Semaphorin SEMA3F and VEGF Have Opposing Effects on Cell Attachment and Spreading (original) (raw)
Abstract
SEMA3F, isolated from a 3p21.3 deletion, has antitumor activity in transfected cells, and protein expression correlates with tumor stage and histology. In primary tumors, SEMA3F and VEGF surface staining is inversely correlated. Coupled with SEMA3F at the leading edge of motile cells, we previously suggested that both proteins competitively regulate cell motility and adhesion. We have investigated this using the breast cancer cell line, MCF7. SEMA3F inhibited cell attachment and spreading as evidenced by loss of lamellipodia extensions, membrane ruffling, and cell-cell contacts, with cells eventually rounding-up and detaching. In contrast, VEGF had opposite effects. Although SEMA3F binds NRP2 with 10-fold greater affinity than NRP1, the effects in MCF7 were mediated by NRP1. This was determined by receptor expression and blocking of anti-NRP1 antibodies. Similar effects, but through NRP2, were observed in the C100 breast cancer cell line. Although we were unable to demonstrate changes in total GTP-bound Rac1 or RhoA, we did observe changes in the localization of Rac1-GFP using time lapse microscopy. Following SEMA3F, Rac1 moved to the base of lamellipodia and — with their collapse — to the membrane. These results support the concept that SEMA3F and VEGF have antagonistic actions affecting motility in primary tumor cell.
Keywords: semaphorin SEMA3F, neuropilin, VEGF, cell spreading, small GTPases
Introduction
Semaphorins are a large family of secreted, transmembrane- and membrane-associated proteins containing a conserved, cystine-rich, 500-amino-acid Sema domain [1]. SEMA3A (Collapsin), as well as SEMA3F and other class 3 semaphorins (SEMA3B, C, D and E), are secreted proteins containing an immunoglobulin-type domain. Originally identified as repulsive molecules for nerve growth cones [2], their widespread expression suggested that they had additional functions outside the nervous system. This was confirmed by a SEMA3A knockout mouse that resulted in abnormal development of somite-derived and visceral tissues, in addition to neural abnormalities [3,4].
SEMA3F was originally isolated from a recurrent 3p21.3 homozygous deletion in small cell lung cancer cell lines, suggesting that it might be a tumor-suppressor gene [5–7]. Similarly, SEMA3B was also identified from the same 3p21.3 deletion [6]. In primary lung tumors, antibody staining against SEMA3F was shown to correlate with both tumor stage and histological subtype [8]. In a human lung cancer cell line NCI-H1299, expressing predominantly NRP1, transfection of SEMA3B and — to a lesser extent — SEMA3F inhibited in vitro colony formation [9]. SEMA3F also inhibited tumorigenesis of A9 cells in a nude mouse model [10]. Likewise, the tumorigenicity of HEY ovarian adenocarcinoma cells was inhibited by SEMA3B [11]. Thus, SEMA3F and SEMA3B demonstrate clear antitumor effects as judged by correlations with staging and by their effects in in vitro and in vivo model systems.
The receptors for class 3 semaphorins are two related proteins, neuropilin-1 (NRP1) and neuropilin-2 (NRP2) [12,13]. In addition, NRP1 in endothelial cells is a coreceptor for vascular endothelial growth factor VEGF165 [14], and NRP2 binds VEGF165 and VEGF145 [15]. In endothelial cells, SEMA3A blocks VEGF165-induced cell motility and lamellipodia formation [16]. In the nervous system, semaphorin signaling involves homo- and heterodimers of NRP1/NRP2, plexins, and small GTPases such as Rac1 (for reviews, see Refs. [17–21]). Other factors affecting SEMA3A signalling include collapsin response mediator protein (CRMP) [22] and the PDZ binding protein NIP [23]. In lung cancer cell lines expressing varying levels of CRMP, their invasive potential was inversely correlated with CRMP expression [24], a finding which is consistent with antitumor effects of class 3 semaphorins. At the cellular level, COS7 cells expressing NRP1 and plexin-1 contract within 5 to 30 minutes of SEMA3A exposure [25]. Similar effects were observed with SEMA3F in COS7 cells expressing NRP2/plexin-1. Other investigators have shown that exposure to semaphorins results in rapid reorganization of actin filaments, normally present in lamellipodia and filopodia [26].
Based on the inverse staining patterns of VEGF and SEMA3F on primary lung cancer cells, SEMA3F staining at the leading edge of motile cells [8], and the reported antagonism between SEMA3A and VEGF in endothelial cells, we suggested that VEGF and SEMA3F might competitively regulate cell motility and adhesion in epithelial cancers. We have studied this in two breast cancer cell lines, MCF7 and C100, which differentially express NRP receptors. We also identified changes in Rac1 localization following exposure to SEMA3F. These results further strengthen the concept that secreted semaphorins compete with VEGF for effects on tumor cells themselves. Thus, the normal balance between VEGF and semaphorins, which is frequently and substantially disrupted in various epithelial cancers, may have important consequences on migration as it does to angiogenesis.
Materials and Methods
Plasmid Constructions
AP-SEMA3F was built by cloning SEMA3F cDNA into pSecTagA vector (Invitrogen, Cergy Pontoise, France) at the 3′ end of the alkaline phosphatase gene and was generously provided by Dr. M. Tessier-Lavigne [13]. AP-pSecTag (AP) expressed alkaline phosphatase as negative control. Rac1-GFP was a gift from Dr. Fort (CRBM, Montpellier, France).
