Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells (original) (raw)
Abstract
The [_URE3_] phenotype in Saccharomyces cerevisiae is caused by the inactive, altered (prion) form of the Ure2 protein (Ure2p), a regulator of nitrogen catabolism. Ure2p has two functional domains: an N-terminal domain necessary and sufficient for prion propagation and a C-terminal domain responsible for nitrogen regulation. We show here that the mRNA encoding Ure2p possesses an IRES (internal ribosome entry site). Internal initiation leads to the synthesis of an N-terminally truncated active form of the protein (amino acids 94–354) lacking the prion-forming domain. Expression of the truncated Ure2p form (94–354) mediated by the IRES element cures yeast cells of the [_URE3_] phenotype. We assume that the balance between the full-length and truncated (94–354) Ure2p forms plays an important role in yeast cell physiology and differentiation.
Keywords: eIF4E/IRES/prion/Ure2p/[_URE3_]
Introduction
The yeast Saccharomyces cerevisiae Ure2p binds the transcription factor Gln3p and thereby blocks the expression of genes involved in the assimilation of poor nitrogen sources, such as arginine, urea, allantoin and ureidosuccinate, when rich nitrogen sources such as glutamine, asparagine or ammonium ion are present in the medium (for a review see Hofman-Bang, 1999). In its prion-like [URE3_] form, Ure2p does not bind Gln3p, leading to enhanced transcription of mRNAs coding for permeases and enzymes involved in alternative nitrogen assimilatory pathways (for a review, see Wickner et al., 2000). Gln3p activates (among others) the transcription of the allantoate permease gene DAL5, which allows the uptake of ureidosuccinic acid (USA), an intermediate in uracil biosynthesis and which is a poor nitrogen source (Hofman-Bang, 1999). This enables Δ_ura2 cells, which are unable to perform the first two steps of pyrimidine biosynthesis leading to USA formation, to grow by utilizing ureidosuccinate (in place of uracil) in the presence of ammonium sulfate. This phenotype is used as a measure of [_URE3_] prion formation (Lacroute, 1971). The mechanism leading to [_URE3_] prion de novo formation is only partially understood. The prion form is able to convert the native form into inactive protein, and overproduction of Ure2p increases 20- to 200-fold the frequency with which a strain becomes [_URE3_] (Wickner, 1994; Masison and Wickner, 1995; Wickner et al., 2000).
URE2 deletion analysis has revealed two functional domains: an N-terminal domain (amino acids 1–65), which is necessary and sufficient for prion propagation, and a C-terminal domain (amino acids 66–354) able to bind Gln3p (Masison and Wickner, 1995; Masison et al., 1997). The N-terminal part of Ure2p, which is rich in asparagine and glutamine residues, is responsible for prion formation and propagation and can form amyloid in vitro (Wickner et al., 2000). Overexpression of an N-terminally truncated Ure2p in cells propagating the prion cures yeast of the [_URE3_] phenotype, probably by disrupting Ure2p aggregates (Edskes et al., 1999; Wickner et al., 2000). However, propagation of [_URE3_] cannot always be attributed to the generation of large Ure2p aggregates, as has been shown recently (Fernandez-Bellot et al., 2002).
Initiation of translation of most eukaryotic cellular mRNAs involves binding of the mRNA 5′ cap structure to a group of proteins referred to as the cap-binding complex or eIF4F (for reviews, see Sachs et al., 1997; Gingras et al., 1999; Hershey and Merrick, 2000), followed by recruitment of the 40S ribosomal subunit and associated initiation factors (43S initiation complex) and movement of the 43S initiation complex along the 5′-untranslated region (UTR) in search of the initiation codon (scanning). Functions of the cap-binding complex formed by initiation factors eIF4G, eIF4E and eIF4A include recognition of the mRNA 5′ cap structure (initiation factor eIF4E), delivery of an RNA helicase (initiation factor eIF4A) to the 5′ mRNA region, and bridging of the mRNA and the ribosome (initiation factor eIF4G via its interaction with 40S-bound eIF3) (Gingras et al., 1999).
During the last decade, it has become clear that a number of viral and eukaryotic cellular mRNAs can be translated via internal initiation, a process directed by a specific mRNA region termed an internal ribosome entry site (IRES). These elements allow direct recruitment of 40S ribosomes to the vicinity of the initiation codon (for reviews, see Jackson and Kaminski, 1995; Belsham and Jackson, 2000; Carter et al., 2000; Jackson, 2000; Hellen and Sarnow, 2001). The subset of initiation factors and associated proteins required for internal initiation varies depending on IRES type and structure. Most of the viral IRES elements do not require either cap-binding factor eIF4E or intact eIF4G (Belsham and Sonenberg, 1996; Belsham and Jackson, 2000; Jackson, 2000). So far, only recruitment to the IRES element of hepatitis A virus is known to require intact initiation factor eIF4G (Borman and Kean, 1997). In contrast, recruitment of 40S ribosomes to hepatitis C virus and swine fever virus IRES-containing mRNAs does not require any of the initiation factors of the eIF4-‘family’ (Pestova et al., 1998), and cricket paralysis virus IRES-containing mRNA directs initiation without a requirement for any of the canonical initiation factors (Wilson et al., 2000). Translation of some cellular capped mRNAs is initiated both 5′-end dependently and internally. These mRNAs carry IRES elements located in the mRNA coding region (Cornelis et al., 2000; Lauring and Overbaugh, 2000; Maier et al., 2002), and internal initiation of translation occurs under physiological conditions when eIF4E activity is downregulated (Johannes et al., 1999<//em>: ,Carter et al., 2000; Sachs, 2000). Translation of such mRNAs results in the production of different protein products, depending on the initiation pathway utilized.
