Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga (original) (raw)
Abstract
Dynamins are a eukaryote-specific family of GTPases. Some family members are involved in diverse and varied cellular activities. Here, we report that the primitive red alga Cyanidioschyzon merolae retains only one dynamin homolog, CmDnm1, belonging to the mitochondrial division subfamily. Previously, the bacterial cell division protein, FtsZ, was shown to localize at the mitochondrial division site in the alga. We showed that FtsZ and dynamin coexist as mitochondrial division-associated proteins that act during different phases of division. CmDnm1 was recruited from 10–20 cytoplasmic patches (dynamin patches) to the midpoint of the constricted mitochondrion-dividing ring (MD ring), which was observed as an electron-dense structure on the cytoplasmic side. CmDnm1 is probably not required for early constriction; it forms a ring or spiral when the outer mitochondrial membrane is finally severed, whereas the FtsZ and MD rings are formed before constriction. It is thought that the FtsZ, MD, and dynamin rings are involved in scaffolding, constriction, and final separation, respectively. In eukaryotes, mitochondrial severance is probably the most conserved role for the dynamin family.
The dynamin family is involved in diverse cellular activities in eukaryotes (reviewed in ref. 1), including endocytosis (2–4), vesicular transport (5, 6), and mitochondrial division (7, 8). Conventional dynamin is a 100-kDa GTPase involved in the final steps of vesicle fission during receptor-mediated endocytosis in a nucleotide-dependent manner (3) and can self-assemble to form rings and spirals (3, 4). During mitochondrial division, it was once thought that dynamin-related proteins caused mitochondrial constriction and fission by forming rings or collars, although it was pointed out that the mitochondrial tubule is too large for a dynamin ring (8).
Most eukaryotes retain mitochondria, which are responsible for aerobic respiration. Mitochondria are believed to be derived from endosymbiont bacteria, although it is still debated how the host cell maintained mitochondrial replication during evolution (reviewed in refs. 9 and 10). Although dynamin-related proteins are necessary for mitochondrial division in yeast (7, 11), nematodes (8), mammals (12), and green plants (13), some primitive eukaryotes retain bacterial FtsZ, which is also associated with mitochondrial division (14). Because FtsZ is not conserved in higher eukaryotes and both FtsZ and dynamin are self-assembling GTPases that localize at the division site, it was thought that dynamin replaced FtsZ in the course of evolution of higher eukaryotes. However, because dynamin seems to be involved in fission of the outer membrane (8), whereas FtsZ localizes to the mitochondrial matrix side (14, 15), the evolutional story of the mitochondrial division system is still a matter of debate.
In the cells of many higher eukaryotes, such as yeast, mitochondrial morphology is so plastic that it is difficult to sample a mitochondrial division event. However, in Cyanidioschyzon merolae, one of the most primitive eukaryotes (16), the single mitochondrion in each cell is round and undergoes orderly division. Previous studies have shown that C. merolae retains mitochondrial FtsZ (15), and an electron-dense structure called the mitochondrion-dividing (MD) ring is observed at the division site under electron microscopy. This is thought to be part of the primitive mitochondrial division system.
This study characterizes a dynamin homolog in C. merolae, CmDnm1, with detailed observation of FtsZ recruitment and MD ring behavior. We provide a model for primitive mitochondrial division involving FtsZ, the MD ring, and dynamin. Among the diverse cellular functions of dynamins, severing the tubular outer mitochondrial membrane is probably the most conserved, and may represent the general function of the dynamin family.
Materials and Methods
Cells.
C. merolae strain 10D-14 (17) was cultured in Allen's medium. For synchronization, the cells were subcultured to a density of <1 × 107 cells per ml and subjected to a 12-h light/12-h dark cycle at 42°C (16). Cells with defective division were obtained by adding 10 μg/ml 5-fluorodeoxiuridine to the growth medium from 10 h into the second light period to 4 h into the second dark period.
Antisera Generation.
Rabbit anti-CmDnm1 antisera were prepared as follows. The total coding region (amino acids 1–768) was cloned into the Escherichia coli expression vector pQE30 (Qiagen, Chatsworth, CA) to produce 6× His-tagged recombinant protein. The recombinant protein was purified with a HisTrap kit (Amersham Pharmacia). The protein was further purified by SDS/PAGE, and the excised protein in the gel slice was used to immunize rabbits.