Cell Lines and Transfections
The human mammary epithelial cell lines utilized were MCF7 and C100, a derivative of MDA-MB-435S [27]. MCF7 cells were grown in RPMI-1640 containing 10% fetal calf serum (FCS) and C100 cells were grown in 50% DMEM/50% Ham's F12 containing 10% FCS. COS7 cells were grown in DMEM plus 10% FCS. Cell lines were transfected with plasmids using Effectene (Qiagen, Courtaboeuf, France) with conditions recommended by the manufacturer. For AP-SEMA3F transfections in COS7 cells, the medium was replaced 2 days after transfection by DMEM, containing 0.5% FCS or OPTIMEM media (Invitrogen). The medium was collected 4 days after transfection and applied to MCF7 and C100 cell cultures in serum-free medium for 14 hours. SEMA3F concentration was estimated by alkaline phosphatase activity (GenHunter, Nashville, TN). Cell ruffling and spreading were measured in cells grown in OPTIMEM for 15 hours before adding AP control media, AP-SEMA3F media, or purified VEGF (Insight Biotechnology, Wembley, UK).
Quantitative RT-PCR
Total RNA and cDNA were prepared as described previously [8]. We assessed levels of SEMA3F, NRP1, NRP2, VEGF, and KDR transcription relative to G3PDH in lung tumors by quantitative real-time RT-PCR carried out using the GeneAmp 5700 (ABI) system with syber green chemistry as described previously [8]. The PCR cycle at which a particular sample reaches an arbitrary threshold fluorescence level (_C_t) is indicative of the input quantity of that template. The PCR was carried out in 50 _µ_l reaction volumes consisting of 1x PCR SYBR Green buffer, 0.25 _µ_M primers, 200 _µ_M dNTPs, and 0.03 U/_µ_l AmpliTaq Gold (Perkin-Elmer, Wellesley, MA). cDNA was amplified as follows: 50°C for 2 minutes, 95°C for 10 minutes followed by 40 cycles at 95°Cx15 seconds, 60°Cx1 minute. SEMA3F, NRP1, NRP2, VEGF, KDR, and FLT-1 cDNA were amplified with the following primers: SEMA3F for 5′ AGCAGACCCAGGACGTGAG 3′ and SEMA3F rev 5′ AAGACCATGCGAATATCAGCC 3′, giving a 112-bp product; VEGF165 for 5′ CAAGACAAGAAAATCCCTGTGG 3′ and VEGF165 rev 5′ CCTCGGCTTGTCACATCTG 3′, giving a 162-bp product; NRP2 for 5′ GGATGGCATTCCACATGTTG 3′ and NRP2 rev 5′ ACCAGGTAGTAACGCGCAGAG 3′, giving a 152-bp product; NRP1 for 5′ATCACGTGCAGCTCAAGTGG 3′ and NRP1 rev 5′ TCATGCAGTGGGCAGAGTTC 3′, giving a 167-bp product; KDR for 5′ TTCTCTTGATCTGCCCAGGC 3′ and KDR rev 5′ AGGCTCCAGTGTCATTTCCG 3′, giving a 182-bp product; FLT-1 for 5′ ATGCCACCTCCATGTTTGATG and FLT-1 rev 5′ GAGGCCTTGGGTTTGCTGTC 3′, giving a 122-bp product.
Immunostaining of Cell Lines
Immunostaining for SEMA3F was performed as described [8], except that cells were finally exposed in the dark to an Alexa488-conjugated goat antirabbit antibody for 30 minutes (1:200) (Molecular Probes, Leiden, Netherlands). Cells were mounted using Vectashield (Vector, Burlingame, CA). Immunostained samples were examined using the blue line (488 nm) of a confocal microscope. For NRP1 and NRP2 immunostaining, cell fixation was performed in 1% paraformaldehyde for 15 minutes without methanol fixation. For anti-NRP staining, we used a NRP1 rabbit polyclonal antibody (1:100) raised against amino acids 583 to 856 of rat NRP1 (gift from Dr. A. L. Kolodkin, Baltimore) [12] and an NRP2 rabbit polyclonal antibody (1:50) raised against amino acids 560 to 858 of human NRP2 (Santa Cruz Biotechnology, Santa Cruz, CA). Cells were exposed in the dark to a second RRX-conjugated goat anti-rabbit antibody diluted at 1:200 (Jackson Immunoresearch Laboratories, West Grove, PA). Immunostained samples were examined using the yellow line (568 nm) of the confocal microscope. NRP1 and NRP2 blocking experiments were performed with a 2-hour preincubation with 10 _µ_g/ml anti-NRP2 polyclonal antibody (Santa Cruz Biotechnology) or/and 10 _µ_g/ml anti-NRP1 polyclonal antibody (Oncogene, Darmstadt, Germany) as described [28].
Confocal Microscopy and Data Analysis
Labelled cells and those observed by transmission light were examined by confocal scanning microscopy using a BioRad MRC 1024 ES (BioRad, Hemel Hempstead, UK) equipped with a 15-mW argon-krypton (Ar-Kr) gas laser. The confocal unit was connected to an inverted microscope (Olympus IX70; Olympus, Tokyo, Japan). Maximal resolution was obtained with Olympus plan apo x60 water, 1.3 numerical aperture objective lens (x–y: 0.3 _µ_m; z: 0.45 _µ_m). RRX fluorochromes were excited with the 568-nm yellow line and the emission of fluorescence was collected through a photomultiplier (PMT1) through a 605-nm band pass filter. The Alexa488 fluorochrome was excited with the 488-nm blue line and the emission of fluorescence was collected through a photomultiplier (PMT2) through a 522-nm band pass filter. Fluorescence signal collection, image construction, and scaling were performed through the control software (Lasersharp 3.2; BioRad). Images and data analysis were performed through the software Lasersharp Processing (BioRad) and Origin 5.0 (Microcal Software, Northampton, MA), respectively. Results were expressed as mean±SEM. Statistical significance between the different mean values was tested using Student's _t_-test.