Little is known about the mechanism of internal initiation in the yeast S.cerevisiae (Altmann et al., 1990; Iizuka et al., 1994; Paz et al., 1999). Internal initiation of reporter mRNAs carrying the poliovirus 5′-UTR was stimulated in yeast extracts when eIF4E activity was limiting (Altmann et al., 1990). Recently, a 148 nucleotide IRES element located in the 5′-UTR of crucifer-infecting tobamovirus (crTMV) coat protein has been shown to be active in yeast cells (Dorokhov et al., 2002). An IRES element located in the genome of cricket paralysis virus has been shown to direct internal initiation when the intracellular concentration of ternary eIF2·GTP·Met-tRNAi complex is low (Thompson et al., 2001). Also, internal initiation has been reported for yeast mRNAs such as those coding for the transcriptional factors TFIID, HAP4 and YAP1, and for translation initiation factor eIF4G1 (Iizuka et al., 1994; Zhou et al., 2001). Further more, an artficial IRES was identified that is activated substantially when yeast cells enter the diauxic phase or when they are artificially starved for carbon sources (Paz et al., 1999).
Here we show that URE2 mRNA possesses an IRES allowing for cap- and eIF4E-independent synthesis of an N-terminally truncated form of Ure2p (amino acids 94–354), which lacks the prion-forming domain and affects [_URE3_] prion propagation in yeast cells. The activity of the URE2 IRES is increased when eIF4E activity is downregulated. To our knowledge, this is the first reported case of an IRES located in the coding region of a yeast mRNA.
Results and discussion
In vivo forms of Ure2p
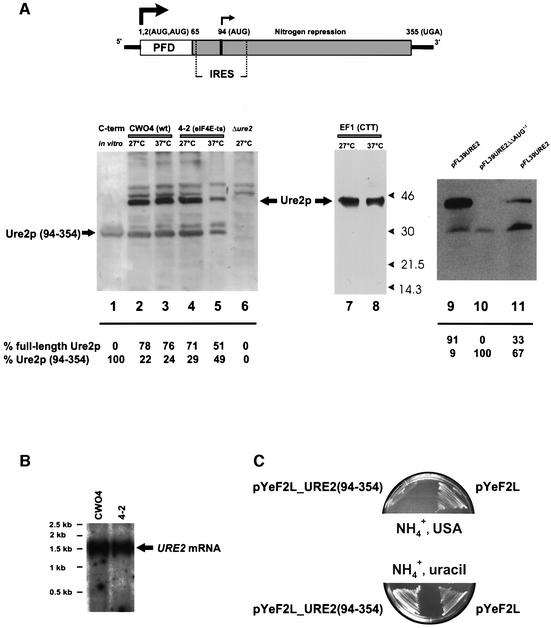
Several forms of Ure2p can be detected in extracts from yeast cells grown at different temperatures and analyzed by immunoblotting with anti-Ure2p antibodies. Among the detected proteins is a major ∼42 kDa product and two minor bands of 31 and 30 kDa (Figure 1A, lanes 2–5). These proteins are derived from the URE2 gene, since they are not detected in a _ure2_Δ strain (Figure 1A, lane 6; strain CC32). The size of the 42 kDa product corresponds to full-length Ure2p (Coschigano and Magasanik, 1991; Thual et al., 1999). The 30 kDa protein co-migrates with the C-terminal Ure2p domain (amino acids 94–354) translated in rabbit reticulocyte lysate from in vitro transcribed mRNA (Figure 1A, lane 1). The 31 kDa product might be an abnormally migrating form of the 30 kDa protein; however, it is not always observed on immunoblots. This product might represent a phosphorylated form of the 30 kDa protein. Phosphorylation of Ure2p has been reported by some laboratories (Cardenas et al., 1999; Hardwick et al., 1999). To verify that the 30 kDa product starts at Met94, we mutagenized the ATG94 in the URE2 open reading frame (ORF) to CTT and replaced the wild-type copy of the URE2 gene with the mutated gene copy in the yeast genome [strain EF1(CTT); see Table I]. Translation of the mutant URE2 mRNA in yeast resulted in the disappearance of the 30 kDa product (Figure 1A, right panel, lanes 7 and 8).
