral ectoderm domain represses a gene encoding another repressor, thereby allowing a gene that is a target of this second repressor, deadringer, to be expressed there. The deadringer gene encodes an activator. The product of this gene in turn feeds back to drive goosecoid expression in the oral ectoderm. The significance of this loop is to ensure the operation of the oral ectoderm regulatory state; expression of deadringer is required for the activity of other oral ectoderm regulatory genes, which operate that state.
These and other examples of which space precludes mention suggest that positively acting, intergenic feedback loops are a general feature of developmental GRNs. Their general role is to drive development forward. They cause the progression of regulatory state, from the initial often transient inputs that set up a new spatial regulatory domain to a (for a while) stable state, that is thereafter independent of the initial inputs. While it lasts, this regulatory state is used to drive activation of new downstream genes. The stabilization device is dynamic, requiring continuous transcriptional activity within the loop and is inactivated when transcription of one of its genes is cut off.
Another recurrent architectural feature of developmental GRNs is the use of transcriptional repression to exclude regulatory states set up in one spatial domain from other spatial domains. Repressive transcriptional interactions are used to establish boundaries in virtually every spatial GRN so far analyzed. Canonical examples appear in the Drosophila pair rule GRN. Here the exact stripes of cells in which given pair rule genes are ultimately expressed is defined for each such gene by repressive cis-regulatory interactions with the products of other pair rule genes (33). The output of the GRN is to set up an exact anterior/posterior pattern of regulatory states within the metameric subunits of the body plan. In the GRN for the initial dorsal/ventral coordinate system of the Drosophila embryo the domain of expression of the Snail repressor is used to set the ventral boundaries of expression of several other genes (34). Expression of transcriptional repressors sets the boundaries of the proneural patches that arise within wing imaginal discs in Drosophila (several examples are reviewed in ref. 1), similarly in the GRN underlying the specification of cardial and pericardial subdomains in the Drosophila heart (35), in the GRN underlying founder cell specification in Caenorhabditis elegans (36), and so forth. The endomesoderm GRN in Fig. 1 affords numerous examples, in which it is possible to see the variety of ways in which repression circuits are actually used. Repression is used within this GRN to preclude alternative regulatory states in the cells expressing the repressor, set spatial boundaries for a domain of given regulatory state (as in most of the other examples cited in this paragraph), and control temporal patterns of expression (37).
Conclusions
The purpose of a developmental GRN is not to provide biochemical insights into how the cell biological functions that actually effect differentiation and development work, but rather to explain why these functions happen when and where they do, how their appropriate execution is organized. Ultimately the GRN is couched in the sequence language of the genomic regulatory code. A GRN is an immensely potent tool for understanding development. It makes it possible to relate developmental processes to the genomic regulatory sequence, at whatever level of biological organization these processes are perceived. We have shown by example how a GRN implies unique logic predictions testable at the level of cell function; and also how it may afford a new understanding of processes identified at the level of cell function.
Intervention at the GRN level offers the only general, canonical approach to experimental alteration of the developmental process. Temporal changes in the amplitude of expression and spatial changes in the locations of the regulatory states controlled by the underlying GRN will cause predictable temporal and spatial changes in cell fate and function. Evolutionary change in body plans can be viewed as just such a process (1): it has occurred in consequence of changes in the genome that result in the “reengineering” of GRNs so as to alter their developmental output. Experimental reorganization of GRNs offer powerful new approaches to the genetic modification of plants and animals, which could have striking implications for agriculture, animal husbandry, and human health.
Fundamental questions of development and evolution will become accessible when it becomes possible to carry out comparative analyses of GRNs. We will learn, for example, what special features of GRN architecture underlie forms of development that are very different from one another. For example, relative to the embryos that have so far been the objects of GRN analysis, in the diversification of cell fates within the migratory cell populations of vertebrate lymphoid systems or neural crest, fixed spatial relations are relatively unimportant for specification: how different is the architecture of the GRNs that control these processes (38)? Comparative study of GRNs will in the not-too-distant future make it possible to approach systematically the identification of a common bilaterian library of architectural GRN motifs. Perhaps it is their common heritage of GRN programming devices that has endowed the Bilateria with their remarkable and definitive ability to execute development.
Acknowledgments
We thank Dr. Ellen Rothenberg for a perspicacious and useful review of a draft of the manuscript. This research was supported by National Institutes of Health Grants HD-37105, GM-61005, RR-06591, and RR-15044, National Aeronautics and Space Administration Grant NAG2-1368, and the Lucille P. Markey Trust.
