Identification of Semaphorin3B as a Direct Target of p53 (original) (raw)
Abstract
A cDNA microarray analysis indicated that Semaphorin3B (Sema3B), a gene whose product is involved in axon guidance and axonal repulsion, is inducible by p53. Introduction of exogenous p53 into a glioblastoma cell line lacking wild-type p53 (U373MG) dramatically induced expression of Sema3B mRNA. An electrophoretic mobility shift assay and a reporter assay confirmed that a potential p53 binding site present in the promoter region had p53-dependent transcriptional activity. Expression of endogenous Sema3B was induced in response to genotoxic stresses caused by adriamycin treatment or UV irradiation in a p53-dependent manner. Ectopic expression of Sema3B in p53-defective cells reduced the number of colonies in colony formation assays. These results suggest that Sema3B might play some role in regulating cell growth as a mediator of p53 tumor-suppressor activity.
Keywords: Semaphorin3B, p53, target gene, growth suppression, apoptosis
Introduction
The tumor-suppressing activity of p53 protein involves elimination of cells that might otherwise become malignant as a result of severe DNA damage or critical cellular stress, through induction of cell cycle arrest and apoptosis [1–3]. The p53 protein initiates these processes by binding to a specific DNA sequence (p53 binding site, p53BS) present in target genes, inducing transcription of molecules that carry out the required antitumorigenic functions [4].
We and others have demonstrated that p53 takes part in many physiological roles by transactivating dozens of target genes such as p53AIP1 [5] in p53-mediated apoptosis, p21/Waf1 [6] in p53-mediated cell cycle arrest, and p53R2 [7] in repair of damaged DNA. In view of the fact that more than 100 potential p53BS are likely to be present in the human genome [8], we assume that many p53 target genes have not yet been identified. To fully understand the entire spectrum of p53 functions including its tumor-suppressive activity, it will be essential to identify all the genes that are targets of this transcription factor. We recently reported several novel p53 target genes using a yeast enhancer trap system [9–12] or a differential display method [5,7,13–15]. To expand the collection of p53 targets further, we have been applying a third approach that exploits cDNA microarray technology.
In neurons, both genetic and epigenetic factors are involved in physiologic and pathophysiologic states [16]. The semaphorin family of proteins is a major agent for axonal guidance, and the axon-repulsive effects of semaphorins have been documented in vitro and in vivo [17]. All semaphorins share a conserved, 500-amino-acid “Sema” domain at their amino terminals, and possess variable regions at their carboxyl ends. More than 20 semaphorins have been identified so far (Semaphorin Nomenclature Committee, 1999) and are classified into eight groups on the basis of domain organization within the primary structure and the species of origin [18]. Vertebrate semaphorins are categorized into five groups, classes 3 through 7; classes 4, 5, and 6 possess transmembrane segments and class 7 is characterized by a GPI anchor motif. The Semaphorin3B (Sema3B) gene encodes a member of class 3, a group of secreted proteins that repulse axonal extension [19]. In the spinal cord, Sema3B is expressed in the floor plate of the midline and does not repel commissural axons that have not yet crossed the floor plate. However, once the commissural axons cross the floor plate, Sema3B inhibits their extension. Targeted inhibition of neurophilin-2, a receptor for Sema3B, leads to defects of midline guidance in mice. Thus, Sema3B appears to be a critical factor in formation of the mammalian neural network [19]. We show evidence here that Sema3B is a direct transcriptional target of p53, and that it might be involved in p53-dependent suppression of cell growth.
Materials and Methods
Cell Lines
Human cancer cell lines U373MG (glioblastoma), H1299 (lung carcinoma), and MCF7 (breast cancer) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). T98G (glioblastoma) was obtained from the Japanese Collection of Research Bioresources (°CRB, Tokyo, Japan). All cells were cultured under conditions recommended by their respective depositors.
Detection of Genes Induced by p53
Adenoviruses containing p53 cDNA (Ad-p53) or β-galactosidase cDNA (Ad-LacZ) under control of the human cytomegalovirus promoter were generated as described previously [20]. U373MG cells, which lack wild-type p53, were infected with Ad-p53 or Ad-LacZ at a multiplicity of infection (MOI) of 80. Cells were collected 0, 6, 12, 24, and 48 hours later, and total RNA was extracted with TRIzol reagent (Life Technologies, Rockville, MD). Poly(A) RNA was purified from each total RNA using an mRNA purification kit (Amersham, Piscataway, NJ). Each poly(A)+ RNA was then amplified using Ampliscribe T7 Transcription Kits (Epicentre Technologies, Madison, WI).