Fig. 1. Ure2p in vivo synthesis. (A) Top: schematic representation of the URE2 mRNA. Solid lines, untranslated regions; boxes, prion-forming (PFD; white) and nitrogen repression (gray) domains (Masison and Wickner, 1995; Masison et al., 1997); arrows, translation start sites. Bottom: Ure2p expression in different yeast strains. CWO4 (lanes 2 and 3), 4-2 (lanes 4 and 5), CC32 (Δ_ure2_) (lane 6) and EF1(CTT) (lanes 7 and 8) were grown for 16 h in minimal medium (SD, supplemented with essential amino acids) at 27°C; subsequently, part of the culture was incubated at 37°C for 5 h. CC32 (Δ_ure2_) transformed with pFL39URE2 (lane 9) and pFL39URE2ΔΔAUG1,2 plasmid (lane 10). CC32 (Δ_ure2_) transformed with pFL39URE2 (lane 11) were grown on SD medium for 48 h to early stationary phase, OD600 = 2.0–2.5. Extracts from equal numbers of cells were prepared, and proteins were separated by SDS–PAGE and blotted to nitrocellulose. The blot was decorated with anti-Ure2p antibodies. Lane 1: the C-terminal (94–354) fragment was synthesized in vitro in the RRL system. The relative ratio of full-length Ure2p to Ure2p (94–354) was determined densitometrically. (B) Northern blot analysis of URE2 mRNA expressed in yeast strains CWO4 and 4-2. For each lane, 20 µg of total yeast RNA was separated on a denaturing agarose gel, transferred onto a BrightStart-Plus Nylon membrane (Ambion) and hybridized to a 32P-labeled DNA URE2 ORF probe. (C) URE2 complementation analysis. Yeast strain CC32 (Δ_ure2_) was transformed with plasmid pYeF2L_URE2(94–354) or with vector pYeF2L and grown either on minimal medium containing 2% galactose, 0.5% ammonium sulfate and USA (15 µg/ml), or on 0.5% ammonium sulfate and uracil (20 µg/ml).
Table I. Strains of S.cerevisiae.
| Strain | Genotype | Source |
|---|---|---|
| CWO4 | MATa, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura3 | P.Linder |
| 4-2 | MATa, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura3, cdc33::LEU2 <cdc33-4-2_; _TRP1, ARSCEN> | M.Altmann |
| CC30 | MATα, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3 | C.Cullin |
| CC32 | MATa, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3, ure2::TRP1 | C.Cullin |
| CC34 | MATα, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3 [URE3] | C.Cullin |
| EF1(CTT) | MATa orα, _ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3, URE2-CTT_94 | This work |
| EF1(CTT)/4-2 | _MATa, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3, cdc33::LEU2 <CDC33_; _TRP1, ARSCEN>URE2-CTT_94 | This work |
| CC34/wt | MATα, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3 cdc33::LEU2 <CDC33_; _TRP1, ARSCEN> | This work |
| CC34/4-2 | MATα, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3 cdc33::LEU2 <cdc33-4-2_; _TRP1, ARSCEN> | This work |
| CC34/4-4 | MATα, ade2-1, leu2-3,112, his3-11,15, trp1-1, ura2::HIS3 cdc33::LEU2 <cdc33-4-4_; _TRP1, ARSCEN> | This work |
| HM6B ρ0 | Mata; trp1, ura2, kar1, can1 [ure-o] [rhoO] | R.Wickner |
| HM6B/EF1(CTT) | Mata; trp1, ura2, kar1, can1, [URE3] [cytoduction from EF1(CTT)] | This work |
| HM6B/CC34/wt | Mata; trp1, ura2, kar1, can1, [URE3] (cytoduction from CC34/wt) | This work |
| HM6B/CC34/4-2 | Mata; trp1, ura2, kar1, can1, [ure-o] (cytoduction from CC34/4-2) | This work |
| HM6B/CC34/4-4 | Mata; trp1, ura2, kar1, can1, [ure-o] (cytoduction from CC34/4-4) | This work |
We also replaced the two first AUGs in the URE2 ORF (ATGATG→GGATCC). As expected, the 42 kDa protein was not expressed, while the 30 kDa product could still be detected (Figure 1A, lanes 9 and 10). This indicates that the 30 kDa protein is not a degradation product of the full-length Ure2p, but derives from initiation of translation of URE2 mRNA at AUG94 either by leaky scanning or by internal initiation. Since initiation by leaky scanning is 5′-end- and most probably eIF4E-dependent, whereas internal initiation is not (Belsham and Jackson, 2000), we tested the dependence of Ure2p synthesis on eIF4E. The yeast strain 4-2 has two point mutations in the eIF4E gene, which result in a temperature-sensitive phenotype (eIF4E-ts). At the non-permissive temperature (37°C), cap-dependent initiation is severely inhibited in this strain (Altmann et al., 1989; Altmann and Trachsel, 1997). We found that accumulation of full-length Ure2p was reduced in strain 4-2 maintained for several hours at the non-permissive temperature, while the accumulation of the 30 kDa product was not, and the ratio of 30:42 kDa product had increased to almost 1:1 (Figure 1A, compare lanes 4 and 5). It should also be noted that the ratio of full-length Ure2p to the 30 kDa product detected by immunoblots might not reflect the real ratio of both Ure2p forms in vivo, since the polyclonal antibody used for the detection may recognize the various domains of Ure2p with different affinity. Interestingly, the amount of full-length Ure2p seems to decrease when cells shift from log phase to stationary phase, while the amount of 30 kDa Ure2p increases (Figure 1A, compare lanes 9 and 11), indicating that the ratio of these two products might vary during different stages of cell growth and that the in vivo physiological response under some conditions might yield the short form of Ure2p.