GST Pull-Down Assays
The GST-PAK-CRIB domain and GST-Rhotekin-RBD were cloned into pGEX-2T fusion (gift of J. G. Collard, Netherlands Cancer Institute, Amsterdam, The Netherlands) [29]. The GST-PAK-CRIB and GST-Rhotekin-RBD recombinant peptides were prepared in Escherichia coli (BL21 strain) and purified using glutathione-sepharose beads (Amersham Pharmacia Biotech, Bucks, UK).
A total of 5x106 C100 and MCF7 cells were washed twice in cold PBS and then lysed in 400 _µ_l of lysis buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5mM MgCl2, 0.05% NP-40 (wt/vol), 1% DOC (wt/vol), 1% Triton X-100, 0.1% SDS, 1.25 mM PMSF, 20 _µ_M leupeptin, 0.8 _µ_M aprotinin, 10 _µ_M pepstatin]. Total amount of protein in each lysate was measured and diluted to 1 _µ_g/µ_l. Lysates were repeatedly passed through a syringe followed by centrifugation at 14,000_g for 1 minute. The supernatant was mixed with 10 to 20 _µ_l of glutathione-sepharose beads corresponding to ∼40 _µ_g of GST fusion protein and incubated at 4°C overnight. Bead-bound complexes were washed once in lysis buffer and three times in the same buffer without DOC, Triton X-100, and SDS. Samples were boiled in Laemmli sample buffer and fractionated by a 12% SDS-PAGE electrophoresis, followed by Western blotting. The presence of RhoA was revealed using a polyclonal anti-RhoA antibody (Santa Cruz Biotechnology). The presence of Rac1 was revealed using a monoclonal anti-Rac1 antibody (Transduction Laboratories, Lexington, KY). As second antibodies, a sheep antimouse IgG, HRP-linked (Amersham Pharmacia Biotech); or a donkey antirabbit IgG, HRP-linked (Amersham Pharmacia Biotech) were used and blots were revealed by ECL detection.
Time Lapse Microscopy
C100 cells were transfected with Rac1-GFP. The medium was replaced 2 days after transfection by medium without FCS for 14 hours. Time lapse microscopy was started after AP-SEMA3F or AP media addition and recorded every 30 seconds for 30 minutes. The GFP was excited with the 488-nm blue line and the emission of fluorescence was collected through a 522-nm band pass filter.
Results
Expression of SEMA3F, Neuropilins, VEGF, KDR, and FLT-1 in MCF7 and C100 Adenocarcinoma Cell Lines
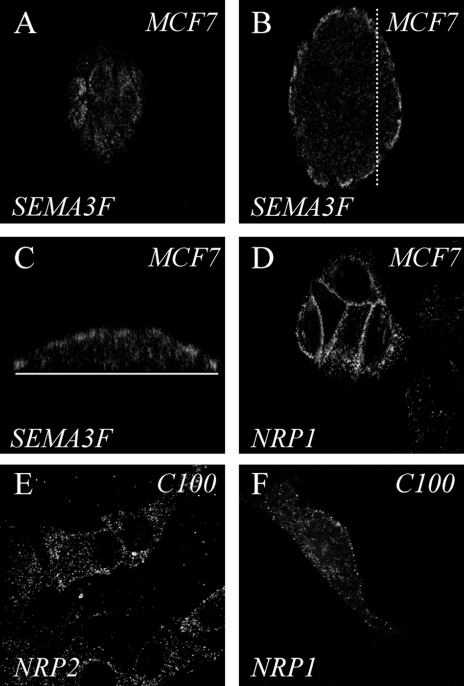
The effects of SEMA3F were investigated in two breast cancer cell lines, which exhibited different biologic phenotypes. MCF7 cells, which grow in islets and exhibit numerous intercellular contacts, provide a model of low metastatic potential. In contrast, C100 cells are highly motile and metastatic. Quantitative RT-PCR demonstrated that MCF7 cells express SEMA3F and NRP1 (Table 1), but barely undetectable NRP2. Immunostaining of MCF7 cells confirmed these results. Using a specific polyclonal antibody raised against SEMA3F [30], cytoplasmic SEMA3F was detected. By laser confocal microscopy, the major distribution of SEMA3F was in near-membrane domains of cells delineating the periphery of the islets (Figure 1, A and B). In a vertical section through the islets, SEMA3F is apically located (Figure 1_C_). Staining with anti-NRP polyclonal antibodies showed that NRP1 was localized at the plasma membrane of all cells within the islets (Figure 1_D_), whereas NRP2 staining was negative (not shown), consistent with the RT-PCR results. The membranous pattern of staining in MCF7 islets for SEMA3F was reminiscent of that obtained in normal bronchial epithelial cells in situ, as well as in cultured adenocarcinoma Calu-3 cells [8].
Table 1.
Relative Expression of SEMA3F, NRP1, NRP2, VEGF, KDR, and FLT-1 for MCF7 and C100 Cell (x1000) Versus G3PDH.
| Cell Lines | SEMA3F | NRP1 | NRP2 | VEGF | KDR | FLT-1 |
|---|---|---|---|---|---|---|
| MCF7 | 27 | 4.2 | 0.0133 | 3 | 0 | 0.025 |
| C100 | 0.017 | 1.5 | 62.5 | 3 | 0.04 | 0 |
Figure 1.
Immunostaining of endogenous SEMA3F and neuropilins in MCF7 and C100 cells. (A–C) MCF7 cells stained for SEMA3F with a polyclonal antibody and observed by laser scanning confocal microscopy (LSCM). (A, B) Horizontal optical section through the surface (A), and the central part (B) of the cell islet. (C) Vertical optical section through the same cell islet obtained along the dotted line shown in (B). The plain white line represents the plate surface. (D) Horizontal optical section obtained by LSCM on MCF7 cells labeled for NRP1 with a polyclonal antibody. (E, F) C100 cells stained for NRP2 (E) and NRP1 (F) with a specific polyclonal antibody and observed by means of LSCM.
