A Plant-Specific Dynamin-Related Protein Forms a Ring at the Chloroplast Division Site (original) (raw)
Abstract
Chloroplasts have retained the bacterial FtsZ for division, whereas mitochondria lack FtsZ except in some lower eukaryotes. Instead, mitochondrial division involves a dynamin-related protein, suggesting that chloroplasts retained the bacterial division system, whereas a dynamin-based system replaced the bacterial system in mitochondria during evolution. In this study, we identified a novel plant-specific group of dynamins from the primitive red alga Cyanidioschyzon merolae. Synchronization of chloroplast division and immunoblot analyses showed that the protein (CmDnm2) associates with the chloroplast only during division. Immunocytochemical analyses showed that CmDnm2 appears in cytoplasmic patches just before chloroplast division and is recruited to the cytosolic side of the chloroplast division site to form a ring in the late stage of division. The ring constricts until division is complete, after which it disappears. These results show that a dynamin-related protein also participates in chloroplast division and that its behavior differs from that of FtsZ and plastid-dividing rings that form before constriction at the site of division. Combined with the results of a recent study of mitochondrial division in Cyanidioschyzon, our findings led us to hypothesize that when first established in lower eukaryotes, mitochondria and chloroplasts divided using a very similar system that included the FtsZ ring, the plastid-dividing/mitochondrion-dividing ring, and the dynamin ring.
INTRODUCTION
Eukaryotic cells contain organelles, such as chloroplasts and mitochondria, that must divide during cell division to be inherited by progeny cells. Chloroplasts and mitochondria probably arose by endosymbiosis of a cyanobacterium and an α-proteobacterium, respectively, with a host cell (Gray, 1992). This concept raises a fundamental question about the establishment of eukaryotes. How did the host cell control replication of the endosymbionts to turn them into organelles? To date, too few components of each division apparatus have been defined to propose a unified model for the division of the two organelles.
Two structures have been identified around the chloroplast division site. The plastid-dividing (PD) ring can be detected by transmission electron microscopy as two or three electron-dense rings: an outer ring on the cytosolic face of the outer envelope (Mita et al., 1986), an inner ring on the stromal face of the inner envelope (Hashimoto, 1986), and occasionally a middle ring in the intermembrane space (Miyagishima et al., 1998; reviewed by Kuroiwa et al., 1998). The outer ring is the main part of the PD ring; it consists of a bundle of unidentified 5-nm filaments (Miyagishima et al., 2001b). A series of molecular genetic studies showed that nucleus-encoded homologs of FtsZ, a self-assembling GTPase originally involved in prokaryotic division (Bi and Lutkenhaus, 1991; reviewed by Bramhill, 1997), are involved in chloroplast division (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998; reviewed by Osteryoung and Pyke, 1998) and form a ring structure at the division site, like bacterial FtsZ (Mori et al., 2001; Vitha et al., 2001). The PD and FtsZ rings have been identified in several photosynthetic eukaryotes and shown to be distinct structures: the FtsZ ring localizes in the stroma and faces the inner PD ring on the stromal side (Miyagishima et al., 2001c; Kuroiwa et al., 2002; summarized by Osteryoung, 2001).
Although nucleus-encoded FtsZ also is found in mitochondria in some lower eukaryotes (Beech et al., 2000; Takahara et al., 2000, 2001; Gilson and Beech, 2001), most eukaryotes lack mitochondrial FtsZ. Recent studies have shown that a member of the dynamin family of self-assembling GTPases (for review, see Hinshaw, 2000) localizes at the cytosolic surface of the mitochondrial division site and is involved in mitochondrial division in animals (Labrousse et al., 1999; Smirnova et al., 2001), fungi (Bleazard et al., 1999; Sesaki and Jensen, 1999), and plants (Arimura and Tsutsumi, 2002). Because dynamin assembles to form a ring or spiral at the neck of a clathrin-coated pit (Hinshaw and Schmid, 1995; Takei et al., 1995), it has been assumed that a similar ring structure formed by a dynamin-related protein is involved in the constriction of the mitochondrial division site. Consequently, it has been suggested that during evolution, a dynamin-based system replaced the FtsZ-based system in mitochondrial division (Beech et al., 2000; Osteryoung, 2001; Arimura and Tsutsumi, 2002). However, the universality of mitochondrial dynamin suggests that it also exists in lower eukaryotes. Therefore, it is expected that both FtsZ and the dynamin-related protein are involved in mitochondrial division in lower eukaryotes. In fact, our recent study of the primitive red alga Cyanidioschyzon merolae showed that both FtsZ and the dynamin-related protein (CmDnm1) form rings at the site of mitochondrial division (Nishida et al., 2003).
Given that a dynamin-related protein also is associated with mitochondrial division in lower eukaryotes and that the system of chloroplast and mitochondrial division is quite similar in primitive red algae, based on the facts that (1) FtsZ forms a ring in both chloroplast (CmFtsZ2) (Miyagishima et al., 2001c) and mitochondrial (CmFtsZ1) (Takahara et al., 2001; Nishida et al., 2003) division, and (2) the mitochondrion-dividing (MD) ring (Kuroiwa et al., 1993, 1998), which is an electron-dense ring structure similar to the PD ring, behaves in a very similar manner to the PD ring (Miyagishima et al., 1999b, 2001a), we postulate that a dynamin-related protein also participates in chloroplast division.
In this study, we identified a plant-specific group of dynamins from the nuclear genome of the primitive red alga Cyanidioschyzon and found that the protein (CmDnm2) forms a ring at the chloroplast division site. Our data suggest that the FtsZ, PD, and dynamin rings orchestrate chloroplast division, that these rings are formed in chloroplast division in this order, and that each acts differently. This system of chloroplast division presumably reflects ancestral mitochondrial division associated with FtsZ, MD, and dynamin rings and leads to the hypothesis that chloroplasts and mitochondria divided in a very similar manner soon after they were established in lower eukaryotes.
RESULTS
CmDnm2 and Two Unknown Arabidopsis Proteins Constitute a Novel Plant-Specific Group of Dynamin-Related Proteins
The genomes of higher plants, such as Arabidopsis, contain many dynamin-like sequences (ADLs in Figure 1A) (Park et al., 1997; Kang et al., 1998; Mikami et al., 2000), and some of these dynamins are known to be involved in activities other than chloroplast division, such as cell plate formation (Gu and Verma, 1996; summarized by Hinshaw, 2000). In addition to these reported sequences, many Arabidopsis sequences similar to dynamin exist in sequence databases. Consequently, it was advantageous to examine a simple alga with a small genome to verify the existence of a dynamin-like protein involved in chloroplast division. We searched for dynamin-like proteins in the complete genome sequences of the mitochondrion (Ohta et al., 1998), chloroplast (Ohta et al., 1999), and nucleus (M. Matsuzaki and T. Kuroiwa, unpublished data; 16.4 Mb) of the unicellular red alga Cyanidioschyzon. This alga, one of the most primitive eukaryotes (Nozaki et al., 2003), contains a single mitochondrion and chloroplast (Suzuki et al., 1994).