Integrity of URE2 mRNA in both CWO4 (wild type) and 4-2 cells was verified by northern blot analysis (Figure 1B). We could not detect any minor URE2 mRNA species that could give rise to 5′-end-dependent translation of the 30 kDa protein. These findings suggest that translation of URE2 mRNA at AUG94 might be initiated internally.
The 30 kDa Ure2p protein is active
Truncated Ure2p (amino acids 66–354) was shown earlier to complement fully the URE2 gene deletion (Masison and Wickner, 1995; Masison et al., 1997). We found that a Δ_ura2_ strain transformed with a yeast plasmid carrying the shortened URE2 (94–354) gene version complements the URE2 gene deletion, as shown by a lack of growth on minimal medium containing USA and ammonium sulfate [Figure 1C; strain CC32 transformed with plasmid pYeF2L_URE2(94–354)]. Our results show that Ure2p (94–354) is active as it prevents ureidosuccinate uptake and cell growth of the Δ_ura2_ strain. The mechanism underlying nitrogen catabolite repression involves association of Ure2p with Gln3p in the cytoplasm, an interaction that prevents Gln3p from reaching its binding sites in the nucleus. Sequestering of Gln3p by Ure2p in the cytoplasm depends on the ratio between Gln3p and Ure2p, and can be affected by under- or overproduction of Ure2p (Cox et al., 2000).
Internal initiation at AUG94 on URE2 mRNA in vitro
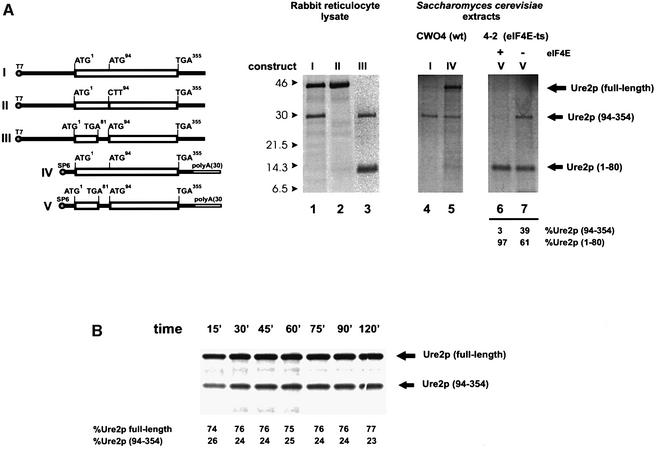
To explore further the possibility that the 30 kDa Ure2p (amino acids 94–354) stems from internal initiation at AUG94 of URE2 mRNA, we carried out a series of in vitro translation experiments using various extracts and in vitro transcribed URE2 mRNAs. We found that translation of the URE2 mRNA in rabbit reticulocyte lysate or wheat germ extract (data not shown) also yielded two proteins of 42 and 30 kDa (Figure 2, lane 1). Translation of mutant URE2 mRNA carrying the ATG94 to CTT substitution gave rise only to full-length Ure2p (Figure 2, lane 2). Introduction of a TGA stop codon at position 81 of the URE2 ORF abolished the synthesis of full-length Ure2p and led to the synthesis of a 14 kDa product resulting from translation of the new upstream ORF (Figure 2, lane 3). This mutation did not affect the synthesis of the 30 kDa protein, suggesting that it is not a degradation product of full-length Ure2p. Possible degradation of full-length Ure2p was analyzed by time-course experiments in the rabbit reticulocyte system. Synthesis of the 30 kDa product did not show a precursor/product relationship with the 42 kDa polypeptide. The ratio of synthesis of these two proteins was constant with time, even after protein synthesis had ceased at 60 min (Figure 2B), indicating that the 30 kDa product (seen in Figure 2A; lanes 1 and 3) is not a degradation product of full-length Ure2p.