Abbreviations
GRN
gene regulatory network
pmc
primary mesenchyme cell
References
- 1.Davidson E H. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic; 2001. [Google Scholar]
- 2.Davidson E H, Cameron R A, Ransick A. Development (Cambridge, UK) 1998;125:3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 3.Angerer L M, Angerer R C. Dev Biol. 2000;218:1–12. doi: 10.1006/dbio.1999.9553. [DOI] [PubMed] [Google Scholar]
- 4.Logan C Y, Miller J R, Ferkowicz M J, McClay D R. Development (Cambridge, UK) 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 5.Sherwood D R, McClay D R. Development (Cambridge, UK) 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- 6.Sweet H C, Gehring M, Ettensohn C A. Development (Cambridge, UK) 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 7.Howard E W, Newman L A, Oleksyn D W, Angerer R C, Angerer L M. Development (Cambridge, UK) 2001;128:365–375. doi: 10.1242/dev.128.3.365. [DOI] [PubMed] [Google Scholar]
- 8.Henry J J, Amemiya S, Wray G A, Raff R A. Dev Biol. 1989;136:140–153. doi: 10.1016/0012-1606(89)90137-1. [DOI] [PubMed] [Google Scholar]
- 9.Davidson E H, Rast J P, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 10.Davidson E H, Rast J P, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- 11.Oliveri P, Carrick D M, Davidson E H. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 12.Yuh C-H, Bolouri H, Davidson E H. Development (Cambridge, UK) 2001;128:617–628. doi: 10.1242/dev.128.5.617. [DOI] [PubMed] [Google Scholar]
- 13.Hörstadius S. Biol Rev Cambridge Philos Soc. 1939;14:132–179. [Google Scholar]
- 14.Rast J P, Amore G, Calestani C, Livi C B, Ransick A, Davidson E H. Dev Biol. 2000;228:270–286. doi: 10.1006/dbio.2000.9941. [DOI] [PubMed] [Google Scholar]
- 15.Rast J P, Cameron R A, Poustka A J, Davidson E H. Dev Biol. 2002;246:191–208. doi: 10.1006/dbio.2002.0654. [DOI] [PubMed] [Google Scholar]
- 16.Ransick A, Rast J P, Minokawa T, Calestani C, Davidson E H. Dev Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- 17.Brown C T, Rust A G, Clarke P J C, Pan Z, Schilstra M J, De Buysscher T, Griffin G, Wold B J, Cameron R A, Davidson E H, et al. Dev Biol. 2002;246:86–102. doi: 10.1006/dbio.2002.0619. [DOI] [PubMed] [Google Scholar]
- 18.Yuh C-H, Brown C T, Livi C B, Rowen L, Clarke P J C, Davidson E H. Dev Biol. 2002;246:148–161. doi: 10.1006/dbio.2002.0618. [DOI] [PubMed] [Google Scholar]
- 19.Li X T, Wikramanayake A H, Klein W H. Dev Biol. 1999;212:425–439. doi: 10.1006/dbio.1999.9360. [DOI] [PubMed] [Google Scholar]
- 20.Ransick A, Davidson E H. Development (Cambridge, UK) 1995;121:3215–3222. doi: 10.1242/dev.121.10.3215. [DOI] [PubMed] [Google Scholar]
- 21.Chuang C, Wikramanayake A H, Mao C, Li X, Klein W. Dev Genet. 1996;19:231–237. doi: 10.1002/(SICI)1520-6408(1996)19:3<231::AID-DVG6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 22.Gross R, McClay D R. Dev Biol. 2001;239:13–147. doi: 10.1006/dbio.2001.0426. [DOI] [PubMed] [Google Scholar]
- 23.Bolouri H, Davidson E H. Dev Biol. 2002;246:2–13. doi: 10.1006/dbio.2002.0617. [DOI] [PubMed] [Google Scholar]
- 24.Cameron R A, Fraser S E, Britten R J, Davidson E H. Development (Cambridge, UK) 1989;106:641–647. doi: 10.1242/dev.106.4.641. [DOI] [PubMed] [Google Scholar]
- 25.Kenny A P, Kozlowski D, Oleksyn D W, Angerer L M, Angerer R C. Development (Cambridge, UK) 1999;126:5473–5483. doi: 10.1242/dev.126.23.5473. [DOI] [PubMed] [Google Scholar]
- Oliveri, P., Davidson, E. H. & McClay, D. R. (2003) Dev. Biol., in press.
- 27.Ransick A, Davidson E H. Science. 1993;259:1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- 28.Zorn A M, Barish G D, Williams B O, Lavender P, Klymkowsky M W, Varmus H E. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 29.Barrow J R, Stadler H S, Capecchi M R. Development (Cambridge, UK) 2000;127:933–944. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- 30.Vesque C, Maconochie M, Nonchev S, Ariza-McNaughton L, Kuroiwa A, Charnay P, Krumlauf R. EMBO J. 1996;15:5383–5396. [PMC free article] [PubMed] [Google Scholar]
- 31.Nonchev S, Vesque C, Maconochie M, Seitanidou T, Ariza-McNaughton L, Frain M, Marshall H, Sham M H, Krumlauf R, Charnay P. Development (Cambridge, UK) 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- 32.Zelzer E, Shilo B-Z. Mech Dev. 2000;91:163–173. doi: 10.1016/s0925-4773(99)00295-6. [DOI] [PubMed] [Google Scholar]
- 33.Nasiadka A, Dietrich B H, Krause H M. Adv Dev Biol Biochem. 2002;12:155–204. [Google Scholar]
- 34.Stathopoulos A, Levine M. Dev Biol. 2002;246:57–67. doi: 10.1006/dbio.2002.0652. [DOI] [PubMed] [Google Scholar]
- 35.Cripps R M, Olson E N. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 36.Maduro M F, Rothman J H. Dev Biol. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- 37.Bolouri H, Davidson E H. BioEssays. 2002;24:1118–1129. doi: 10.1002/bies.10189. [DOI] [PubMed] [Google Scholar]
- 38.Rothenberg E V, Anderson M K. Dev Biol. 2002;246:29–44. doi: 10.1006/dbio.2002.0667. [DOI] [PubMed] [Google Scholar]