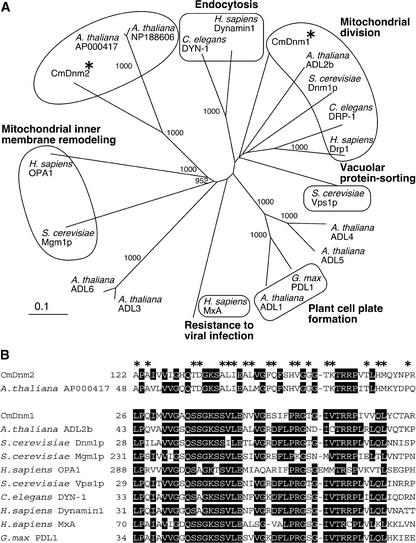
Figure 1.
CmDnm2 and Two Unknown Arabidopsis Proteins Form a Plant-Specific Monophyletic Group in the Phylogenetic Tree of the Dynamin Family of Proteins.
(A) Phylogenetic tree constructed using the N-J method (Saitou and Nei, 1987). Bootstrap values of >900 based on 1000 replications are shown, and some known or predicted functions are annotated according to Hinshaw (2000). Asterisks indicate CmDnm1 and CmDnm2.
(B) Comparison of partial amino acid sequences of CmDnm2 and other proteins. Identical amino acids present in more than eight sequences are boxed in black, and asterisks indicate amino acids identical in CmDnm2 and AP000417.
Before this study, we found a single gene that encodes a protein similar to known dynamin family members (the expectation in the BLAST [Basic Local Alignment Search Tool] search was set to <1e−50). The gene Cmdnm1 encodes a protein that is most similar to Dnm1p (39.6% identical; Figure 1) and that is involved in mitochondrial division in budding yeast (Bleazard et al., 1999; Sesaki and Jensen, 1999). Recently, CmDnm1 was shown to form a ring at the mitochondrial division site in Cyanidioschyzon (Nishida et al., 2003). By weakening the stringency of the search, we found another gene that encodes a dynamin-like protein (the expectation of the BLAST search was limited to <1e−4), Cmdnm2 (Figure 1). There are not any more genes similar to dynamin family genes in the Cyanidioschyzon genome. Cmdnm2 encodes a protein that is most similar (40% identity) to two Arabidopsis proteins with unknown functions. (Accession numbers for the nucleoticde sequences are AP00417 and NP_188606. These protein IDs are BAB02559.1 and At3g19720, respectively.) Among characterized proteins, Cmdnm2 also is partially similar (at the N-terminal half) to human dynamin2 (Hinshaw, 2000) (the expectation by BLAST search was 5e−15). Cmdnm1 and Cmdnm2 are predicted to encode proteins of 768 and 962 residues, respectively, which share 26% identity at amino acid positions 119 to 474 of CmDnm2.
Phylogenetic analyses demonstrated that CmDnm2 and the two Arabidopsis proteins form a monophyletic group that is distinct from the group involved in mitochondrial division, endocytosis, and other activities (Figure 1A). In databases of ESTs, we found sequences of algae and plants with partial sequences specific to this group (data not shown). In this tree, the group that includes CmDnm2 is close to the group that includes human OPA1 and budding yeast Mgm1p, which are imported into mitochondria (reviewed by Hinshaw, 2000). CmDnm2 and the two Arabidopsis proteins have no N-terminal extensions, no predicted transit peptides, and no membrane-spanning domains, suggesting that they are cytosolic proteins unlike Mgm1p and OPA1. Therefore, we concluded that this group of dynamins is specific to eukaryotes that contain chloroplasts.
CmDnm2 Associates with Chloroplasts Only during the Division Phase
To determine the relationship between CmDnm2 and chloroplasts, we generated antiserum to bacterially expressed protein. We obtained antiserum specific to a protein corresponding to the predicted molecular mass of CmDnm2 (106.6 kD) (Figure 2A). Immunoblot analyses showed that some of the CmDnm2 associated with isolated chloroplasts (Figure 2B), that it associated with the insoluble fraction of chloroplasts after osmotic bursting, and that it was solubilized with 0.5% Nonidet P-40 and 0.1 mg/mL DNaseI (Figure 2C). In Cyanidioschyzon, cell and organelle division can be synchronized using a 12-h-light/12-h-dark cycle, and the organelles divide in M-phase (the dark period) (Suzuki et al., 1994). In synchronous cultures, little CmDnm2 was detected in interphase cells in the light period. By contrast, CmDnm2 was detected at the start of the dark period, and the level decreased as cell and organelle division progressed (Figure 2D). When the cell cycle was arrested at S phase by adding 5-fluorodeoxyuridine, mitochondrial division was arrested, whereas chloroplast division occurred continuously, producing four or eight chloroplasts per cell (Itoh et al., 1996; Miyagishima et al., 1999a). In this case, the level of CmDnm2 remained constant after the onset of the dark period (Figure 2D). These results suggest that some CmDnm2 associates with the chloroplast specifically during the division phase.
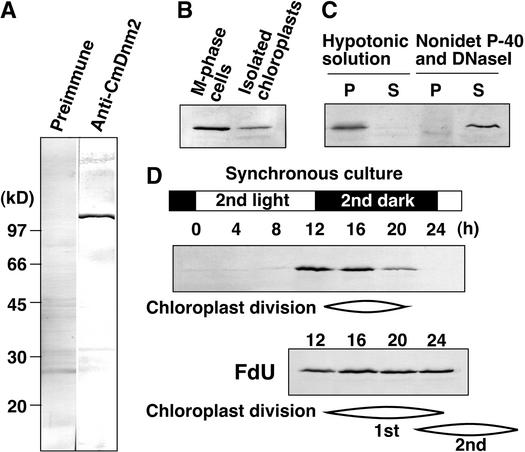
Figure 2.
CmDnm2 Associates with Chloroplasts Only during the Division Phase.
Immunoblot analyses using anti-CmDnm2 antibodies. Twenty micrograms of protein in each sample was separated in each lane, except for in (C), in which samples were obtained from isolated chloroplasts containing 100 μg of protein.
(A) Total protein from synchronized M-phase cells was blotted with preimmune antisera or anti-CmDnm2 antibodies.