Western Analysis.
Cells were centrifuged at 1,000 × g for 10 min and suspended in homogenizing buffer (20 mM Tris⋅HCl, pH 7.6/1 mM EDTA/0.1 mM PMSF/1 mM DTT, complete protease inhibitor mixture; Roche Molecular Biochemicals). Cell suspensions were homogenized by a Parr cell disruption bomb at 1,500 psi (Parr Instruments, Moline, IL). Samples were separated into supernatant and pellet fractions by centrifugation at 30,000 × g for 20 min. All procedures were performed on ice or at 4°C. Protein samples were separated by SDS/PAGE and transferred to poly(vinylidene difluoride) membrane. Membranes were blocked with 5% gelatin in TBS and incubated with anti-CmDnm1 or anti-CmFtsZ1 antiserum at a dilution of 1:1,000 in TBS as the primary antibody reaction, then in goat anti-rabbit antibody conjugated with alkaline phosphatase at a dilution of 1:1,000. Color was developed with an alkaline phosphatase conjugate substrate kit (Bio-Rad).
Fluorescence Microscopy.
Cells were stained with 500 nM Mito Tracker CMX-Ros (Molecular Probes) in growth medium for 45 min, fixed in chilled methanol containing 2% (wt/vol) paraformaldehyde and 10% (vol/vol) dimethyl sulfoxide for 5 min at −20°C, washed with methanol, and then washed with PBS. The following antibody reactions were performed in microtubes at 30°C. Fixed cells were incubated in 5% (wt/vol) BSA in PBS for 1 h, treated with a 1:50 dilution of rabbit anti-CmDnm1 or anti-CmFtsZ1 antiserum in 5% BSA in PBS for 1 h, and washed twice for 15 min. Cells were then treated with a 1:100 dilution of FITC-conjugated goat anti-rabbit secondary antibody (BioSource) in 5% (vol/vol) BSA in PBS for 1 h, and washed twice for 10 min. Cells were placed on glass slides and observed with a CSU10 confocal laser scanning system (Yokogawa) and IPLAB SPECTRUM software (Solution Systems). Images were processed digitally with PHOTOSHOP software (Adobe Systems, Mountain View, CA).
Electron Microscopy.
As previously described (18), cells were rapidly frozen in liquid propane chilled in liquid nitrogen, transferred to dried acetone containing 1% osmium tetroxide (OsO4) at −80°C, embedded in Spurr's resin, cut into 90-nm serial thin sections, and stained with uranyl acetate and lead citrate. For immunogold labeling, cells were frozen rapidly in liquid propane chilled in liquid nitrogen, fixed in dried acetone containing 2% (wt/vol) glutaraldehyde, embedded in LR-white resin, and cut into 90-nm serial thin sections. The sections were immunostained with anti-CmDnm1 antiserum at a dilution of 1:40, then with 10 nm of gold particle-conjugated goat anti-rabbit antibody (British BioCell International, Cardiff, U.K.), and stained with uranyl acetate. All samples were examined with a JEM-1200EX electron microscope (JEOL). Images were processed digitally with PHOTOSHOP software (Adobe Systems).
Results
CmDnm1 Belongs to a Subfamily Involved in Mitochondrial Division.
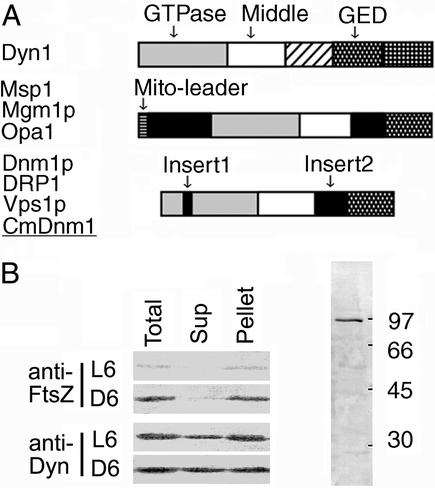
In C. merolae, the complete sequences of the mitochondrial (19), plastid (20), and nuclear (unpublished) genomes have revealed that Cmdnm1 on chromosome 5 is the only dynamin homolog that matches known dynamin family members with an E value less than 1e-50 in a blast database search. The deduced amino acid sequence of CmDnm1 is closest to that of Dnm1p of budding yeast (11) (39.6% identity). Dnm1p is required for mitochondrial division (11) and is thought to constrict mitochondria throughout division. The domain structure of CmDnm1 is also quite similar to that of the Dnm1p subfamily (Fig. 2A).