Fig. 2. (A) Ure2p in vitro synthesis. In vitro transcribed and capped URE2 mRNAs (Komar et al., 1997) were translated in the presence of 0.3 mCi/ml [35S]methionine (Amersham) in either rabbit reticulocyte lysate or yeast extract (autoradiograms are shown). I, URE2 mRNA with 204 nucleotide 5′-UTR [no poly(A)]; II, URE2 mRNA with ATG94-Met substituted for CTT-Leu, 204 nucleotide 5′-UTR [no poly(A)]; III, URE2 with TTA81-Leu substituted by TGA-Stop, 204 nucleotide 5′-UTR [no poly(A)]; IV, URE2 mRNA with short 5′-UTR (25 nucleotides) and poly(A) tail; V, URE2 mRNA with TTA81-Leu substituted by TGA-Stop, short 5′-UTR (25 nucleotides) and poly(A) tail. Autoradiograms: rabbit reticulocyte lysate (lanes 1–3), yeast extracts (lanes 4–7), wild type (lanes 4 and 5) or 4-2 extract (lanes 6 and 7). Lane 6, no eIF4E added; lane 7, 2 pmol eIF4E added. (B) Time course of Ure2p synthesis in a rabbit reticulocyte lysate cell-free system using Ure2p mRNA construct I. The relative ratio of full-length Ure2p to Ure2p (94–354) was determined densitometrically.
We also tested translation of URE2 mRNA in yeast translation extracts that are known to be very sensitive to the presence of hairpin structures in the mRNA 5′-UTR (Sagliocco et al., 1993). Translation of the URE2 mRNA carrying a 5′-UTR of 204 bp (construct I) in an extract obtained from wild-type yeast cells yielded hardly any full-length Ure2p (Figure 2, lane 4). However, the 30 kDa protein was synthesized. Shortening of the 5′-UTR from 204 to 25 bp and addition of a poly(A) tail to the 3′-UTR of URE2 mRNA strongly enhanced the production of full-length Ure2p without affecting the synthesis of the 30 kDa protein (Figure 2, lane 5).
In an extract derived from strain 4-2 (eIF4E-ts), synthesis of the 30 kDa protein (amino acids 94–354) from construct V was only efficient in the absence of functional eIF4E (Figure 2, lanes 6 and 7), indicating that the synthesis of the 30 kDa protein could proceed via an eIF4E-independent pathway (in these experiments, the activity of endogenous eIF4E was shut off by incubating the extracts for 7.5 min at 37°C prior to translation). Ure2p (1–80) synthesis, which was surprisingly high in the absence of functional eIF4E, was mildly enhanced (∼2-fold) by the addition of eIF4E (Figure 2A; lane 6), which is to be expected for cap-dependent translation.
URE2 AUG94 directs internal initiation in vivo
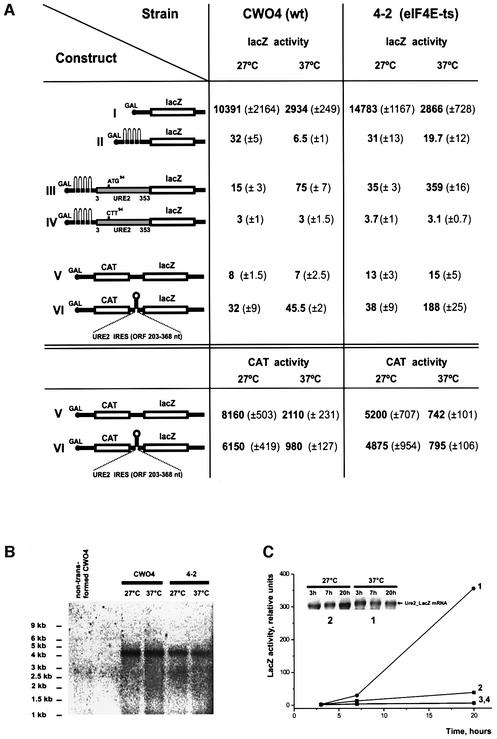
To probe for the existence of an IRES in the URE2 mRNA and to measure further its efficiency in vivo, we produced DNA constructs encoding inducible lacZ mRNAs (Figure 3A) and transformed them into isogenic yeast wild-type (CWO4) and eIF4E-ts strain 4-2. Construct I encoding lacZ mRNA with a 5′-UTR derived from the GAL1 gene (Mueller et al., 1987) is translated very efficiently at 27°C, in both wild-type and eIF4E-ts strain 4-2 (Figure 3A). A significant decrease in lacZ expression of construct I was observed for both strains at 37°C. We assume that in both strains, eIF4E activity becomes limiting (due to its inactivation) but the effect is more pronounced in strain 4-2, as expected. Insertion of a stable hairpin structure in the 5′-UTR (Altmann et al., 1993) reduces lacZ expression almost completely in both strains (construct II). To test for internal initiation of the putative URE2 IRES, we inserted the URE2 ORF (amino acids 3–353) in-frame with lacZ behind the stable hairpin structure and compared lacZ expression from constructs carrying at position 94 either ATG-Met (construct III) or CCT-Leu (construct IV). Whereas there was almost no expression from construct IV, a significantly increased level of β-galactosidase was observed from construct III in both the wild type and eIF4E-ts strain at the non-permissive temperature (37°C). The lacZ activity was significantly higher in strain 4-2 (359 versus 35 U), indicating that expression driven by Met94 in the URE2 ORF is eIF4E independent and proceeds in vivo via a cap-independent mechanism. We also tested the URE2 IRES (URE2 ORF amino acids 67–122) in a bicistronic CAT–lacZ construct (construct VI): significantly higher expression of lacZ was detected at 37°C in strain 4-2 when compared with cells transformed with the bicistronic construct devoid of an IRES (188 versus 15 U). Enhanced translation of construct VI at 37°C was also observed in wild-type cells, but the effect was less pronounced (45.5 U) than in the eIF4E-deficient strain. In contrast, translation of the first cistron (CAT) was downregulated 6- to 7-fold for both bicistronic constructs (V and VI), indicating that its translation is cap-dependent. Northern blot analysis of mRNA from cells transformed with construct III (which shows the highest level of internal initiation) revealed a predominant mRNA band of the expected size of ∼4.2 kb in both yeast strains (Figure 3B). No shorter mRNA band could be detected that would give rise to 5′-end-dependent translation of the reporter gene. No lacZ activity was observed when cells expressing construct III were grown on glucose-containing media, proving cryptic promoters to be absent in the DNA constructs used (data not shown). Moreover, after cells were shifted from glucose- to galactose-containing medium, no lacZ activity was observed during the first 3 h after the shift, although, the URE2-lacZ mRNA was readily detected (Figure 3C). It has been shown previously that translation in yeast is inhibited rapidly when glucose is replaced with an alternative carbon source such as galactose, and that this inhibition is maintained for >2 h (Ashe et al., 2000). Our results are consistent with these data. The observation that the expression of lacZ is dependent on the downregulation of eIF4E activity (taken together with the data on northern blotting) is consistent with lacZ being expressed from an IRES-containing mRNA rather than from an mRNA derived from a cryptic promoter. The absence of lacZ expression from construct IV carrying the ATG94 to CTT94 substitution in URE2 indicates that lacZ expression from the URE2 IRES is, indeed, driven by AUG94. It should be noted that the URE2 ORF cloned in front of lacZ carries seven other AUGs (downstream of AUG94) that are in-frame, but do not give rise to lacZ expression.
Fig. 3. (A) Expression of reporter constructs under the control of the GAL1/10 promoter transformed into wild-type strain CWO4 and strain 4-2. β-galactosidase activity (relative units) was determined following the protocol described in the Clontech Yeast Protocols Handbook after an 18 h induction of the cells in 2% galactose at the indicated temperature. CAT activity was measured using the FAST CAT Green (deoxy) Chloramphenicol Acetyltransferase Assay Kit (Molecular Probes). (B) Northern blot analysis of construct III mRNA expressed in CWO4 and 4-2 yeast strains at 27 and 37°C. For each lane, 15 µg of total yeast RNA was separated on a denaturing agarose gel, transferred onto a BrightStart-Plus Nylon membrane (Ambion) and hybridized to a 32P-labeled DNA lacZ probe. The first lane on the left shows RNA isolated from non-transformed CWO4 cells. (C) Time course of β-galactosidase activity (relative units) expressed from constructs III and IV transformed into strain 4-2. LacZ activities at 27°C (construct III, plot line 2; construct IV, plot line 3) and 37°C (construct III, plot line 1; construct IV, plot line 4) are shown. Cells were grown on minimal SD medium containing 2% glucose fo 18 h, washed with water and then transferred to minimal SD medium with 2% galactose. Sample aliquots were taken at the indicated times for lacZ activity measurements and RNA isolation. Insert: northern blot analysis of construct III mRNA expressed in strain 4-2.
We conclude from the above data that URE2 mRNA possesses an IRES, which allows for cap-independent translation initiation at Met94 of the URE2 ORF. However, this IRES element promotes translation less efficiently than the cap-dependent process; downregulation of eIF4E activity enhances IRES-directed translation. A similar result was obtained previously for the p110PITSLRE protein kinase IRES: two PITSLRE protein kinase isoforms, p110PITSLRE and p58PITSLRE, are translated from a single transcript by initiation at alternative in-frame AUG codons. p110PITSLRE was shown to be produced by cap-dependent translation, whereas p58PITSLRE resulted from internal initiation of translation. The expression of p58PITSLRE was enhanced upon downregulation of eIF4E in the G2/M phase of the cell cycle (Cornelis et al., 2000).