(B) and (C) Dividing chloroplasts were isolated from M-phase synchronous culture (B) and fractionated further into the pellet (P) and supernatant (S) by osmotic bursting or treatment with 0.5% Nonidet P-40 and 0.1 mg/mL DNaseI and then centrifugation (C).
(D) Aliquots of the synchronous culture were collected at the indicated times, and total proteins were separated. 5-Flurodeoxyuridine (FdU) was added to the culture at a concentration of 10 μg/mL at 2 h before the onset of the second dark period. The cell cycle was arrested at S-phase by 5-flurodeoxyuridine, whereas chloroplast division occurred continuously, producing four or eight chloroplasts per cell.
CmDnm2 Is Recruited from Cytosolic Patch Structures to Form a Ring at the Chloroplast Division Site at the Late Stage of Division
To examine the localization of CmDnm2 in dividing chloroplasts, we performed immunofluorescence microscopy. In M-phase cells, CmDnm2 was localized at the constricted region of the dividing chloroplast as a closed ring, and some was detected in the cytosol as patch structures (Figure 3A). Preimmune antisera were used as controls for antibody labeling, and they produced little signal (data not shown). These results indicate that some CmDnm2 exists in the cytosol and the rest associates with chloroplast at the division site.
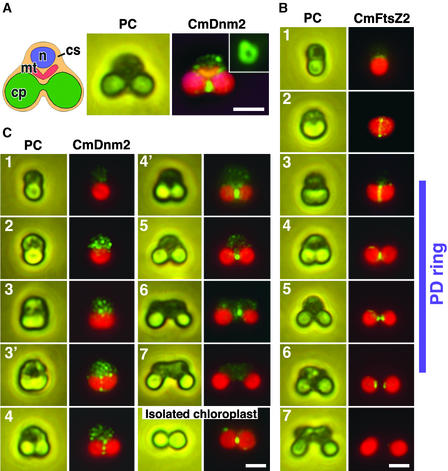
Figure 3.
Immunofluorescence Images of CmDnm2 Accumulating to Form a Ring at the Chloroplast Division Site Only during the Late Stage of Division.
(A) Scheme of Cyanidioschyzon cells during M-phase (left), and a phase-contrast image (PC; middle) and an immunofluorescent image (CmDnm2; right) of the same cell showing CmDnm2 localization. When cells were squashed, CmDnm2 ring structures were observed as a closed ring (inset). cp, chloroplast, cs, cytosol; mt, mitochondrion; n, nucleus.
(B) Immunofluorescence (CmFtsZ2) and phase-contrast (PC) images of the chloroplast FtsZ ring. The chloroplast division cycle is separated into seven phases. The acorn-shaped chloroplast in the interphase cell (1) changes to a cup (2), trapezoid (3), and dumbbell (4 and 5) shape in M-phase, divides (6), and the resulting chloroplasts are inherited by the daughter cells during cytokinesis (7). The phases in which the PD ring is present are indicated at right.
(C) Immunofluorescence (CmDnm2) and phase-contrast (PC) images showing the behavior of CmDnm2 during the chloroplast division cycle and the presence of CmDnm2 on an isolated chloroplast.
Green fluorescence indicates CmDnm2 in (A) and (C) and CmFtsZ2 in (B). Red fluorescence indicates the autofluorescence of chlorophyll. Orange fluorescence in (A) shows the mitochondrion stained by MitoTracker. Bars = 2 μm.
To further examine the behavior of CmDnm2 during chloroplast division, we compared changes in the localization of CmDnm2 with the shape of the chloroplast and the localization of chloroplast FtsZ (CmFtsZ2) (Takahara et al., 2000; Miyagishima et al., 2001c). The uniform cell size in culture and previous time-lapse observations of chloroplast and cell division allow us to order the images of fixed cells (Miyagishima et al., 1999b). In Cyanidioschyzon, the acorn-shaped chloroplast in the interphase cell (phase 1) becomes cup shaped (phase 2), trapezoid shaped (phase 3), and then dumbbell shaped (phases 4 and 5) in M-phase. It divides (phase 6), and the resulting chloroplasts are inherited by the daughter cells during cytokinesis (phase 7) (Figure 3B). The FtsZ ring forms before the onset of division site constriction in the cup-shaped chloroplast (phase 2), and the total fluorescence intensity decreases with constriction (phases 3 and 4) (Figure 3B) (Miyagishima et al., 2001c). At the final stage of constriction (phase 5), the FtsZ ring disassembles and the proteins separate toward the two future daughter chloroplasts (Figure 3B) (Miyagishima et al., 2001c). A split fluorescent signal is detected after chloroplast division (phase 6) and disappears before cytokinesis (phase 7) (Figure 3B). The cytosolic outer PD ring forms after the FtsZ ring in the trapezoidal chloroplast (phase 3) and disappears in the cytosol after chloroplast division (phase 6) (Figure 3B) (Miyagishima et al., 2001a, 2001c).
Although CmDnm2 was not detected in interphase cells (phase 1), many labeled patches appeared in the cytosol at the onset of M-phase (phase 2) (Figure 3C). At the start of chloroplast division site constriction, patches also were detected around the division site in approximately half of the cells that contained trapezoidal chloroplasts (phases 3 and 3′) (Figure 3C). Most of the patches in the cytosol and some of those around the chloroplast division site were detected during early constriction of the division site (phase 4) (Figure 3C). In these cells, the intensity of the fluorescence of patches in the cytosol and at the chloroplast division site was similar. At the late stage of division site constriction, the cytosolic patches diminished, and there was a corresponding accumulation of fluorescent labeling to form a ring at the chloroplast division site, as in Figure 3A (phase 4′) (Figure 3C). A more constricted signal was observed at the final phase of chloroplast division (phase 5) (Figure 3C). After division, labeling was seen on one side of the daughter chloroplasts in many cases (phase 6) (Figure 3C), as reported for dynamin-related proteins in mitochondrial division (Bleazard et al., 1999; Labrousse et al., 1999; Arimura and Tsutsumi, 2002). It then disappeared before cytokinesis (phase 7) (Figure 3C).
These sequential immunofluorescence images (Figure 3), together with the immunoblot results (Figure 2D), suggest that CmDnm2 is translated specifically just before the onset of chloroplast division and is assembled into cytosolic patches that are recruited to the site of chloroplast division during constriction and after the formation of the FtsZ and PD rings. Consequently, CmDnm2 patches accumulate to form a ring structure only during the late stage of constriction. The ring constricts, and proteolysis occurs after chloroplast division. It is estimated that the CmDnm2 ring is present for 1 of the 2 hours required for total constriction of the chloroplast-dividing site in Cyanidioschyzon (Miyagishima et al., 1999b). The CmDnm2 ring was conserved in isolated chloroplasts from M-phase synchronous culture (Figure 3C), confirming the immunoblot result (Figure 2B) that only a portion of the CmDnm2 was associated with the chloroplast during M-phase and the rest was located in the cytosol. These results demonstrate that the CmDnm2 ring forms from cytosolic patch structures containing CmDnm2 that are grouped around the chloroplast division site.