Figure 2.
(A) Simplified protein domains of representative dynamin family members (1). Dyn1 (dynamin 1) is involved in endocytosis (3, 4) in animals and is a prototype member of the family. It has a GTPase domain for GTP binding and hydrolysis, a middle conserved domain, pleckstrin homology, a GTPase effecter domain (GED) for self-assembly, and a proline-rich domain. Fission yeast _Msp_I, budding yeast Mgm1p, and human Opa1 are involved in the maintenance of mitochondrial morphology and localize at the mitochondrial inner membrane (reviewed in refs. 1 and 10). Dnm1p is required for mitochondrial division in yeast (7, 11). Nematode DRP1 was shown to be required for mitochondrial outer membrane fission (8). Yeast Vps1p is involved in vesicular transport (5, 6) but not in mitochondrial division. (B) Western blotting with anti-CmDnm1 and anti-CmFtsZ antisera. The numbers to the right of the right lane indicate molecular masses (kDa). L6 and D6 indicate the hours after the start of the second light or dark period, respectively. Total synchronous cultures (Total) were separated into supernatant (Sup) and pellet (Pellet) fractions by centrifugation at 30,000 × g for 20 min (left lanes).
Mitochondrial Division Can Be Separated into Three Phases.
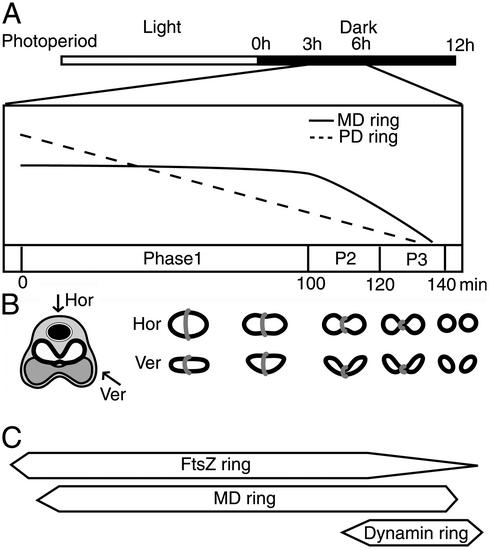
C. merolae cells contain a single mitochondrion and chloroplast that divide synchronously during light and dark cycles (16) (Fig. 1A). A previous study revealed the existence of electron-dense components at the mitochondrial and plastid division sites, named the MD and plastid-dividing (PD) rings (21), respectively. Three-dimensional reconstruction from serial thin sections of C. merolae cells using electron microscopy (22) and real-time analysis of chloroplast division in living cells (23) revealed that mitochondria and chloroplasts divide as the MD and PD rings, respectively, contract. The decreasing relative circumferences of the MD and PD rings, which reflect the progression of organelle constriction (23), were examined in time-course studies (Fig. 1A). The disk-shaped C. merolae mitochondrion becomes dumbbell-shaped and divides, accompanied by bending at the dividing plane, in a regular order (Fig. 1B). Based on previous studies (22, 23), mitochondrial division can be separated into three phases based on differences in the circumference, thickness and width of the MD ring (Fig. 1A). In phase 1, the MD ring is in place but does not contract for ≈100 min; in phase 2, the MD ring begins to contract; and in phase 3, the MD ring becomes thickened (23), and over a period of ≈20 min the mitochondrion is divided. The dynamin ring appears in phase 3, preceded by both the FtsZ and MD rings (Fig. 1C), shown below.
Figure 1.