Genes induced by exogenous p53 were identified by hybridizing the RNA to a cDNA microarray. Preparation of probes, hybridization procedures, and scanning were performed as described previously [21]. The fluorescent intensities of Cy5 (Ad-p53) and Cy3 (Ad-LacZ) for each target spot were adjusted so that the mean Cy5/Cy3 ratio of 52 housekeeping genes for each slide became equal. One induced gene, Sema3B, was selected from the microarray for further investigation.
Northern Blotting
One microgram of each poly(A)+ RNA was separated on 1% agarose gels containing 1x 4-morpholinepropane-sulfonic acid and 2% formaldehyde, and transferred onto nylon membranes. The membranes were hybridized with random-primed, 32P-labeled Sema3B or actin cDNA, washed with 0.1x SSC, 0.1% SDS at 65°C, and exposed for autoradiography at -80°C.
Electrophoretic Mobility Shift Assay (EMSA)
The EMSA was performed as described previously [7] using oligonucleotides corresponding to a putative p53 binding sequence present in the promoter of Sema3B (5′-TCA CTT GCA TGC CCA GAG ACA TGT CT-3′ for the sense strand and 5′-AGA CAT GTC TCT GGG CAT GCA AGT GA-3′ for the antisense strand).
Luciferase Assay
One or two copies of oligonucleotides 5′-CGCGTTCACTTGCATGCCCAGAGA CATGTCTC-3′ (sense) and 5′-TCGAGAGACATGTCTCTGGGCATGCAAGTGA A-3′ (antisense) were annealed, ligated into pGL3 promoter vector (Promega, Madison, WI), and designated p53BSx1 or p53BSx2. One or two copies of 5′-TCACTTTCATTCCCAGAGACATGTCTTC-3′ (sense) and 5′-AGACATGTCTCTGGGAATGAAAGTGAAA-3′ (antisense) were also annealed, ligated into pGL3 promoter vector, and designated p53BSx1.mt or p53BSx2.mt. An additional reporter plasmid was constructed by cloning into pGL3 Basic vector (Promega) an 880-bp genomic sequence corresponding to part of the promoter region, using PCR primers, 5′-AAAACGCGTTCAAGCTCAAATGCAGAGGC-3′ and 5′-TTTCTCGAG CCACAATCACAGACACACCC-3′. A clone containing a C-to-T substitution at the fourth nucleotide in the p53BS was generated using a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Luciferase assays were performed as described previously [15].
Colony Formation Assay
The entire coding sequence of Sema3B cDNA, with or without a FLAG tag at the 3′-end, was amplified by PCR using KOD-plus-PCR Polymerase (Toyobo, Osaka, Japan) and inserted into pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA). As a control, the entire coding sequence of Sema3B cDNA was inserted into pcDNA3.1(-) vector (Invitrogen). Plasmid mixtures were preincubated for 30 minutes with 25 _µ_l of FuGENE reagent (Roche, Indianapolis, IN). Seven microgams of each Sema3B expression vector, with or without a FLAG tag, and control vector were transfected into approximately 1x104 cells of H1299 or 5x104 cells of T98G and U373MG. The transfected cells were cultured in G418-containing culture media (0.6 _µ_g/ml for U373 and 0.8 _µ_g/ml for T98G and H1299) and stained with crystal violet 10 to 21 days later.
DNA-Damaging Treatments
MCF7 (p53+/+) and H1299 (p53-/-) cells were UV-irradiated at different dosages (J/m2) using a UV crosslinker (Stratagene), or treated with adriamycin (doxorubicin; Kyowa Hakko Kogyo, Tokyo, Japan) at different dosages. Total RNA was isolated from the cells at selected times after treatment and expression of Sema3B was examined by Northern blotting.
Results
To identify additional p53 targets, we applied a cDNA microarray to screen for p53-inducible transcripts [21]. We confirmed that this approach could detect the elevated expressions of several well-known p53 target genes (Table 1). Among dozens of genes that were transactivated in a time-dependent manner after introduction of exogeneous p53 to a p53-deficient cell line, we confirmed significant induction of Sema3B (GenBank NM004636) by a Northern blot analysis that identified its major 3.4-kb transcript (Figure 1_A_). To investigate whether Sema3B was a direct target of p53, we searched for one or more p53 binding sequences (p53BS) within the gene and found a possible p53BS in its promoter region (Figure 1_B_), which revealed a 90% match to the consensus p53 binding sequence [4] with a spacer of two nucleotides (Figure 1_C_).