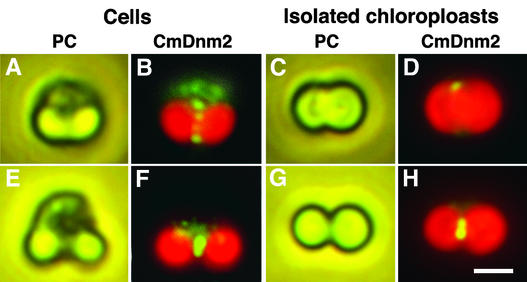
To examine when CmDnm2 binds to the chloroplast, we compared the localization of CmDnm2 in whole cells and isolated chloroplasts (Figure 4). During the early stage of chloroplast division (corresponding to phase 4 in Figure 3C), CmDnm2 patch structures were observed in the cytosol and around the chloroplast division site in whole cells (Figures 4A and 4B), whereas few signals were detected around isolated chloroplasts (Figures 4C and 4D). During the late stage of chloroplast division (corresponding to phase 4′ in Figure 3C), similar accumulations of CmDnm2 signals were detected in whole cells (Figures 4E and 4F) and isolated chloroplasts (Figures 4G and 4H). These results suggest that during the early stage of constriction, CmDnm2 patch structures do not bind tightly to the chloroplast and that CmDnm2 associates with the chloroplast at the late stage of constriction, when most CmDnm2 accumulates at the chloroplast division site.
Figure 4.
Immunofluorescence Images Showing That CmDnm2 Binds to Chloroplasts Only during the Late Stage of Division.
(A) to (D) Phase-contrast (PC) and immunofluorescence (CmDnm2) images showing the localization of CmDnm2 in a cell ([A] and [B]) or an isolated chloroplast ([C] and [D]) during the early stage of chloroplast division corresponding to phase 4 in Figure 3.
(E) to (H) Phase-contrast (PC) and immunofluorescence (CmDnm2) images showing the localization of CmDnm2 in a cell ([E] and [F]) or an isolated chloroplast ([G] and [H]) during the late stage of chloroplast division corresponding to phase 4′ in Figure 3.
Bar in (H) = 2 μm for (A) to (H).
The CmDnm2 Ring Is Distinct from the Outer PD Ring and Associates with the Outer Envelope at the Division Site Only at the Final Phase of Division
Because CmDnm2 is predicted to be a cytosolic protein, the CmDnm2 ring probably is on the cytosolic face of the constricted region of the dividing chloroplast. Chloroplast FtsZ has transit peptides and forms a ring inside the chloroplast (McAndrew et al., 2001; Miyagishima et al., 2001c; Mori et al., 2001; Kuroiwa et al., 2002; reviewed by Osteryoung, 2001). The PD ring is a double or triple ring structure, and the main part (the outer PD ring) faces the chloroplast on the cytosolic face of the outer envelope (Hashimoto, 1986; Mita et al., 1986; Kuroiwa et al., 1998; Miyagishima et al., 1998, 1999a, 1999b, 2001a, 2001b, 2001c) (Figures 5A to 5C). Therefore, the predicted location of the CmDnm2 ring is similar to that of the outer PD ring. We reported previously that the outer PD ring is insoluble in Nonidet P-40 and that the electron density and dimensions of the outer PD ring were preserved after Nonidet P-40 treatment (Miyagishima et al., 2001b). Immunoblot analysis showed that Nonidet P-40 solubilized CmDnm2 (Figure 2C), unlike the outer PD ring. Immunofluorescence results (Figures 3 and 4) showed that CmDnm2 forms a closed ring only during the late stage of division, unlike the PD ring, and that during early constriction isolated chloroplasts possess a PD ring (Miyagishima et al., 1999a) but do not contain CmDnm2. These results suggest that CmDnm2 is not a major component of the outer PD ring and is distinct from the 5-nm filaments that constitute the outer PD ring.
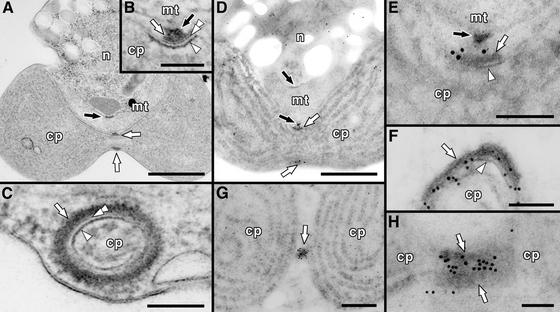
Figure 5.
Electron and Immunoelectron Micrographs Showing the Location of the CmDnm2 Ring.
(A) to (C) Electron micrographs of a Cyanidioschyzon cell containing a dividing chloroplast and mitochondrion.
(A) A whole cell showing the PD ring and the MD ring.
(B) and (C) Magnified cross-sectional (B) and circumferential (C) views of the PD ring showing that it is composed of three rings.
(D) to (H) Immunoelectron micrographs showing the localization of CmDnm2.
(D) Whole image of a cell containing a chloroplast during the late stage of division, as in (A).
(E) Magnified image of the region around the PD ring in (D), as in (B).
(F) Circumferential view of the PD ring, as in (C). From its diameter, this ring is thought to be constricted more than the ring shown in (D) and (E).
(G) Whole image of a chloroplast at the final stage of division.
(H) Magnified image of the region around the PD ring at the final stage of chloroplast division.
Gold particles indicate the location of CmDnm2. White arrows, arrowheads, and double arrowheads indicate the outer (at the cytosolic face of the outer envelope), inner (at the stromal face of the inner envelope), and middle (in the intermembrane space) PD rings, respectively. Black arrows indicate the MD ring. cp, chloroplast; mt, mitochondrion; n, nucleus. Bars in (A), (D), and (G) = 500 nm; bar in (B) = 50 nm; bars in (C), (E), (F), and (H) = 100 nm.