Relationship among the cell division cycle, organelle division, and the apparatus associated with mitochondrial division. (A) Diagrams of the organelle division cycle. Open and filled images in the top bar indicate 12-h light/12-h dark photoperiods of synchronous culture (22), respectively. Hours after the beginning of the dark period are indicated above. Changes in the relative circumferences of the PD and MD rings between 3 and 6 h are drawn with broken and solid lines, respectively (23). The mitochondrial division phases defined in this study are indicated as phase 1, P2, and P3. (B) A simplified representation of a dividing C. merolae cell and mitochondrion with an MD ring. Major cell components are indicated with images as follows: cytoplasm, pale gray; nucleus, filled black with envelope; dumbbell-shaped mitochondrion, outlined; plastid, dark gray with outline. Images of mitochondria and MD rings at each division stage corresponding to A are represented by simple outline drawings and gray images, respectively. Hor and Ver indicate images viewed in the horizontal and vertical planes of the cell (the upper side is the cytosol), respectively. (C) The formation of the apparatus associated with mitochondrial division, revealed in this and previous (22) studies, corresponding to A. The width of the boxes indicates the intensity of localization of a putative formed ring.
CmDnm1 Is Recruited Dynamically Between Cytoplasmic Patches and the Mitochondrial Constriction Site, but Is Not Required for Early Constriction.
For biochemical and cytological analyses of CmDnm1, we generated antisera against recombinant CmDnm1 protein (Fig. 2B). Western analysis was performed at each cell division stage by using total, supernatant, and pellet fractions with anti-CmDnm1 or anti-CmFtsZ1 (15) antisera. With anti-CmDnm1 antisera, a band of ≈90 kDa was specifically detected in the total lysate; this was consistent with the expected size of CmDnm1 (Fig. 2B, right lane), although in some cases the band split into two, depending on the sample preparation conditions. Little difference in the CmDnm1 signals of interphase and dividing cells was observed overall, or in the proportions in the supernatant and pellet fractions (Fig. 2B, anti-Dyn). In the case of CmFtsZ1, signals were consistently detected only in the total and pellet fraction of dividing cells (Fig. 2B, anti-FtsZ D6 Pellet), whereas little or no signal was detected from cells in interphase (Fig. 2B, anti-FtsZ L6) or from the supernatant fraction of dividing cells (Fig. 2B, anti-FtsZ D6 Sup), indicating that most of the mitochondria were in the pellet fraction.
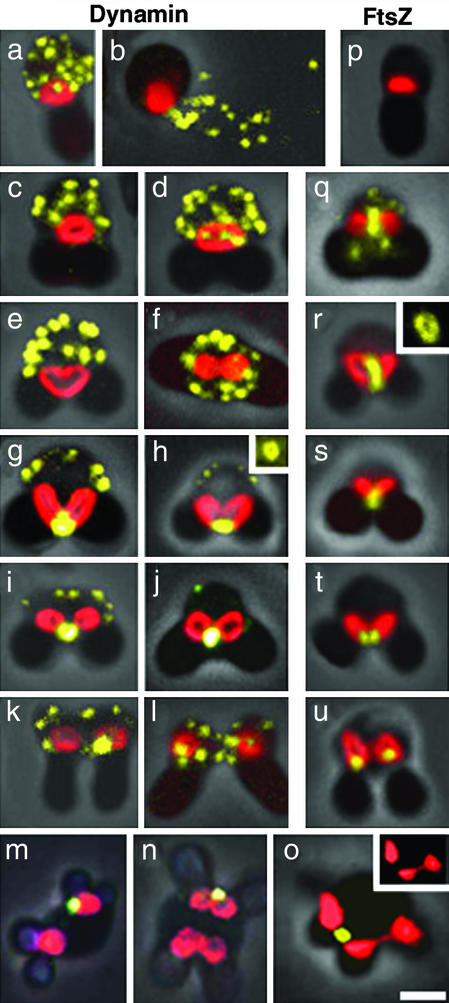
To examine the subcellular localization of CmDnm1 and to compare it with FtsZ, cells were stained with MitoTracker CMXRos (Molecular Probes) to visualize mitochondria, then fixed and immunolabeled with anti-CmDnm1 or anti-CmFtsZ1 antisera. CmDnm1 signals appeared as 10–20 cytoplasmic patches (dynamin patches) in interphase cells (Fig. 3a), even after the cells had been thoroughly squashed (Fig. 3b). The patches remained in the cytoplasm during phase 2 of mitochondrial division, when the mitochondria began to contract at the center and become heart or dumbbell shaped (Fig. 3 c–f). When viewed in the horizontal plane, the mitochondria appeared constricted (Fig. 3f). In the final phase, the cytoplasmic dynamin patches were reduced and correspondingly strong immunolabeling accumulated to form a ring (Fig. 3h Inset) at the site of mitochondrial constriction (Fig. 3 g–j). After mitochondrial division, CmDnm1 labeling was located on one side of the divided daughter mitochondria in many cases and was then dispersed in the cytoplasm (Fig. 3 k and l).