In contrast, the C100 cells contain very low levels of SEMA3F transcripts, about 1600 times less than MCF7 (Table 1), and immunostaining with the polyclonal antibody was negative (not shown). Quantitative RT-PCR showed that C100 cells had high levels of NRP2 receptor mRNA, and immunostaining against NRP2 showed a cytoplasmic vesicular pattern together with a significant distribution of NRP2 at the surface membrane (Figure 1_E_). NRP1 was also expressed in C100, but at levels that were approximately one-third that in MCF7 (Table 1, Figure 1_F_). Both C100 and MCF7 expressed moderate levels of VEGF (compared to various other cell lines not shown) and low to absent levels of the VEGF receptor, KDR. In addition, FLT-1 was barely detectable in MCF7 and absent in C100 cells.
Rapid Effects of Extracellular SEMA3F on Cell Morphology
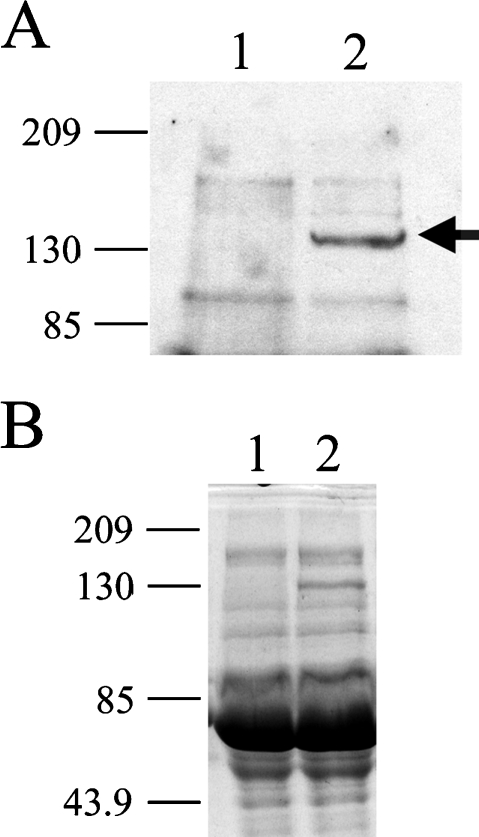
Previous studies have shown that class 3 semaphorins have a repulsive effect on developing axons and in COS7 cells transfected with neuropilins and plexins [25]. To explore the effects of SEMA3F in MCF7 and C100 cells, we transfected COS7 cells with alkaline phosphatase-tagged SEMA3F (AP-SEMA3F), and the cultured supernatant was used as a source of SEMA3F. As a negative control, COS7 cells were transfected with the AP vector only. This approach was first described by two groups [12,31], and further widely used by others because bacterial or Baculovirus-produced semaphorins are insoluble, presumably due to their highly disulfide-linked nature. Secretion of AP-SEMA3F was confirmed by Western blot (Figure 2) and the enzymatic alkaline phosphatase activity was used to determine the amount of SEMA3F protein applied to cells.
Figure 2.
Western blot analysis of AP-SEMA3F expression. Media from transfected AP (1) or AP-SEMA3F (2) COS7 cells were concentrated on Microcon 10 (Amicon, Guyancourt, France) and subjected to electrophoresis on a 12% polyacrylamide gel. Immunodetection was performed with a polyclonal AP antibody 1/2500 (GenHunter) in (A) and loading controls stained by Coomassie blue are shown in (B).
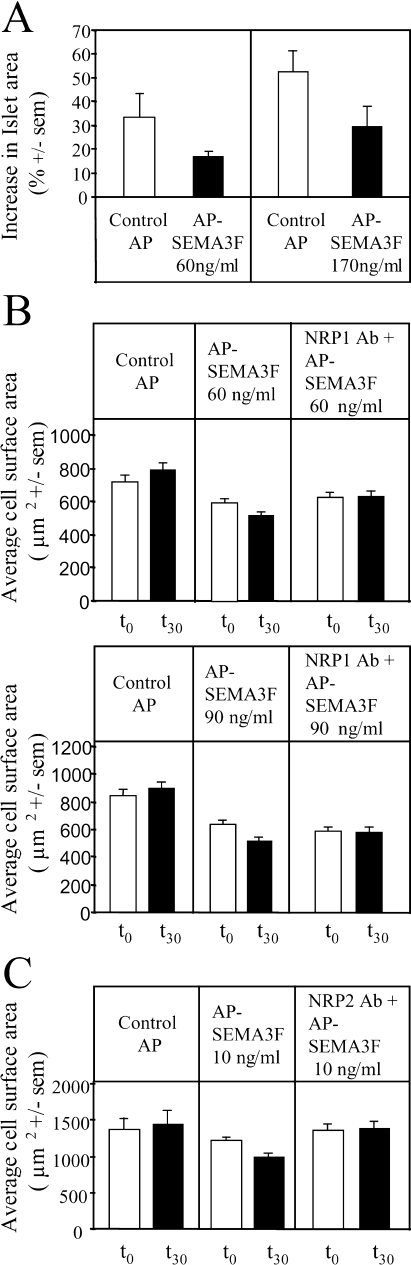
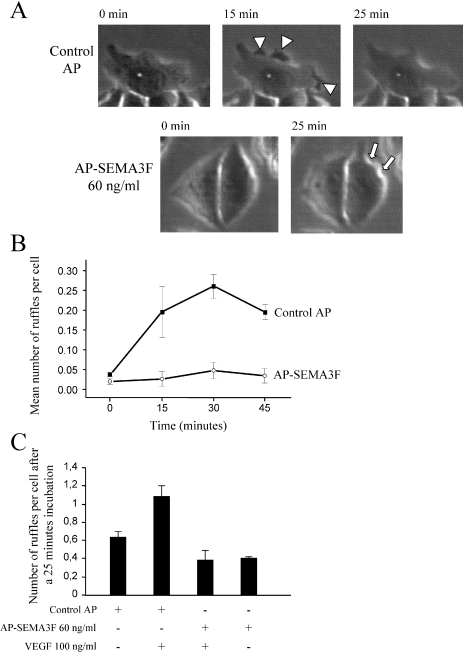
After a 60-minute incubation with control AP medium, MCF7 cells were observed to spread and form membrane ruffles and lamellipodia, which resulted in an increase in the average surface area of cell islets and individual cells (Figure 3, A and B). This cell spreading was significantly inhibited after incubation with 60, 90, and 170 ng/ml AP-SEMA3F (Figure 3, A and B). The ruffling activity of MCF7 cells was recorded by microcinematography (Figure 4_A_). The number of ruffles was increased over a 45-minute period by the AP control medium, suggesting that COS7 cells secrete one or more factors that positively affect membrane ruffling. Alternatively, the effect of the AP control medium could be due to the 0.5% serum from the transfected COS cells because the MCF7 cells were placed in serum-free medium before treatment (Figure 4_B_). This background effect was also dose-dependent (Figure 3_A_). Nevertheless, membrane ruffling was strongly inhibited by AP-SEMA3F (Figure 4_B_). When 100 ng/ml VEGF was added to control AP medium, the ruffling activity was promoted (Figure 4_C_). AP-SEMA3F at 60 ng/ml was