To further confirm the location of the CmDnm2 ring at higher resolution and examine its positional relationship with the PD ring and the envelope membrane, we performed immunoelectron microscopy. At the late stage of chloroplast division, some gold particles accumulated on the cytosolic side of the chloroplast at the constricted region, and others were dispersed in the cytosol (Figures 5D and 5E; corresponding to phase 4′ in Figure 3C), confirming that CmDnm2 is a cytosolic protein and that it forms a ring outside of the chloroplast. During this stage, CmDnm2 localized at the cytosolic face of the outer PD ring (on the far side of the outer envelope at the division site) (Figure 5E). As constriction progressed, the CmDnm2 label migrated to a position between the outer PD ring and the outer envelope (Figure 5F). At the final stage of division, increased labeling was detected in the constricted region (Figure 5G; corresponding to phase 5 in Figure 3C), and most was located between two deposits of the outer PD ring (on the near side of the outer envelope) (Figure 5H). In all cases, little gold labeling overlaid the electron-dense PD ring, although it was seen on the edges of the deposits, except in tangential sections of the PD ring, as in Figure 5G. By arranging these images in temporal order according to the stage of constriction, these observations suggest that the CmDnm2 ring first faces the outer PD ring at the far side from the outer envelope and then migrates to the space between the outer PD ring and the outer envelope, recruiting CmDnm2 from the cytosol. Thus, the CmDnm2 ring can directly access the outer envelope at the division site only in the final phase of chloroplast division.
DISCUSSION
The primitive red alga Cyanidioschyzon has only two dynamin family proteins. We reported here that one of them, CmDnm2, localizes at the chloroplast division site, showing a mechanism common to chloroplast and mitochondrial division. The other protein, CmDnm1, was shown previously to associate in mitochondrial division (Nishida et al., 2003). Although it is unclear whether endocytosis occurs in Cyanidioschyzon, one Golgi apparatus, many vesicles, lysosome-like structures (S. Miyagishima and T. Kuroiwa, unpublished observation), thylakoids, and cristae exist, and one peroxisome proliferates by binary fission (Miyagishima et al., 2001a). Although dynamin-related proteins are associated with several membrane systems (Hinshaw, 2000), the generation of membrane compartments other than the chloroplast and mitochondrion likely occurs without dynamin-related protein in this primitive red alga.
Role of the Dynamin-Related Protein in Chloroplast Division
Although punctate structures formed by dynamin-related proteins are observed at the mitochondrial division site in several species, the stage of their formation has not been examined thoroughly, probably because of the complicated morphology of mitochondria, which changes with division and fusion in many cells. The role of dynamin in endocytosis during the fission of vesicles from the plasma membrane via ring formation led to the hypothesis that the ring of dynamin-related protein plays a role in the constriction of the mitochondrial division site. Although both vesicle formation and organelle division are common phenomena based on membrane fission, there is a difference in the dimensions involved. The diameter of a clathrin-coated vesicle is <100 nm. In vitro rings of dynamin (Hinshaw and Schmid, 1995) and dynamin-related proteins (e.g., human Drp1 for mitochondrial division; Smirnova et al., 2001) and in vivo dynamin rings (reviewed by Hinshaw, 2000) are <50 nm in size. This is far smaller than the diameters of chloroplasts and mitochondria without constriction or during the early division stage. However, this study showed that CmDnm2 associates with the chloroplast and forms a ring only during the late stage of division site constriction. CmDnm1 also forms a ring structure at the mitochondrial division site only during the late stage of division (Nishida et al., 2003). Therefore, dynamin rings form after the division site has constricted to an appropriate diameter. This idea is consistent with the involvement of dynamins in endocytosis, during which a dynamin ring forms after the formation of a constricted coated pit with a neck (Warnock and Schmid, 1996). The ring-like localization observed in this study does not necessarily mean that CmDnm2 self-assembled as a higher order structure. After ring formation, CmDnm2 still accumulated with the progression of the late stage constriction. It is possible that a higher order oligomer of CmDnm2 forms only at the final pinching-off stage.
Dynamin plays an essential role in clathrin-mediated endocytosis, but its precise function remains controversial (discussed by Sever et al., 2000a). Several models explain how dynamin severs membranes by a mechanical-chemical means based on the formation and constriction of a self-assembled ring (Warnock and Schmid, 1996; Sever et al., 2000a). By contrast, recent studies have raised the possibility that dynamin acts as a regulatory GTPase rather than as a mechanical-chemical enzyme (Sever et al., 2000a, 2000b). The overexpression of dynamin that is defective in GTPase activity stimulated by self-assembly (K694A cannot self-assemble and R725A can self-assemble) accelerated the formation of constricted coated pits. Subsequent vesicle release from coated pits is increased by the K694A mutation but inhibited by the R725A mutation (Sever et al., 2000b). These results suggest that self-assembly is not a prerequisite for efficient endocytosis. Instead, dynamin:GTP controls the formation of constricted pits and recruits additional components that more directly mediate membrane fission. A high rate of GTP hydrolysis stimulated by self-assembly is required for the disassembly of dynamin so that the subsequent fission step can occur (Sever et al., 2000a, 2000b). Therefore, in this scenario, dynamin is turned off by self-assembly and after GTP hydrolysis, contrary to the hypothesis that its mechanical-chemical function is turned on by ring formation. A similar result was obtained for Dnm1p in mitochondrial division in yeast (Fukushima et al., 2001).
By either model, CmDnm2 oligomerization at the late stage of chloroplast division would be required for further constriction or pinching off of the outer envelope by monitoring the diameter of the division site or force generation. Although some patches involving CmDnm2 were detected near the division site during the initial constriction of the division site, they did not bind to the chloroplast. CmDnm2 begins to associate with the chloroplast during the late stage of division. The interaction between dynamin and the membrane is thought to be essential for its role (reviewed by Sever et al., 2000a). Therefore, if CmDnm2 regulates chloroplast division before its oligomerization, the function would be restricted to the late stage of chloroplast division. This speculation is consistent with the observation that in Caenorhabditis elegans, muscle cells with mutant DRP-1 have constrictions in the mitochondrial outer membranes, suggesting that DRP-1 is involved only during the late stage of division site constriction (Labrousse et al., 1999). To determine whether dynamin is involved only in the late stage of chloroplast division, as well as the universality of the process, it is necessary to determine at which stage the expression of the appropriate dominant-negative proteins or the disruption of the gene arrests chloroplast division.
Although methods for a genetic approach have not been established in Cyanidioschyzon, Arabidopsis proteins most related to CmDnm2 may be useful for further studies. We obtained a T-DNA insertional line of Arabidopsis (Garlic_71_D11. b.1a.Lb3Fa) from the Torrey Mesa Research Institute (Syngenta, San Diego, CA) (McElver et al., 2001). The sequence around the left border of the T-DNA indicates that the left border is located just before nucleotide position 2702 of AP000417 (from the estimated start codon), which corresponds to intron 8 (the gene is estimated to consist of 16 exons). In a preliminary study, we found no remarkable difference in leaf chloroplast morphology in 20 plants compared with wild-type plants. No differences in chloroplast morphology might be caused by the lethality of the homozygous T-DNA insertion or by redundancy between AP000417 and NP_188606. Nevertheless, further study of the genotypes of the plants and the expression of the two genes in each tissue are needed.