Figure 3.
Immunofluorescence images of CmDnm1 and CmFtsZ1 in C. merolae from confocal laser scanning microscopy. Mitochondria stained with MitoTracker CMXRos (Molecular Probes) are shown in red; signals detected with fluorescein-conjugated secondary antibody are shown in yellow; and cell outlines are shown with overlaying phase-contrast images. (a–l) Localization of CmDnm1 during the sequence of cell division. (p–u) Localization of CmFtsZ1 during the sequence of cell division. (m_–_o) Localization of CmDnm1 in cells with defective cell division. Insets in h and r, respectively, show immunofluorescence of the dynamin and FtsZ rings from other samples that were squashed to obtain spread images. Inset in o shows images of mitochondria at half scale without overlaying. All of the cell images are displayed with the cytoplasmic region uppermost and chloroplasts below, except f and m–o, which are displayed as viewed from the cytoplasmic side. (Bar = 1 μm.)
Cells with cell division defects were also immunostained. After pulse treatment with 5-fluorodeoxiuridine, the cells contained two mitochondria and multiple chloroplasts, as the cell cycle was arrested at S phase, whereas organelle division continued (24). After binary fission of mitochondria with CmDnm1 immunolabeling on one side of the mitochondrion (Fig. 3m), the two daughter mitochondria began to divide simultaneously and became dumbbell shaped, but only one was immunoreactive to CmDnm1 (Fig. 3n). Mitochondrial division would probably have been completed only at the site with CmDnm1 accumulation (Fig. 3o). Furthermore, quantitative analyses of dynamin signals using a video-intensified photon-counting system (25) showed that the total immunolabeling count from the cytoplasmic dynamin patches of an interphase cell (30 × 106 photons) was close to that of the labeling at the bridge of the two daughter mitochondria in an M phase cell (28 × 106 photons).
FtsZ Ring Formation Precedes Mitochondrial Constriction.
Because mitochondrial constriction precedes dynamin ring formation, we suspect that the dynamin ring behaves differently from the mitochondrial FtsZ ring. FtsZ is also a self-assembling GTPase that was originally involved in bacterial cell division (reviewed in ref. 26). FtsZ was introduced into plastids (27) and mitochondria (24) via endosymbiosis, although the ftsZ genes were transferred into the host nucleus (14). In a previous immunoelectron microscopy study (15), the mitochondrial FtsZ ring was shown to localize on the matrix side of the inner membrane at the predicted division site. We performed immunofluorescence microscopy throughout the cell division cycle by using the method established in this study. Unlike dynamin, FtsZ is absent in cells at interphase (Fig. 3p). The FtsZ ring (Fig. 3r, Inset) appears early in phase 1 of mitochondrial division (Fig. 3 q and r). In some of these cells, weak signals that were not associated with mitochondria were observed; they were not as clear as the dynamin patches (Fig. 3q). The FtsZ ring contracts, constricting the mitochondrion (Fig. 3 q–s), until the final split into daughter mitochondria (Fig. 3 t and u).
CmDnm1 Is Recruited to the Space Between the Contracted MD Ring and Outer Membrane for Final Severance.