able to block the positive effect of VEGF (Figure 4_C_). These results are consistent with competing actions of SEMA3A and VEGF on endothelial cells [16]. Retraction of the cell body and extensions could be observed in MCF7 cells that had separated from the islet. After exposure to SEMA3F for 30 minutes, these cells rounded-up and eventually detached from adjacent cells. This resulted in an overall decrease in the number of cells within a microscope field, whereas with control COS7 supernatant, the number of cells increased from cell division.
Figure 3.
AP-SEMA3F reduced the ability of MCF7 cell islets and C100 cells to expand. (A) Total MCF7 islets areas were measured before and after 60 minutes of treatment at 37°C, with COS7 cells culture media containing 0.5% FCS and secreted AP (white columns: control) or AP-SEMA3F (black columns) at two different concentrations (60 and 170 ng/ml). A volume of AP media, equivalent to that of AP-SEMA3F media, is added for the control. The percentage of increase in islet areas was calculated and reported on bar graph. Error bars, SEM. (B) Changes in cell surface area when MCF7 cells are cultured for 15 minutes with control AP or AP-SEMA3F, 60 and 90 ng/ml, with and without a blocking anti-NRP1 antibody. (C) Mean C100 cells areas were calculated before (white columns) and after treatment for 30 minutes (black columns) with COS7 cells media containing 0.5% FCS and control AP (left panel) or AP-SEMA3F at 10 ng/ml (middle panel). AP-SEMA3F was added in the presence of an anti-NRP2 antibody (right panel). Error bars, SEM.
Figure 4.
MCF7 cell ruffling is stimulated by VEGF but is inhibited by SEMA3F. MCF7 cells were grown with control AP or AP-SEMA3F and ruffling was recorded by microcinematography (A). Ruffles are shown by arrow heads and retraction by arrows. Mean number of ruffles per cell is shown in (B). A total of 100 ng/ml VEGF was tested on cell ruffling in the presence or absence of control AP or AP-SEMA3F (C). Number of ruffling is expressed at time of 25 minutes. Error bars, SEM.
Responses in C100 cells were more complex because of morphologic heterogeneity. These cells could be divided in three groups representing small round cells with few extensions, spindle-shaped cells with neurite-like cell processes, and extended cells with large lamellipodia. Whereas the first and second groups of cells did not appear to respond to SEMA3F, cell spreading was significantly inhibited after 30 minutes by 10 ng/ml AP-SEMA3F in the third subset of cells having an extended morphology. Thus, the average cell surface area decreased as a result of lamellipodia retraction (Figure 3_C_), and other cells were observed to round-up and detach from the substrate.
SEMA3F Signal Transduction
As MCF7 and C100 cells exhibited different patterns of NRP1 and NRP2 expression, we tested antibodies for their ability to block SEMA3F action. In MCF7, the SEMA3F effects on cell spreading were inhibited by anti-NRP1 antibodies (Figure 3_B_), whereas in C100 cells (Figure 3_C_), anti-NRP2 blocked SEMA3F inhibition of cell spreading and anti-NRP1 had no effect. These results are consistent with the levels of NRP1 and NRP2 expression in MCF7 and C100 cells (Table 1) and also demonstrate that NRP1 can function in SEMA3F signalling.