Table 1.
Induction of Sema3B and Other p53 Target Genes in Ad-_p53_-Infected U373MG Cells.
| Gene | UniGene Number | Cy5/Cy3* |
|---|---|---|
| p21/WAF1 | hs.179665 | >15.0 |
| FRACTALKINE | hs.80420 | 10.4 |
| GADD45A | hs.80409 | 10.0 |
| IGFBP3 | hs.77326 | 9.7 |
| SEMA3B | hs.82222 | 7.8 |
| p53DINP1 | hs.75497 | 3.0 |
| MDM2 | hs.170027 | 2.4 |
Figure 1.
Induction of Sema3B mRNA by exogenous p53 and detection of a potential p53 binding site in the promoter region. (A) Northern blot analysis of Sema3B and p21WAF1 mRNA. p53-deficient U373MG cells were infected with Ad-p53 or Ad-LacZ at MOI of 80, and mRNA were isolated at the indicated times after infection. Hybridization of the same blot with an actin probe served as a quantitative control for the amount of mRNA loaded in each lane. (B) Nucleotide sequence of the promoter region of the human Sema3B gene. A potential p53BS and putative TATA boxes are underlined. The locations of the primers used to amplify the native promoter DNA are indicated by arrows. (C) Comparison of sequences between the p53BS of the Sema3B gene and the p53 binding consensus sequence. R, purine; Y, pyrimidine; W, A or T.
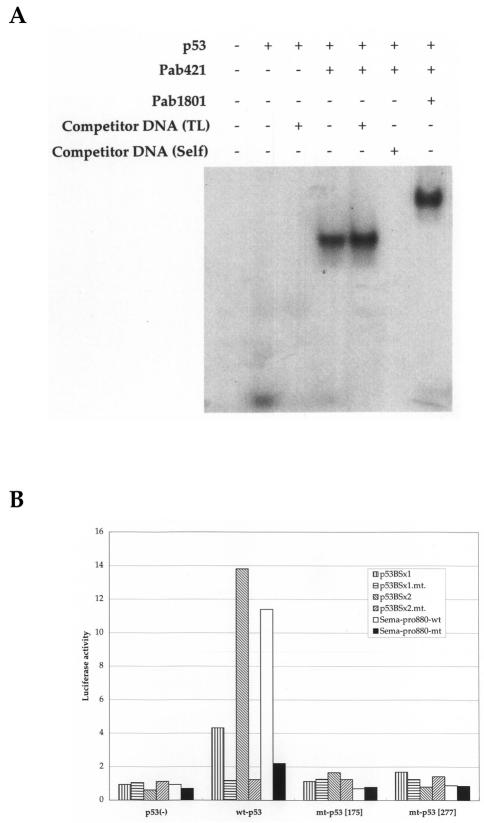
To verify whether p53 was able to bind to oligonucleotides corresponding to this candidate p53BS, we performed EMSA (Figure 2_A_) using nuclear extracts purified from H1299 (p53-null) cells infected with Ad-p53. A shifted band was observed after addition of the nuclear extract containing wild-type p53, and was supershifted when anti-p53 antibody (Pab421) was added. This supershifted band was supersupershifted when another anti-p53 antibody (Pab1801) was added. Unlabeled self-oligonucleotides (Self), but not nonspecific oligonucleotides (TL), inhibited the binding, indicating that p53 protein could in fact bind to this p53BS in vitro.
Figure 2.
Sema3B as a direct target of p53. (A) EMSA, using purified nuclear extracts from H1299 (p53-/-) cells infected with Ad-p53. The probe was a 32P-labeled oligonucleotide designed according to the p53BS sequence present in the gene's promoter. Interaction between p53 protein and DNA was inhibited by unlabeled oligonucleotides corresponding to the binding site of the Sema3B gene (Self) but not by nonspecific oligonucleotides (TL). (B) Reporter assay. Vectors containing either one (p53BSx1) or two (p53BSx2) copies of the p53 binding site, or an 880-bp genomic sequence in the promoter encompassing the p53BS (Sema-prom.) and their mutants (p53BSx1.mt., p53BSx2.mt., and Sema-prom.mt.) were constructed. These vectors were cotransfected into H1299 cells with wild-type p53 (wt-p53) expression vector, mutant p53 (mt-p53[ 175 ] or mt-p53[277]) expression vector, or mock vector (p53(-)). Luciferase activity is indicated relative to the activity of either pGL3 promoter or pGL3 basic vector.