Internal initiation affects [URE3] prion propagation in yeast cells
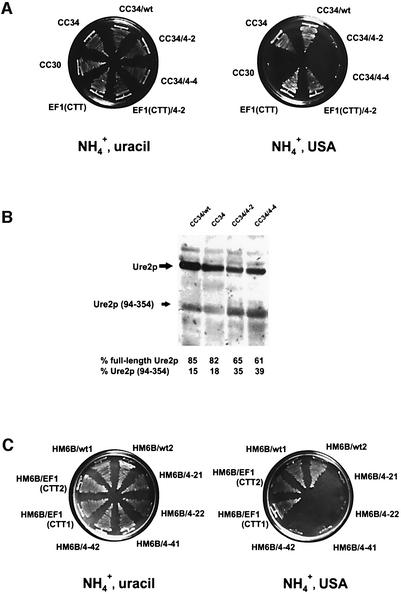
Since expression of an N-terminally truncated Ure2p (amino acids 66–354) can cure yeast cells of the [_URE3_] phenotype (probably by disrupting Ure2p aggregates; Edskes et al., 1999) and is unable to give rise to [_URE3_] (Masison and Wickner, 1995; Masison et al., 1997), the IRES found in URE2 mRNA may allow the synthesis of an [_URE3_]-resistant Ure2p form. To test this possibility, we replaced the genomic eIF4E gene by the temperature-sensitive eIF4E alleles (4-2 and 4-4) and expressed them from a centromeric plasmid (Altmann et al., 1989; Altmann and Trachsel, 1997) in the [_URE3_] yeast strain CC34 (Fernandez-Bellot et al., 2000), which allows for ureidosuccinate uptake in the presence of ammonium sulfate (Figure 4A). This cured the cells of the [_URE3_] phenotype, since CC34 strains bearing the eIF4E-ts mutations were not able to grow at 32°C on selective medium containing USA and ammonium sulfate, while they did grow on minimal medium containing uracil and ammonium sulfate (Figure 4A). An isogenic strain carrying the wild-type eIF4E allele on the same plasmid (CC34/wt) was not able to cure the cells of the [_URE3_] phenotype. The altered ratio of full-length Ure2p to Ure2p (94–354) in 4-2 and 4-4 ts strains in comparison with carrying the wild-type eIF4E was demonstrated by western blotting (Figure 4B).
Fig. 4. (A) URE2 IRES and [_URE3_] phenotype: mutations in eIF4E prevent the uptake of ureidosuccinate on ammonium-containing media. Mutation of AUG to CTT at codon 94 of URE2 [EF1(CTT strain), a derivative of CC30 (Table I)] gives rise de novo to a [_URE3_] phenotype. Left: SD medium, containing 0.5% ammonium sulfate and uracil (20 µg/ml). Right: minimal medium containing 0.5% ammonium sulfate and USA (150 µg/ml). eIF4E-wt and ts mutations 4-2 and 4-4 were introduced into the haploid strain CC34, yielding strains CC34/wt, CC34/4-2 and CC34/4-4; CC30, CC34 and EF1(CTT) were used as controls (Table I). The ts mutation 4-2 was introduced into haploid strain EF1(CTT), rendering EF1(CTT)/4-2 (Table I). (B) Ure2p in vivo synthesis: western blot analysis. Cells were grown for 20 h in minimal medium (SD, supplemented with essential amino acids) at 32°C. Extracts from equal numbers of cells were prepared, proteins separated by SDS–PAGE and blotted to nitrocellulose. The blot was decorated with anti-Ure2p antibodies. The relative ratio of full-length Ure2p to Ure2p (94–354) was determined densitometrically. (C) Cytoduction experiments: the cytoplasm from CC34/4-2 and CC34/4-4 cannot transmit [_URE3_], while the cytoplasm from CC34/wt and the EF1(CTT) strain can transform to the prion phenotype. Growth of cytoductant strains HM6B/wt; HM6B/4-2, HM6B/4-4; HM6B/EF1(CTT) (two clones for each) is shown after the transfer of cytoplasm from donor strains CC34/wt, CC34/4-2, CC34/4-4 or EF1(CTT). Left: SD medium, containing 0.5% ammonium sulfate and uracil (20 µg/ml). Right: minimal medium containing 0.5% ammonium sulfate and USA (125 µg/ml). The cytoduction procedure (Wesolowski and Wickner, 1984; Moriyama et al., 2000) is described in Materials and methods.