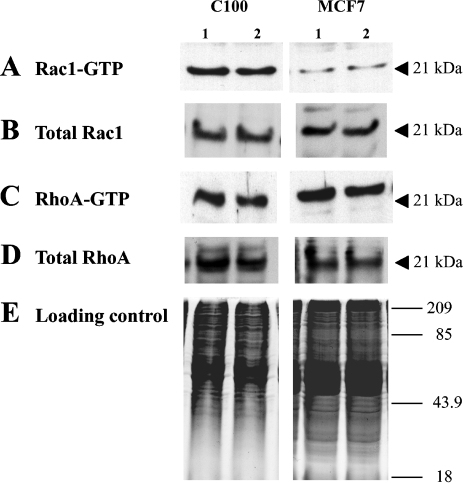
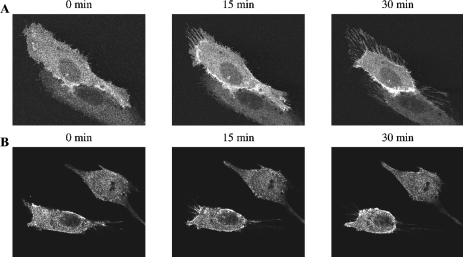
As neuropilin signal transduction has been shown to involve small GTPases in neural cells, we looked for changes in the amount of active GTP-bound Rac1 and RhoA using GST fusion proteins expressing either the GST-PAK-CRIB domain or GST-Rhotekin-RBD. In a pull-down experiment using extracts of C100 and MCF7 cells treated with AP-SEMA3F, we did not identify any difference in levels of GTP-bound Rac1 and RhoA (Figure 5). Also, we did not find any difference in the amount of total Rac1 and RhoA upon AP-SEMA3F treatment. As this negative result might have been due to insufficient sensitivity, we examined the subcellular distribution of Rac1 using C100 cells transfected with Rac1-GFP. Time lapse analysis by confocal laser scanning microscopy showed that Rac1 was predominant at intercellular junctions before the addition of AP-SEMA3F. Upon addition of AP-SEMA3F, Rac1 was observed first at the base of lamellipodia and subsequently at the membrane (Figure 6_A_). Isolated cells became spherical at the end of the retraction process and were decorated with Rac1 around the membrane (Figure 6_B_). These results indicate that SEMA3F induces changes in Rac1 activation, at least at a local level, and that the presence of Rac1 at the membrane is associated with the retraction.
Figure 5.
Levels of Rac1-GTP and RhoA-GTP were not affected by AP-SEMA3F. Levels of active Rac1-GTP (A) and RhoA-GTP (C) were estimated by GST pull-down assay in C100 and MCF7 cells after treatment with AP (1) or AP-SEMA3F (2). Total amount of total Rac1 (B) and RhoA (D) was determined by Western blot after electrophoresis on a 12% SDS polyacrylamid gel, and a loading control is shown in (E).
Figure 6.
Rac1 moved to the base of lamellipodia and later to the membrane upon AP-SEMA3F treatment. Time lapse analysis by confocal laser scanning microscopy showed Rac-GFP localization in connected (A) and in isolated C100 cells (B) at time 0, 15, and 30 minutes after AP-SEMA3F addition.
Discussion
Semaphorins were initially identified on the basis of their inhibition of nerve growth cone migration; however, they clearly affect non-neural tissues. On the basis of its localization at 3p21.3, a region that undergoes frequent loss of heterozygosity in lung tumors, and because of the activity of genomic clones containing SEMA3F in suppressing tumorigenicity of mouse fibrosarcoma A9 cells [32], SEMA3F was hypothesized to be a tumor-suppressor gene. Subsequent studies demonstrated that SEMA3F protein expression was decreased or absent in high-grade lung cancers [8]. These studies also suggested, based on complementary staining patterns, that SEMA3F might compete with VEGF for binding to their common neuropilin receptors. More recent experiments using single gene transfectants have confirmed that SEMA3F has tumor-suppressor gene activity [10].
Our results demonstrate that SEMA3F and VEGF have opposing actions on primary tumor cells. Whereas VEGF promotes cell spreading and membrane ruffling, activities associated with increased cell migration and metastasis, SEMA3F inhibits these changes and is able to block the effects of VEGF. During a brief treatment with AP-SEMA3F, MCF7 cell spreading and lamellipodia extension were inhibited and membrane ruffling was reduced. Even though MCF7 cells do not express NRP2 (or express very little), they express NRP1 and responded to the cultured supernatant of AP-SEMA3F expressing COS7 cells. These effects were mediated by NRP1, as shown by the use of an anti-NRP1 antibody, which blocked the activity. In contrast, C100 cells express NRP2 at high levels, and anti-NRP2 (but not anti-NRP1) antibodies were able to block the SEMA3F effects. These results demonstrate that both NRP1 and NRP2 are capable of binding and transmitting SEMA3F signals. Although MCF7 cells express SEMA3F, they still responded to exogenous SEMA3F. This may simply represent a dosage effect in the presence of excess NRP1 receptor. As noted, anti-SEMA3F staining was detected predominantly at the periphery of the islets whereas NRP1 staining was detected throughout. Also, SEMA3F may have been in competition with VEGF present in the cultured supernatant.
Initially considered as an endothelial-specific mitogen, there is now evidence that VEGF stimulates nonendothelial cell types. For example, VEGF stimulates osteoblast migration [33,34] and positively influences melanoma cell migration in vitro through an integrin-dependent mechanism [35]. We have not investigated whether SEMA3F affects integrin activation. However, our findings do suggest that SEMA3F affects cell adhesion as evidenced by the separation of cells, their rounding-up, and subsequent detachment from the substrate. These responses are likely comparable to the effects seen in NP/plexin-transfected COS7 cells following exposure to SEMA3A or SEMA3F [25]. In these cells, SEMA3F led to cytoskeleton perturbations similar to those described in nerve growth cones. This suggests that SEMA3F has a common action on different cell types that may involve small GTP binding proteins like Rho family GTPases because lamellipodia were generally affected. Although we were unable to detect changes in total GTP-bound Rac1 or Rho, we did detect changes in Rac1-GFP localization. The Rho family of small GTPases is the central regulator of cytoskeletal dynamics and controls the organization of actin filaments and cellular morphology [36]. In growth cones, SEMA3A (Collapsin) has been shown to initiate clustering of neuropilin and plexin receptors. This occurred in a CRMP-dependent manner and was Rac1-dependent (for review, see Ref. [20]). Similarly, plexin-A1, a coreceptor for class 3 semaphorins, interacts not only with Rnd1 but also with RhoD, and these GTPases have antagonistic effects on the activity of plexin-A1 [37]. These authors suggested that interaction of Rnd1 results in a conformational change that ultimately activates downstream signal transduction cascades, including Rac1, RhoA, LIM kinase 1, and cofilin that mediate growth cone collapse [38]. Indeed, we demonstrated in epithelial tumor cells a clear recruitment of Rac1 to retraction fibers upon AP-SEMA3F treatment.