To clarify the p53-dependent, transcription-enhancing activity of this p53 binding sequence, we performed a reporter assay with a luciferase gene that was fused to one (p53BSx1) or two copies (p53BSx2) of the p53BS inserted upstream of the SV40 promoter in a pGL3 promoter vector. These constructs were cotransfected with a wild-type p53 (wt-p53) expression plasmid that strongly enhanced luciferase activity. No enhancement was seen when the reporter plasmid was cotransfected with mutant p53 (mt-p53 [175] or mt-p53 [277]; Figure 2_B_). We also constructed a luciferase vector containing an 880-bp DNA fragment corresponding to the promoter region of the Sema3B gene (Sema-pro880-wt) including the p53BS or the same DNA fragment with a DNA substitution within the p53BS (Sema-pro880-mt). Luciferase activity of Sema-pro880-wt, but not Sema-pro880-mt, was enhanced by cotransfection of the plasmid designed to express wt-p53. This enhancement was not observed after cotransfection with plasmids designed to express mt-p53. The reporter gene assay, combined with the result of EMSA, clearly indicated that Sema3B is a bonafide target of wild-type p53.
We then examined transcriptional activation of Sema3B by endogenous p53 in response to cellular stresses causing DNA damage (UV irradiation and adriamycin treatment), using MCF7 (p53+/+) and H1299 (p53-/-) cells. As shown in Figure 3, expression of Sema3B mRNA increased remarkably in MCF7 cells after exposure to either genotoxic stress, in a time-dependent manner. However, expression was unchanged in H1299 cells (data not shown). We noted that induction of p21WAF1 in response to these treatments reached a maximum 12 and 24 hours after treatment, whereas expression of Sema3B mRNA increased up to 72 hours after the exposure. We had previously observed the same induction pattern of p53AIP1 mRNA after these exposures; this similarity implies that modification of p53 protein is required for induction of Sema3B.
Figure 3.
Induction of Sema3B and p21WAF1 mRNA in MCF7 (p53 +/+) cells in response to genotoxic stresses, shown by Northern blot analysis. mRNA were isolated at the indicated times after treatment with adriamycin or UV radiation. Expression of the actin gene served as a quantity control.
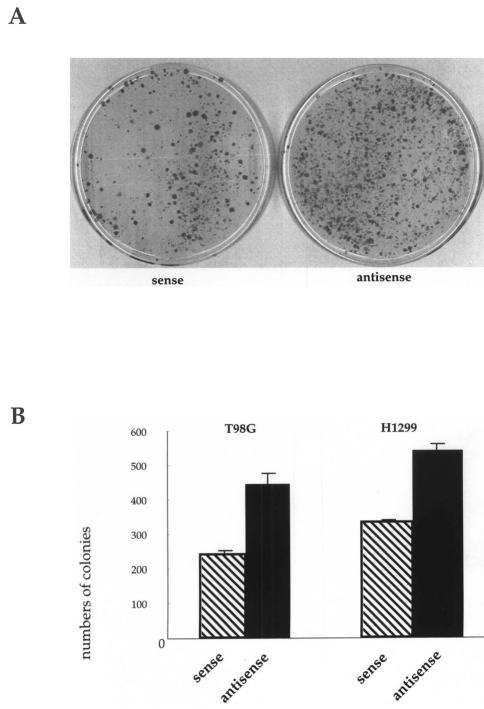
We performed colony formation assays to investigate the effect of Sema3B on cell growth, using expression vectors (sense-_Sema3B_-FLAG or sense-Sema3B) designed to express Sema3B protein with or without a FLAG tag at the C-terminal. As a negative control, we also prepared an expression vector in which the entire coding region of Sema3B was inserted in the antisense direction (antisense-Sema3B). Each of these vectors was transfected into T98G (glioblastoma), U373MG (glioblastoma), or H1299 (lung cancer) cells, all of which lack wild-type p53. Expression of Sema3B protein after transfection with sense-_Sema3B_-FLAG vector was confirmed by immunoblotting, using mouse anti-FLAG monoclonal antibody (Kodak; data not shown). After 2 weeks of incubation in geneticin-containing media, ectopic expression of Sema3B had reduced the numbers of colonies in all three cell lines (Figure 4; data for U373MG not shown). Similar results were obtained when we used sense-_Sema3B_-FLAG expression vector (data not shown), suggesting that Sema3B might play a role in either apoptosis or cell cycle arrest.