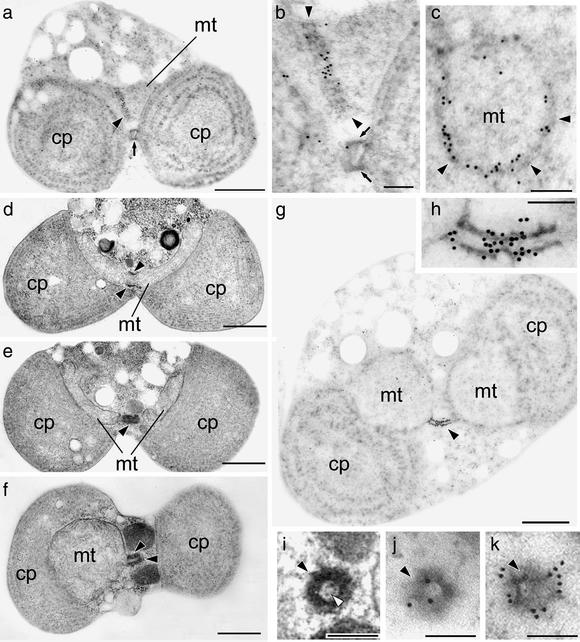
To examine how the dynamin ring is arranged in association with the MD ring and mitochondrial membrane, we performed immunoelectron microscopy against CmDnm1 (Fig. 4). The labeling density at each subcellular site and in the background was quantified (28) (Table 1). Corresponding high-contrast images without immunolabeling are also shown (Fig. 4 d–f and i). The MD ring was negative for CmDnm1 immunoreactivity in the early stage of mitochondrial division, when it had constricted to a circumference of just over 2.2 μm, as estimated from cross sections (Table 1). As constriction proceeded, until the circumference was <2.2 μm in late phase 2, gold particles appeared on the MD ring (Table 1, Fig. 4 a and b). Round sections of the MD ring showed CmDnm1 lining signals inside the MD ring (Fig. 4c). In phase 3, when the circumference was <1.5 μm, the MD ring elongated and became cylindrical (Fig. 4 e and f, filled arrowheads), and antibody labeling was specifically concentrated at the center of the cylinder (Fig. 4g, magnified in h). As MD ring labeling increased, cytoplasmic labeling decreased correspondingly (Table 1). Serial round sections of these cylinders showed that at the constricted center of the cylinder, antibody labeling was located inside the MD ring (Fig. 4j). In the next section, beside the constriction site, the outside of the MD ring was labeled (Fig. 4k). The corresponding high-contrast image shows the inner division apparatus, presumably attaching to the inner membrane (Fig. 4i, open arrowhead).
Figure 4.
Serial thin sections viewed by transmission electron microscopy. (a) Immunoelectron micrographs of a cell at late phase 2 of mitochondrial division against CmDnm1. Sections grazing the MD and PD rings at late phase 2. (b) A ×3 magnified image of a. (c) A round section of an MD ring in early phase 3. (d–f) Thin-section electron microscopy showing higher contrast images; d shows an earlier stage with a larger MD ring circumference than in e or f. (g) Immunoelectron micrographs of an MD ring bridging a dividing mitochondrion at late phase 3. (h) A ×5 magnified image of g. (i–k) Round section of the constricted MD ring site at late phase 3, which is predicted to correspond to the midpoint of the MD ring in h. (i) Thin-section electron microscopy showing higher contrast images. (j and k) Immunoelectron micrographs of two serial sections. The inner membrane (open arrowhead) can be observed in i but is invisible in j. pt, plastid; mt, mitochondrion; arrow, PD ring; filled arrowhead, MD ring. (Bars = 500 nm: a, and d–g; 100 nm: b, c, and h–k).
Table 1.
Quantitative calculation of immunolabeling density at each subcellular site against CmDnm1
| Phase | 1 | 2 | 3 |
|---|---|---|---|
| MD ring* | X > 2.2 | 2.2 ≧ X > 1.5 | 1.5 ≧ X |
| Cells† | 40 | 22 | 25 |
| Site‡ | Labeling density§ | ||
| Cytoplasm | 19.3 ± 3.5 | 15.0 ± 2.9 | 3.7 ± 3.7 |
| Mitochondria | 1.6 ± 0.9 | 1.5 ± 1.0 | 1.0 ± 0.6 |
| MD ring | 0 | 6.9 ± 9.1 | 146.8 ± 88.4 |
| Background | 0.4 ± 0.5 | 0.1 ± 0.3 | 0 |
Discussion
Dynamic Recruitment of CmDnm1.