Finally, we have some additional observations regarding the viability of the detached cells following SEMA3F exposure. These cells were not able to reattach and the number of cells decreased over time, suggesting that they underwent apoptosis or anoikis. An apoptotic effect was reported for SEMA3A in sensory neurons [39] and in neural progenitors [40]. This apoptotic effect was shown to be mediated by NRP1 and was antagonized by VEGF165 [40]. We also performed additional experiments showing that C100 cells undergo apoptosis in response to transfected SEMA3F as evidenced by annexin and propidium iodine staining (data not shown).
In summary, we have shown that mammary adenocarcinoma cells stimulated with SEMA3F lose lamellipodia extensions and cell-cell contacts, and eventually detach with subsequent apoptosis or anoikis. These effects can be mediated by either NRP1 or NRP2 receptors and appear to involve Rac1 redistribution.
Acknowledgements
We are very grateful to M. Tessier-Lavigne and Kolodkin for providing us with the AP-SEMA3F construct and neuropilin antibodies, respectively. We thank P. Fort for the Rac-GFP vector and J. Collard for GST-Rhotekin-RBD and GST-PAK-CRIB constructs. We thank A. Cantereau for technical assistance in the confocal microscopy studies performed in the confocal microscopy core of the Federative Research Institute IFR59 at the University of Poitiers. We thank J. Habrioux and J. P. Poindessault for edition of the figures.
Footnotes
1
Present address: Institut de Biotechnologies, 123 avenue Albert Thomas, Limoges Cédex 87060, France.
References
- 1.Semaphorin Nomenclature Committee, author. Unified nomenclature for the semaphorins/collapsins. Cell. 1999;97:551–552. doi: 10.1016/s0092-8674(00)80766-7. [DOI] [PubMed] [Google Scholar]
- 2.Luo Y, Raible D, Raper A. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 3.Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature. 1996;383:525–528. doi: 10.1038/383525a0. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi M, Yuasa S, Fujisawa H, Naruse I, Saga S, Mishina M, Yagi T. Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron. 1997;19:519–530. doi: 10.1016/s0896-6273(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 5.Roche J, Boldog F, Robinson M, Robinson L, Varella-Garcia M, Swanton M, Waggoner B, Fishel R, Franklin W, Gemmill R, Drabkin H. Distinct 3p21.3 deletions in lung cancer, analysis of deleted genes and identification of a new human semaphorin. Oncogene. 1996;12:1289–1297. [PubMed] [Google Scholar]
- 6.Sekido Y, Bader S, Latif F, Chen JY, Duh FM, Wei MH, Albanes JP, Lee CC, Lerman MI, Minna JD. Human semaphorins A (V) and (IV) reside in the 3p21.3 small cell lung cancer deletion region and demonstrate distinct expression patterns. Proc Natl Acad Sci USA. 1996;93:4120–4125. doi: 10.1073/pnas.93.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang R, Hensel C, Garcia D, Carlson H, Kok K, Daly M, Kerbacher K, Van Den Berg A, Veldhuis P, Buys C, Naylor S. Isolation of the human semaphorin III/F gene (SEMA3F) at chromosome 3p21, a region deleted in lung cancer. Genomics. 1996;32:39–48. doi: 10.1006/geno.1996.0074. [DOI] [PubMed] [Google Scholar]
- 8.Brambilla E, Constantin B, Drabkin H, Roche J. Semaphorin SEMA3F localization in malignant human lung and cell lines: a suggested role in cell adhesion and cell migration. Am J Pathol. 2000;156:939–950. doi: 10.1016/S0002-9440(10)64962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomizawa Y, Sekido Y, Kondo M, Gao B, Yokota J, Roche J, Drabkin H, Lerman M, Gazdar A, Minna J. Inhibition of lung cancer cell growth and induction of apoptosis following re-expression of 3p21.3 candidate tumor suppressor gene SEMA3B. Proc Natl Acad Sci USA. 2001;98:13954–13959. doi: 10.1073/pnas.231490898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang R, Davalos AR, Hensel CH, Zhou XJ, Tse C, Naylor SL. Semaphorin 3F gene from human 3p21.3 suppresses tumor formation in nude mice. Cancer Res. 2002;62:2637–2643. [PubMed] [Google Scholar]
- 11.Tse C, Xiang RH, Bracht T, Naylor SL. Human semaphorin 3B (SEMA3B) located at chromosome 3p21.3 suppresses tumor formation in an adenocarcinoma cell line. Cancer Res. 2002;62:542–546. [PubMed] [Google Scholar]
- 12.Kolodkin A, Levengood D, Rowe E, Tai Y, Giger R, Ginty D. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Chédotal A, He Z, Goodman CS, Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron. 1997;19:547–559. doi: 10.1016/s0896-6273(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 14.Soker S, Takashima S, Miao H, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 15.Gluzman-Poltorak Z, Cohen T, Herzog Y, Neufeld G. Neuropilin-2 and neuropilin-1 are receptors for VEGF165 and PLGF-2, but only neuropilin-2 functions as a receptor for VEGF145. J Biol Chem. 2000;275:18040–18045. doi: 10.1074/jbc.M909259199. [DOI] [PubMed] [Google Scholar]
- 16.Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura F, Kalb RG, Strittmatter SM. Molecular basis of semaphorin-mediated axon guidance. J Neurobiol. 2000;44:219–229. doi: 10.1002/1097-4695(200008)44:2<219::aid-neu11>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 18.Goshima Y, Sasaki Y, Nakayama T, Ito T, Kimura T. Functions of semaphorins in axon guidance and neuronal regeneration. Jpn J Pharmacol. 2000;82:273–279. doi: 10.1254/jjp.82.273. [DOI] [PubMed] [Google Scholar]