The fact that dynamin-related proteins associate with both the chloroplast and the mitochondrion during division suggests that factors related to these proteins are conserved in chloroplast and mitochondrial division. In Saccharomyces cerevisiae, two proteins that bind to Dnm1p have been identified and shown to be involved in mitochondrial division. One protein, Mdv1p/Fis2p/Gag3p/Net2p, is a WD-repeat protein that colocalizes with Dnm1p (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001). The other protein, Fis1p/Mdv2p, is an integral component of the mitochondrial membrane that is distributed evenly on the surface of the mitochondrion (Mozdy et al., 2000; Tieu and Nunnari, 2000). There are no obvious homologs of these proteins in Cyanidioschyzon or Arabidopsis, suggesting that these partners are not involved in chloroplast division, although other common partners may await discovery.
The Chloroplast Division Apparatus Is Composed of FtsZ, PD, and Dynamin Rings
The solubilization of CmDnm2 by Nonidet P-40 and the formation of a ring during the late division stage that differs from the PD ring (Miyagishima et al., 2001b) indicate that CmDnm2 is distinct from the 5-nm filaments that constitute the outer PD ring. It needs to be determined whether CmDnm2 is an integral component of the outer PD ring or forms a separate structure. Under immunoelectron microscopy, the gold particles showing the localization of CmDnm2 did not overlay the outer PD ring, although they were seen around the ring. Because of the technical limitations of immunoelectron microscopy, such as antigen accessibility, this observation may be insufficient to conclude that CmDnm2 forms a ring that is separate from the outer PD ring. Our previous ultrastructural study showed that the outer PD ring is composed mainly of 5-nm filaments arranged in rows at 6.4-nm intervals. The ring constricts while maintaining this interval (Miyagishima et al., 2001b). Given the fact that the size of one dynamin molecule is ∼10 nm (Zhang and Hinshaw, 2001), it is unlikely that CmDnm2 or a higher order structure resulting from self-assembly can locate between the 5-nm filaments. Therefore, CmDnm2 probably forms a ring that is separate from the PD ring and faces the membrane directly, like other members of the dynamin family, as shown by immunoelectron microscopy.
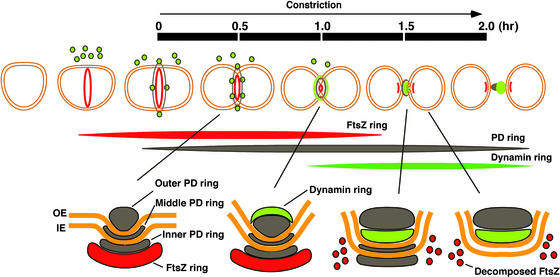
Chloroplast division now can be understood as a sequence of steps involving multiple ring structures that appear at different times (Figure 6). Because the PD ring consists of three rings that behave differently during chloroplast division (Miyagishima et al., 2001a), at least five ring structures are involved in chloroplast division and each ring behaves differently. The FtsZ ring forms first, inside the chloroplast, and then the PD ring forms before division site constriction (Miyagishima et al., 2001c). The FtsZ ring localizes at the stromal side of the inner PD ring (Miyagishima et al., 2001c). During the late constriction stage, the dynamin ring forms on the cytosolic side of the outer PD ring from cytoplasmic patches and then migrates to the space between the outer PD ring and the outer envelope. In addition, during this stage, the FtsZ ring disassembles and FtsZ is excluded from the division site toward two future daughter chloroplasts (Miyagishima et al., 2001c). The middle and inner PD rings disappear just before the completion of division (Miyagishima et al., 2001a), whereas the remnant of the outer PD ring remains between the daughter chloroplasts and the remnants of the dynamin ring cling to either daughter chloroplast. Of the five rings, only the outer PD ring exists throughout division site constriction. Constriction starts just after the formation of the outer PD ring (Miyagishima et al., 2001a). Consequently, the outer PD ring and the 5-nm filaments that make up the ring (Miyagishima et al., 2001b) likely are most associated with generating the constriction force from the initial stage.
Figure 6.
Scheme of Chloroplast Division by the FtsZ, PD, and Dynamin Rings.
Sequential events during chloroplast division are illustrated at top. A cross-section of the division site is shown at bottom. The time scale from trapezoidal chloroplast to just after division (from the start to the end of constriction) (Miyagishima et al., 1999a) is indicated. Red indicates the FtsZ ring; green indicates patches or rings of dynamin-related protein; and black indicates the PD ring (only the cytosolic outer PD ring is shown at top). IE, inner envelope; OE, outer envelope. See text for details.
Evolutionary Relationship between Chloroplast and Mitochondrial Division
The FtsZ, PD, and dynamin rings form in chloroplast division in that order. Several lines of evidence suggest that a similar mechanism exists in the mitochondria of lower eukaryotes and explain its change during evolution. These are as follows. (1) A dynamin-related protein also is involved in mitochondrial division in diverse eukaryote species (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999; Smirnova et al., 2001; Arimura and Tsutsumi, 2002), although these species lack mitochondrial FtsZ. (2) In lower eukaryotes, including Cyanidioschyzon, FtsZ also participates in mitochondrial division (Beech et al., 2000; Takahara et al., 2000; Gilson and Beech, 2001; Nishida et al., 2003). (3) In mitochondrial and chloroplast division in Cyanidioschyzon, the MD and PD ring structures change in a very similar manner during contraction and disassembly (Miyagishima et al., 1999b, 2001a). (4) In the true slime mold Physarum polycephalum, a MD ring–like structure is observed in dividing mitochondria (T. Kuroiwa, unpublished observation). (5) Our recent study showed that the FtsZ, MD, and dynamin rings form in that order in mitochondria in Cyanidioschyzon (Nishida et al., 2003). These results suggest that when first established in lower eukaryotes, mitochondria and chloroplasts divided using a very similar mechanism that was orchestrated by the FtsZ, PD/MD, and dynamin rings. During eukaryotic evolution, this system was conserved in principle in chloroplasts, whereas the FtsZ ring was lost in mitochondria; the MD ring might have been lost or simplified, and the rings involved from the start of constriction could have been replaced by unknown mechanisms. This hypothesis will enable further investigation of the chloroplast division mechanism based on information regarding the mitochondrial division mechanism and vice versa. This study is an example of work that has been promoted on the basis of this concept.