Western blot analyses suggested that CmDnm1 is partially soluble in the cytosol, and more than half is associated with membranous structures, including mitochondria, throughout the cell division cycle. Immunofluorescence studies have indicated that there is dynamic recruitment of CmDnm1 to between 10 and 20 cytoplasmic dynamin patches and the constricted mitochondrial division site, and suggest that CmDnm1 accumulation is not required for the equatorial contraction of mitochondria during the early and middle division stages. Immunoelectron microscopy further indicated that CmDnm1 is recruited to the cytoplasmic surface of the tubular outer membrane, which is surrounded by a constricted cylindrical MD ring that bridges the future two daughter mitochondria. This implicates CmDnm1 in the final severance of the tubular mitochondria, but not in early constriction. In addition, galactose-induced expression of CmDnm1 in Saccharomyces cerevisiae resulted in serious defects in mitochondrial division, similar to the dnm1 mutant (data not shown). CmDnm1 might antagonize Dnm1p because their functions seem quite similar. We postulate that the function of dynamin in mitochondrial division is common to other organisms. These studies do not conflict with the basic function of dynamin, which is to pinch off membrane systems at either the final stage of membrane fission in mitochondrial division or in vesicle formation during endocytosis (3, 4). Here, a problem arises. Why does CmDnm1 exist as dynamin patches during interphase? We observed a few patches remaining near the cell surface in the cytoplasm, even when most of the CmDnm1 had accumulated at the mitochondrial constriction site (Fig. 3 g–j). Given that the function of dynamin is common to most organisms, CmDnm1 may have other roles, such as that of Vps1p in yeast (5, 6); CmDnm1 might be a multifunctional dynamin.
Mitochondrial Division Model for Primitive Eukaryotes and the Conserved Function of Dynamin.
Dynamin-related proteins were thought to play a major role throughout mitochondrial constriction and to have replaced the role of FtsZ in higher eukaryotes. Our results, however, suggest that the dynamin ring is involved only in the final separation of the mitochondrion, after the FtsZ and MD rings have constricted, and that each ring has a different function, is localized differently, and operates in different phases. We propose that mitochondrial division has three phases: scaffolding is built at the division site, which involves FtsZ ring formation; the MD ring constricts; and the membrane is severed, which involves dynamin (Fig. 5). FtsZ localizes to form a ring on the matrix side of the mitochondrial inner envelope (15), as it does in plastids (18, 27). Following FtsZ ring formation, the MD ring forms on the cytoplasmic side of the mitochondrion and thickens as it contracts (23) in phases 1 and 2. During phase 3, CmDnm1 is recruited from cytoplasmic patches to the inside of the constricting MD ring, and to the cytosolic surface of the outer membrane, before final severance of the mitochondrion produces two daughter mitochondria (Fig. 5).
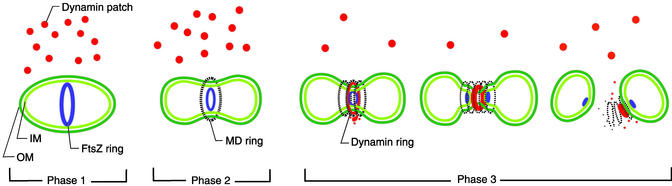
Figure 5.
Model for mitochondrial division in a primitive eukaryote. The mitochondrial outer (OM) and inner (IM) membranes are drawn in dark and light green, respectively. The localization of mitochondrial FtsZ and dynamin is indicated in blue and red, respectively. Images of the MD ring are represented with black broken lines. In phase 1, the FtsZ ring forms from the matrix side, and the division site is determined. Dynamin is located in cytoplasmic patches. After FtsZ ring formation, the MD ring appears on the cytosolic side of the outer membrane. In phase 2, the FtsZ and MD rings begin to constrict the mitochondrion. Dynamin is not involved in early constriction. When the mitochondrion has constricted sufficiently to become tubular at the division site, dynamin is recruited from the patches to a space inside the thickened MD ring and outside the outer membrane in the first step of phase 3. In the second step, division of the mitochondrial matrix and inner membrane is complete, and FtsZ is split to form two divided matrices, followed by outer membrane fission. As dynamin forms a ring or spiral, the outer membrane is pinched off. Dynamin then sticks to one side of the daughter mitochondria and returns to the patches in the third step of phase 3.
Evolution of the Mitochondrial Division Apparatus and Eukaryotes.
We suggest that in the course of evolution dynamin has had a universal function in severing the mitochondrion, even in the endosymbiont event, which involves severing the envelope from the plasma membrane (29). FtsZ, which may be involved in division site placement or scaffolding, might have been lost or replaced by another factor on the matrix side. The MD ring, which may cause mitochondrial constriction, might have been simplified or degenerated because mitochondria have a plastic form rather than a bacteria-like shape in higher eukaryotes and thus require other division machinery. In the case of another endosymbiont organelle, the plastid, FtsZ and the PD ring are also involved in the division event (18, 27). If the two components work in a manner similar to mitochondrial FtsZ and the MD ring, respectively, it is possible that dynamin is required for the final stage of plastid division. Because the endosymbiont event is one of the major factors characterizing eukaryotes, dynamins might have played an essential role in developing eukaryotes in the course of evolution.