- 19.Tamagnone L, Comoglio PM. Signalling by semaphorin receptors: cell guidance and beyond. Trends Cell Biol. 2000;10:377–383. doi: 10.1016/s0962-8924(00)01816-x. [DOI] [PubMed] [Google Scholar]
- 20.Liu BP, Strittmatter SM. Semaphorin-mediated axonal guidance via Rho-related G proteins. Curr Opin Cell Biol. 2001;13:619–626. doi: 10.1016/s0955-0674(00)00260-x. [DOI] [PubMed] [Google Scholar]
- 21.He Z, Wang KC, Koprivica V, Ming G, Song HJ. Knowing how to navigate: mechanisms of semaphorin signaling in the nervous system. Sci STKE. 2002;2002:RE1. doi: 10.1126/stke.2002.119.re1. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, Ihara Y. Evidence that collapsin response mediator protein-2 (CRMP-2) is involved in the dynamics of microtubules. J Biol Chem. 2000;275:17917–17920. doi: 10.1074/jbc.C000179200. [DOI] [PubMed] [Google Scholar]
- 23.Cai H, Reed RR. Cloning and characterization of neuropilin-interacting protein: a PSD-95/Dlg/ZO-1 domain-containing protein that interacts with the cytoplasmic domain of neuropilin-1. J Neurosci. 1999;19:6519–6527. doi: 10.1523/JNEUROSCI.19-15-06519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih JY, Yang SC, Hong TM, Yuan A, Chen JJ, Yu CJ, Chang YL, Lee YC, Peck K, Wu CW, Yang PC. Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. J Natl Cancer Inst. 2001;93:1392–1400. doi: 10.1093/jnci/93.18.1392. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi T, Fournier A, Nakamura F, Wang LH, Murakami Y, Kalb RG, Fujisawa H, Strittmatter SM. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell. 1999;99:59–69. doi: 10.1016/s0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 26.Fan J, Raper R. Localized collapsing cues can steer growth cones without inducing their full collapse. Neuron. 1995;14:263–274. doi: 10.1016/0896-6273(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 27.Kantor JD, McCormick B, Steeg PS, Zetter BR. Inhibition of cell motility after nm23 transfection of human and murine tumor cells. Cancer Res. 1993;53:1971–1973. [PubMed] [Google Scholar]
- 28.Henke-Fahle S, Beck KW, Püschel A. Differential responsiveness to the chemorepellent semaphorin 3A distinguishes ipsi- and contralaterally projecting axons in the chick midbrain. Dev Biol. 2001;237:381. doi: 10.1006/dbio.2001.0376. [DOI] [PubMed] [Google Scholar]
- 29.Sander EE, van Delft S, ten Klooster JP, Reid T, van der Kammen RA, Michiels F, Collard JG. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998;143:1385–1398. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch E, Hu L-J, Prigent A, Constantin B, Agid Y, Drabkin H, Roche J. Distribution of semaphorin IV in adult human brain. Brain Res. 1999;823:67–79. doi: 10.1016/s0006-8993(99)01103-8. [DOI] [PubMed] [Google Scholar]
- 31.He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell. 1997;90:739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 32.Todd M, Xiang R, Garcia D, Kerbacher K, Moore S, Hensel C, Liu P, Siciliano M, Kok K, van den Berg A, Veldhuis P, Buys C, Killary A, Naylor S. An 80 kb P1 clone from chromosome 3p21.3 suppresses tumor growth in vivo. Oncogene. 1996;13:2387–2396. [PubMed] [Google Scholar]
- 33.Deckers MM, Karperien M, van der Bent C, Yamashita T, Papapoulos SE, Lowik CW. Expression of vascular endothelial growth factors and their receptors during osteoblast differentiation. Endocrinology. 2000;141:1667–1674. doi: 10.1210/endo.141.5.7458. [DOI] [PubMed] [Google Scholar]
- 34.Midy V, Plouet J. Vasculotropin/vascular endothelial growth factor induces differentiation in cultured osteoblasts. Biochem Biophys Res Commun. 1994;199:380–386. doi: 10.1006/bbrc.1994.1240. [DOI] [PubMed] [Google Scholar]
- 35.Byzova TV, Goldman CK, Pampori N, Thomas KA, Bett A, Shattil SJ, Plow EF. A mechanism for modulation of cellular responses to VEGF: activation of the integrins. Mol Cell. 2000;6:851–860. [PubMed] [Google Scholar]
- 36.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 37.Zanata SM, Hovatta I, Rohm B, Puschel AW. Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in Semaphorin 3A-induced cytoskeletal collapse. J Neurosci. 2002;22:471–477. doi: 10.1523/JNEUROSCI.22-02-00471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aizawa H, Wakatsuki S, Ishii A, Moriyama K, Sasaki Y, Ohashi K, Sekine-Aizawa Y, Sehara-Fujisawa A, Mizuno K, Goshima Y, Yahara I. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat Neurosci. 2001;4:367–373. doi: 10.1038/86011. [DOI] [PubMed] [Google Scholar]
- 39.Gagliardini V, Fankhauser C. Semaphorin III can induce death in sensory neurons. Mol Cell Neurosci. 1999;14:301–316. doi: 10.1006/mcne.1999.0787. [DOI] [PubMed] [Google Scholar]
- 40.Bagnard D, Vaillant C, Khuth ST, Dufay N, Lohrum M, Puschel AW, Belin MF, Bolz J, Thomasset N. Semaphorin 3A-vascular endothelial growth factor-165 balance mediates migration and apoptosis of neural progenitor cells by the recruitment of shared receptor. J Neurosci. 2001;21:3332–3341. doi: 10.1523/JNEUROSCI.21-10-03332.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]