Figure 4.
Colony formation assay. (A) Growth suppression by ectopically expressed Sema3B. Sense: sense-Sema3B expression vector; antisense: antisense-Sema3B expression vector. (B) Numbers of G-418-resistant colonies from each of two p53-deficient cell lines (T98G and H1299) transfected with either sense-Sema3B expression vector (sense; hatched bars) or antisense-Sema3B expression vector (antisense; solid bars). Each experiment was repeated at least three times using triplicate samples, and the average scores are shown along with error bars.
Discussion
In addition to the specific functions of semaphorins in axon guidance and axonal repulsion, emerging evidence indicates their involvement in neuronal apoptosis [22]. Semaphorin3A (Sema3A), a member of the same class as Sema3B, induces neuronal apoptosis by binding to its receptor, neurophilin-1 (NRP1), which is known as a receptor for Sema3B [23,24]. On the other hand, VEGF165 (vascular endothelial growth factor) can block Sema3A-NRP1 signaling for neuronal apoptosis by directly competing with Sema3A for binding to NRP1 [23,24]. Therefore, the balance between Sema3A and VEGF165 might determine a cell's fate, survival, or apoptosis. We speculate that Sema3B is also involved in regulating apoptosis via binding to NRP1, by cooperating with Sema3A or competing with VEGF165.
Several groups have reported a possible role of p53 in neuronal apoptosis; for one thing, expression of p53 mRNA precedes neuronal apoptosis in neurons of injured brain and spinal cord [25]. Second, neuronal cells from _p53_-/-knockout mice are resistant to apoptosis induced by neuronal injury, both in vitro and in vivo [26]. As a third line of evidence, experimental overexpression of p53 in neuronal cells can induce apoptosis [26,27]. These data indicate that p53 plays an important role in regulating neuronal apoptosis, probably through the transcriptional regulation of target gene(s). Indeed, in our experiments, overexpression of Sema3B suppressed the growth of neuronal and nonneuronal cancer cells. Taken together, the results support a notion that Sema3B might be one of the mediators involved in p53-dependent apoptosis for not only neuronal cells but also non-neuronal ones. The information presented here should shed light toward a fuller understanding of the physiological functions of p53.
Acknowledgements
We thank C. C. Ng, C. Tanikawa, K. Matsui, and S. Onoue for their excellent technical assistance.
Abbreviations
p53BS
p53 binding site
Sema3B
Semaphorin3B
Sema3A
Semaphorin3A
NRP1
neurophilin-1
VEGF
vascular endothelial growth factor
Footnotes
1
This work was supported by Grant #13216031 from the Ministry of Education, Cullture, Sports, Science and Technology to H.A., and by “Research for the Future” Program Grant #00L01402 from the Japan Society for the Promotion of Science to Y.N.
References
- 1.Oren M. Relationship of p53 to the control of apoptotic cell death. Semin Cancer Biol. 1994;5:221–227. [PubMed] [Google Scholar]
- 2.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 3.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 4.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 5.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 8.Tokino T, Thiagalingam S, el-Deiry WS, Waldman T, Kinzler KW, Vogelstein B. p53 tagged sites from human genomic DNA. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 9.Furuhata T, Tokino T, Urano T, Nakamura Y. Isolation of a novel GPI-anchored gene specifically regulated by p53; correlation between its expression and anti-cancer drug sensitivity. Oncogene. 1996;13:1965–1970. [PubMed] [Google Scholar]
- 10.Urano T, Nishimori H, Han H, Furuhata T, Kimura Y, Nakamura Y, Tokino T. Cloning of P2XM, a novel human P2X receptor gene regulated by p53. Cancer Res. 1997;57:3281–3227. [PubMed] [Google Scholar]