METHODS
Synchronization and Isolation of Dividing Chloroplasts
Cyanidioschyzon merolae 10D was cultured in Allen's medium (Allen, 1959). For synchronization, the cells were subcultured to <1 × 107 cells/mL and subjected to a 12-h-light/12-h-dark cycle at 45°C (Suzuki et al., 1994). To arrest the cell cycle at S-phase, 5-fluorodeoxyuridine (10 μg/mL; Sigma) was added to the culture 2 h before the onset of the second dark period. Dividing chloroplasts were isolated as described previously (Miyagishima et al., 1999a). Isolated chloroplasts were suspended in isolation medium (20 mM Tris, pH 7.6, 5 mM MgCl2, 5 mM KCl, 5 mM EGTA, and 300 mM Suc) and kept on ice until use. Further fractiona-tion of isolated chloroplasts was performed as described previously (Miyagishima et al., 2001b, 2001c). Isolated dividing chloroplasts were lysed at a concentration of 1 mg total protein/mL in Suc-free isolation medium with or without 0.5% Nonidet P-40 and 0.1 mg/mL DNaseI (D4527; Sigma) for 1 h on ice. The soluble and insoluble fractions were separated by centrifugation at 20,000_g_ for 15 min at 4°C. The supernatants were enriched 20-fold by ultrafiltration with Microcon-30 (30-kD cutoff; Millipore, Bedford, MA) and mixed with a one-third volume of 4 × Laemmli sample buffer (50 mM Tris, pH 6.8, 6% 2-mercaptoethanol, 2% sodium dodecyl sulfate, 10% glycerol) for immunoblot analyses.
Antibodies and Immunoblot Analyses
The anti-CmFtsZ2 antibody used for this study was generated previously (Takahara et al., 2000). ESTs showed that Cmdnm2 lacked introns. The region representing amino acids 71 to 457 of CmDnm2 was amplified by PCR from nuclear DNA with primers 5′-CACCGCCGAAGAGAGAACGATGAACC-3′ and 5′-TTAGACGCGCCCCTTTTCTTTTAG-3′ and cloned into pET100 expression vector (Invitrogen, Carlsbad, CA). Six His fusion proteins were expressed in Escherichia coli (strain BL21 Star DE3; Invitrogen), purified using nickel–nitrilotriacetic acid agarose resin (HisTrap; Amersham Pharmacia), and separated electrophoretically. Gel slices were homogenized and were injected into rabbits to stimulate antibody production. Each rabbit received 1.5 mg of purified polypeptide. Immunoblot analyses were performed as described previously (Miyagishima et al., 2001c) using 12.5% acrylamide gels. The primary antibody was diluted 1:1000.
Immunofluorescence and Immunoelectron Microscopy
Immunofluorescence microscopy was performed according to a protocol described by Nishida et al. (2003). Synchronous interphase and M-phase cultures of Cyanidioschyzon cells were fixed in 1% paraformaldehyde and 10% DMSO in methanol at −20°C for 5 min, washed with methanol, and washed with PBS. The cells were labeled with primary antibody diluted 1:100 in blocking buffer (PBS containing 5% BSA) at 30°C for 2 h. Primary antibodies were detected with Alexa Fluoro 488 goat anti-rabbit IgG for CmDnm2 or goat anti-mouse IgG (Molecular Probes, Eugene, OR) for CmFtsZ2 at a dilution of 1:400 at 30°C for 1 h, and cells were observed as described previously (Miyagishima et al., 2001c). Preimmune antisera were used as controls for antibody labeling, and they produced little signal. To visualize mitochondria, cells were labeled with 100 nM MitoTracker CMX-Ros (Molecular Probes) for 45 min before fixation.
Immunoelectron microscopy was performed as described previously (Itoh et al., 1999). In brief, cells were fixed by rapid freezing/freeze substitution in dried acetone and embedded in LR White resin (London Resin Co., London, UK). Thin sections of ∼70 nm were labeled with primary antibody diluted 1:100 and then labeled with 10-nm gold particle–conjugated secondary antibody (British BioCell International, Cardiff, UK) diluted 1:80.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the nucleotide sequences described in this article are AY162473 (Cmdnm1) and AB094119 (Cmdnm2). The accession numbers for the sequence data used in Figure 1 are as follows: Arabidopsis thaliana ADL1 (L36939), ADL2b (AB072375), ADL3 (AB026987), ADL4 (NP_567094), ADL5 (AAF22293), and ADL6 (AAF22291); Homo sapiens Drp1 (AF000430); Caenorhabditis elegans DRP-1 (AF166274); Saccharomyces cerevisiae Dnm1p (L40588); C. elegans DYN-1 (AF167982); H. sapiens Dynamin1 (NP_004399); S. cerevisiae Mgm1p (X62834); H. sapiens MxA (P20591); H. sapiens OPA1 (AB011139); Glycine max PDL1 (U25547); and S. cerevisiae Vps1p (M33315).
Acknowledgments
We thank C. Saito (Nara Institute of Science and Technology, Ikoma, Nara, Japan) for careful reading of the manuscript. This work was supported by a research fellowship from the Japanese Society for the Promotion of Science for Young Scientists (7498) to S.M. and by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (12446222 and 12874111) and from the Program for the Promotion of Basic Research Activities for Innovative Biosciences to T.K.