Acknowledgments
We thank Akio Toh-e (University of Tokyo) and his laboratory members for helping us with the yeast experiments. This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 12446222 and 12874111 (to T.K.) and by a grant from the Program for the Promotion of Basic Research Activities for Innovative Bioscience (to T.K.).
Abbreviations
MD
mitochondrion-dividing
PD
plastid-dividing
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AY162473 (CmDnm1)].
References
- 1.van der Bliek A M. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen M S, Obar R A, Schroeder C C, Austin T W, Poodry C A, Wadsworth S C, Vallee R B. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 3.Takei K, McPherson P S, Schmid S L, De Camilli P. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 4.Hinshaw J E, Schmid S L. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 5.Rothman J H, Raymond C K, Gilbert T, O'Hara P J, Stevens T H. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- 6.Wilsbach K, Payne G S. EMBO J. 1993;8:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleazard W, McCaffery J M, King E J, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw J M. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labrousse A M, Zapaterra M, Rube D A, van der Bliek A M. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 9.Gray M W, Burger G, Lang B F. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe M P. Science. 1999;283:1493. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- 11.Otsuga D, Keegan B R, Brisch E, Thatcher J W, Hermann G J, Bleazard W, Shaw J M. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova E, Griparic L, Shurland D L, van der Bliek A M. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arimura S, Tsutsumi N. Proc Natl Acad Sci USA. 2002;99:5727–5731. doi: 10.1073/pnas.082663299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beech P L, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson P R, McFadden G I. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- 15.Takahara M, Kuroiwa H, Miyagishima S, Mori T, Kuroiwa T. Cytologia. 2001;66:421–425. [Google Scholar]
- 16.Suzuki K, Ehara T, Osafune T, Kuroiwa H, Kawano S, Kuroiwa T. Eur J Cell Biol. 1994;63:280–288. [PubMed] [Google Scholar]
- 17.Toda K, Takano H, Miyagishima S, Kuroiwa H, Kuroiwa T. Biochim Biophys Acta. 1998;1403:72–84. doi: 10.1016/s0167-4889(98)00031-7. [DOI] [PubMed] [Google Scholar]
- 18.Miyagishima S, Takahara M, Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. Plant Cell. 2001;13:2257–2268. doi: 10.1105/tpc.010185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohta N, Sato N, Kuroiwa T. Nucleic Acids Res. 1998;26:5190–5298. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohta N, Sato N, Kuroiwa T. In: Enigmatic Microorganisms and Extreme Environments. Seckbach J, editor. Dordrecht, The Netherlands: Kluwer; 1999. pp. 139–149. [Google Scholar]
- 21.Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. Int Rev Cytol. 1998;181:1–41. doi: 10.1016/s0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- 22.Miyagishima S, Kuroiwa H, Kuroiwa T. Planta. 2001;212:517–528. doi: 10.1007/s004250000426. [DOI] [PubMed] [Google Scholar]
- 23.Miyagishima S, Itoh R, Toda K, Kuroiwa H, Kuroiwa T. Planta. 1999;207:343–353. doi: 10.1007/s004250050645. [DOI] [PubMed] [Google Scholar]
- 24.Itoh R, Takahashi H, Toda K, Kuroiwa H, Kuroiwa T. Eur J Cell Biol. 1996;71:303–310. [PubMed] [Google Scholar]
- 25.Matsuzaki M, Kikuchi T, Kita K, Kojima S, Kuroiwa T. Protoplasma. 2001;218:180–191. doi: 10.1007/BF01306607. [DOI] [PubMed] [Google Scholar]
- 26.Bramhill D. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 27.Osteryoung K W, McAndrew R S. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:315–333. doi: 10.1146/annurev.arplant.52.1.315. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K, Oikawa K, Ohta N, Kuroiwa H, Kuroiwa T, Takahashi H. Science. 1996;272:1932–1935. doi: 10.1126/science.272.5270.1932. [DOI] [PubMed] [Google Scholar]
- 29.van der Bliek A M. J Cell Biol. 2000;151:F1–F4. doi: 10.1083/jcb.151.2.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]