- 11.Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K, Yoshida S, Ono M, Kuwano M, Nakamura Y, Tokino T. A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene. 1997;15:2145–2150. doi: 10.1038/sj.onc.1201542. [DOI] [PubMed] [Google Scholar]
- 12.Han HJ, Tokino T, Nakamura Y. CSR, a scavenger receptor-like protein with a protective role against cellular damage caused by UV irradiation and oxidative stress. Hum Mol Genet. 1998;7:1039–1046. doi: 10.1093/hmg/7.6.1039. [DOI] [PubMed] [Google Scholar]
- 13.Takei Y, Ishikawa S, Tokino T, Muto T, Nakamura Y. Isolation of a novel TP53 target gene from a colon cancer cell line carrying a highly regulated wild-type TP53 expression system. Genes, Chromosomes Cancer. 1998;23:1–9. [PubMed] [Google Scholar]
- 14.Ng CC, Koyama K, Okamura S, Kondoh H, Takei Y, Nakamura Y. Isolation and characterization of a novel TP53-inducible gene, TP53TG3. Genes, Chromosomes Cancer. 1999;26:239–335. [PubMed] [Google Scholar]
- 15.Shiraishi K, Fukuda S, Mori T, Matsuda K, Yamaguchi T, Tanikawa C, Ogawa M, Nakamura Y, Arakawa H. Identification of fractalkine, a CX3C-type chemokine, as a direct target of p53. Cancer Res. 2000;60:3722–3726. [PubMed] [Google Scholar]
- 16.Ochi K, Hata J-I, Nozaki H, Tanaka F, Kato S, Fukuzawa R, Sobue G, Fukuuchi Y, Toyama Y, Umezawa A. Specific bisulfite modification of CTG triplet repeats of the androgen receptor gene, a gene associated with the triplet repeat disease X-linked spinal and bulbar muscular atrophy (SBMA) Neurosci Res Commun. 2001;28:1–10. [Google Scholar]
- 17.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura F, Kalb RG, Strittmatter SM. Molecular basis of semaphorin-mediated axon guidance. J Neurobiol. 2000;44:219–229. doi: 10.1002/1097-4695(200008)44:2<219::aid-neu11>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 19.Zou Y, Stoeckli E, Chen H, Tessier-Lavigne M. Squeezing axons out of the gray matter: a role for slit and semaphorin proteins from midline and ventral spinal cord. Cell. 2000;102:363–375. doi: 10.1016/s0092-8674(00)00041-6. [DOI] [PubMed] [Google Scholar]
- 20.McGrory WJ, Bautista DS, Graham FL. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 21.Ono K, Tanaka T, Tsunoda T, Kitahara O, Kihara C, Okamoto A, Ochiai K, Takagi T, Nakamura Y. Identification by cDNA microarray of genes involved in ovarian carcinogenesis. Cancer Res. 2000;60:5007–5011. [PubMed] [Google Scholar]
- 22.Shirvan A, Ziv I, Fleminger G, Shina R, He Z, Brudo I, Melamed E, Barzilai A. Semaphorins as mediators of neuronal apoptosis. J Neurochem. 1999;73:961–671. doi: 10.1046/j.1471-4159.1999.0730961.x. [DOI] [PubMed] [Google Scholar]
- 23.Bagnard D, Vaillant C, Khuth ST, Dufay N, Lohrum M, Puschel AW, Belin MF, Bolz J, Thomasset N. Semaphorin 3A-vascular endothelial growth factor-165 balance mediates migration and apoptosis of neural progenitor cells by the recruitment of shared receptor. J Neurosci. 2001;21:3332–3341. doi: 10.1523/JNEUROSCI.21-10-03332.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi T, Nakamura F, Jin Z, Kalb RG, Strittmatter SM. Semaphorins A and E act as antagonists of neurophilin-1 and agonists of neurophilin-2 receptors. Nat Neurosci. 1998;1:487–493. doi: 10.1038/2203. [DOI] [PubMed] [Google Scholar]
- 25.Hughes PE, Alexi T, Schreiber SS. A role for the tumour suppressor gene p53 in regulating neuronal apoptosis. NeuroReport. 1997;8:v–xii. [PubMed] [Google Scholar]
- 26.Xiang H, Hochman DW, Saya H, Fujiwara T, Schwartzkroin PA, Morrison RS. Evidence for p53-mediated modulation of neuronal viability. J Neurosci. 1996;16:6753–6765. doi: 10.1523/JNEUROSCI.16-21-06753.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan J, Galindo MF, Prehn JH, Weichselbaum RR, Beckett M, Ghadge GD, Roos RP, Leiden JM, Miller RJ. p53 expression induces apoptosis in hippocampal pyramidal neuron cultures. J Neurosci. 1997;17:1397–1405. doi: 10.1523/JNEUROSCI.17-04-01397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]