References
- Allen, M.B. (1959). Studies with Cyanidium caldarium, an anomously pigmented chlorophyte. Arch. Microbiol. 32**,** 270–277. [DOI] [PubMed] [Google Scholar]
- Arimura, S., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99**,** 5727–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech, P.L., Nheu, T., Schultz, T., Herbert, S., Lithgow, T., Gilson, P.R., and McFadden, G.I. (2000). Mitochondrial FtsZ in a chromophyte alga. Science 287**,** 1276–1279. [DOI] [PubMed] [Google Scholar]
- Bi, E., and Lutkenhaus, J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354**,** 161–164. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1**,** 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill, D. (1997). Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13**,** 395–424. [DOI] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel _DNM1_-interacting protein, Net2p. Mol. Biol. Cell 12**,** 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes, P., Shepard, K.A., and Yaffe, M.P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151**,** 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, N.H., Brisch, E., Keegan, B.R., Bleazard, W., and Shaw, J.M. (2001). The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol. Biol. Cell 12**,** 2756–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson, P.R., and Beech, P.L. (2001). Cell division protein FtsZ: Running rings around bacteria, chloroplasts and mitochondria. Res. Microbiol. 152**,** 3–10. [DOI] [PubMed] [Google Scholar]
- Gray, M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141**,** 233–357. [DOI] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15**,** 695–704. [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H. (1986). Double-ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma 135**,** 166–172. [Google Scholar]
- Hinshaw, J.E. (2000). Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 16**,** 483–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw, J.E., and Schmid, S.L. (1995). Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374**,** 190–192. [DOI] [PubMed] [Google Scholar]
- Itoh, R., Takahashi, H., Toda, K., Kuroiwa, H., and Kuroiwa, T. (1996). Aphidicolin uncouples the chloroplast division cycle from the mitotic cycle in the unicellular red alga Cyanidioschyzon merolae. Eur. J. Cell Biol. 71**,** 303–310. [PubMed] [Google Scholar]
- Itoh, R., Takano, H., Ohta, N., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (1999). Two ftsH-family genes encoded in the nuclear and chloroplast genomes of the primitive red alga Cyanidioschyzon merolae. Plant Mol. Biol. 41**,** 321–337. [DOI] [PubMed] [Google Scholar]
- Kang, S.G., Jin, J.B., Piao, H.L., Pih, K.T., Jang, H.J., Lim, J.H., and Hwang, I. (1998). Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol. Biol. 38**,** 437–447. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, H., Mori, T., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2002). Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215**,** 185–190. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Kuroiwa, H., Sakai, A., Takahashi, H., Toda, K., and Itoh, R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181**,** 1–41. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Suzuki, K., and Kuroiwa, H. (1993). Mitochondrial division by an electron-dense ring in Cyanidioschyzon merolae. Protoplasma 175**,** 173–177. [Google Scholar]
- Labrousse, A.M., Zappaterra, M.D., Rude, D.A., and van der Bliek, A.M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4**,** 815–826. [DOI] [PubMed] [Google Scholar]
- McAndrew, R.S., Froehlich, J.E., Vitha, S., Stokes, K.D., and Osteryoung, K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127**,** 1656–1666. [PMC free article] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159**,** 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami, K., Iuchi, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). A novel Arabidopsis thaliana dynamin-like protein containing the pleckstrin homology domain. J. Exp. Bot. 51**,** 317–318. [DOI] [PubMed] [Google Scholar]
- Mita, T., Kanbe, T., Tanaka, K., and Kuroiwa, T. (1986). A ring structure around the dividing plane of the Cyanidium caldarium chloroplast. Protoplasma 130**,** 211–213. [Google Scholar]
- Miyagishima, S., Itoh, R., Aita, S., Kuroiwa, H., and Kuroiwa, T. (1999. a). Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga Cyanidioschyzon merolae. Planta 209**,** 371–375. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Kuroiwa, H., and Kuroiwa, T. (1999. b). Real-time analyses of chloroplast and mitochondrial division and differences in the behavior of their dividing rings during contraction. Planta 207**,** 343–353. [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Takahashi, H., Kuroiwa, H., and Kuroiwa, T. (1998). Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J. Electron Microsc. 47**,** 269–272. [Google Scholar]
- Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001. a). The timing and manner of disassembly of the apparatuses for chloroplast and mitochondrial division in the red alga Cyanidioschyzon merolae. Planta 212**,** 517–528. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S., Takahara, M., and Kuroiwa, T. (2001. b). Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13**,** 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima, S., Takahara, M., Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2001. c). Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13**,** 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2001). Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 42**,** 555–559. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151**,** 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, K., Takahara, M., Miyagishima, S., Kuroiwa, H., Matsuzaki, M., and Kuroiwa, T. (2003). Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- Nozaki, H., Matsuzaki, M., Takahara, M., Misumi, O., Kuroiwa, H., Hasegawa, M., Shin-i, T., Kohara, Y., Ogasawara, N., and Kuroiwa, T. (2003). The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J. Mol. Evol., in press. [DOI] [PubMed]
- Ohta, N., Sato, N., and Kuroiwa, T. (1998). Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 26**,** 5190–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, N., Sato, N., and Kuroiwa, T. (1999). The organellar genomes of Cyanidioschyzon merolae. In Enigmatic Microorganisms and Extreme Environments, J. Seckbach, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 139–149.
- Osteryoung, K.W. (2001). Organelle fission in eukaryotes. Curr. Opin. Microbiol. 4**,** 639–646. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Pyke, K.A. (1998). Plastid division: Evidence for a prokaryotically derived mechanism. Curr. Opin. Plant Biol. 1**,** 475–479. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., Stokes, K.D., Rutherford, S.M., Percival, A.L., and Lee, W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10**,** 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Vierling, E. (1995). Conserved cell and organelle division. Nature 376**,** 473–474. [DOI] [PubMed] [Google Scholar]
- Park, J.M., Kang, S.G., Pih, K.T., Jang, H.J., Piao, H.L., Yoon, H.W., Cho, M.J., and Hwang, I. (1997). A dynamin-like protein, ADL1, is present in membranes as a high-molecular-mass complex in Arabidopsis thaliana. Plant Physiol. 115**,** 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4**,** 406–425. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147**,** 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000. a). Garrotes, spring, ratchets, and whips: putting dynamin models to the test. Traffic 1**,** 385–392. [DOI] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000. b). Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 150**,** 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Griparic, L., Shurland, D.-L., and van der Bliek, A.M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12**,** 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp, R., Scholz, S., Kruse, S., Speth, V., and Reski, R. (1998). Plant molecular gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95**,** 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Ehara, T., Osafune, T., Kuroiwa, H., Kawano, S., and Kuroiwa, T. (1994). Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur. J. Cell Biol. 63**,** 280–288. [PubMed] [Google Scholar]
- Takahara, M., Kuroiwa, H., Miyagishima, S., Mori, T., and Kuroiwa, T. (2001). Localization of the mitochondrial FtsZ protein in a dividing mitochondrion. Cytologia 66**,** 421–425. [Google Scholar]
- Takahara, M., Takahashi, H., Matsunaga, S., Miyagishima, S., Sakai, A., Kawano, S., and Kuroiwa, T. (2000). A putative mitochondrial ftsZ gene is encoded in the unicellular primitive red alga Cyanidioschyzon merolae. Mol. Gen. Genet. 264**,** 452–460. [DOI] [PubMed] [Google Scholar]
- Takei, K., McPherson, P.S., Schmid, S.L., and De Camilli, P. (1995). Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature 374**,** 186–190. [DOI] [PubMed] [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151**,** 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha, S., McAndrew, R.S., and Osteryoung, K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153**,** 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock, D.E., and Schmid, S.L. (1996). Dynamin GTPase, a force-generating molecular switch. BioEssays 18**,** 885–893. [DOI] [PubMed] [Google Scholar]
- Zhang, P., and Hinshaw, J.E. (2001). Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 3**,** 922–926. [DOI] [PubMed] [Google Scholar]