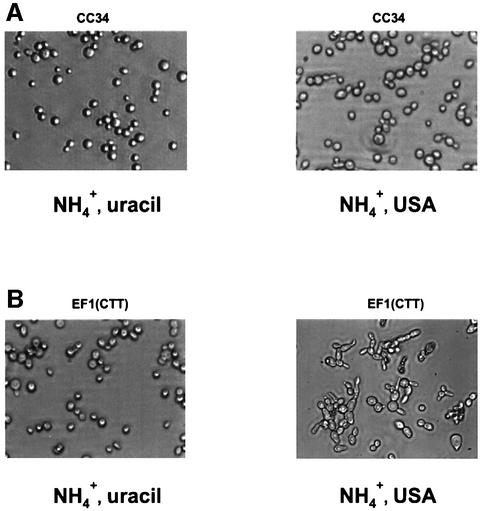
To test whether eIF4E-ts mutations result in the loss of [_URE3_] rather than in loss of the ability to convert USA to uracil, cytoplasm from CC34/wt, CC34/4-2 and CC34/4-4 was transferred to the [ure-o] [rhoO] HM6B strain. The ability to donate [_URE3_] with the transfer of cytoplasm is considered to be one of the characteristic features of prion-propagating cells (Moriyama et al., 2000). Indeed, cytoplasm from CC34/4-2 and CC34/4-4 strains could not transmit [_URE3_], while cytoplasm from the CC34/wt strain did (Figure 4C). This shows that HM6B/CC34/4-2 and HM6B/CC34/4-4 mutants have lost the [_URE3_] phenotype. We also found that the EF1(CTT) strain [URE2 (ATG94 to CTT) mutation integrated in the yeast genome of strain CC30] gave rise de novo to the [_URE3_] prion phenotype (Figure 4C), while the ancestor wild-type CC30 strain (Fernandez-Bellot et al., 2000) was not able to grow on USA (Figure 4A). To test whether Ure2p is its prion form in EF1(CTT) cells, cytoplasm from these cells was transferred to the [ure-o] [rhoO] HM6B strain. Cytoplasm from the EF1(CTT) strain was able to donate [_URE3_] to recipient [ure-o] cells, as shown by growth on selective medium containing USA and ammonium sulfate (Figure 4C). This indicates that Ure2p is present in EF1(CTT) cells in its prion form and that in the absence of Ure2p (94–354), full-length Ure2p is more prone to aggregation. In order to rule out the possibility that the eIF4E mutation could have an indirect effect on [_URE3_] propagation, we introduced the temperature-sensitive eIF4E allele (4-2) into strain EF1(CTT) [EF1(CTT)/4-2; Table I]. This did not cure the EF1(CTT) cells of the [_URE3_] phenotype (Figure 4A). Strain EF1(CTT)/4-2 grew on a selective medium containing USA and ammonium sulfate [although at a slower rate in comparison with the ancestor EF1(CTT) strain]. Interestingly, both haploid EF1(CTT) and EF1(CTT)/4-2 strains, but not the CC34 or CC32 (_ure2_Δ) strains, exhibit morphological abnormalities on a selective medium containing USA and ammonium sulfate, i.e. filamentous growth (Figure 5B). These abnormalities are very similar to those observed under conditions of yeast cell treatment with fusel alcohols (for references, see Lorenz et al., 2000). Fusel alcohols are the products of amino acid catabolism under nitrogen-limiting conditions and are known to promote aberrant elongated morphology in S.cerevisiae (Lorenz et al., 2000). Fusel alcohols also have been shown to inhibit translation initiation rapidly by altering the activity of translation initiation factor eIF2B (Ashe et al., 2001). Since these morphological changes are not observed in CC34 [_URE3_] (Figure 5A) or CC32 _ure2_Δ (data not shown), we propose that Ure2p (94–354) might play an as yet unknown cellular role.
Fig. 5. Yeast cell morphology. Yeast strains CC34 (A) and EF1(CTT) (B) were grown on a solid SD medium. Left: SD medium, containing 0.5% ammonium sulfate and uracil (20 µg/ml). Right: min imal medium, containing 0.5% ammonium sulfate and USA (125 µg/ml). Cells were scraped off the plate and resuspended in 10 µl of water on microscope slides. Images were taken using a Zeiss TELAVAL 31 microscope equipped with a Spot Diagnostic Instrument Inc. digital camera at ×40 magnification.
What is the physiological role of the URE2 mRNA IRES element?
Proteins whose synthesis is initiated internally are involved in a variety of processes, including development, differentiation, stress, growth/cancer and programmed cell death (reviewed in Carter et al., 2000; Hellen and Sarnow, 2001). The presence of an IRES element in a cellular mRNA may reflect the need to produce the corresponding protein at a particular stage of cell growth under conditions when 5′-end translation is downregulated (Paz et al., 1999; Van der Velden and Thomas, 1999; Carter et al., 2000). It is shown here that the mRNA encoding Ure2p possesses an IRES leading to the synthesis of an N-terminally truncated and active form of the protein (amino acids 94–354) lacking the prion-forming domain. Our findings suggest that alteration of eIF4E activity affects the ratio of full-length to 30 kDa Ure2p and [_URE3_] prion propagation in yeast cells. This indicates that yeast cells may have developed a mechanism to impede or at least alleviate Ure2p aggregation by producing an N-terminally truncated form of the protein, and that [_URE3_] propagation might be regulated by a balance between 5′-end-dependent and internal initiation mechanisms. Interestingly, it was found that the activity of URE2 IRES in a bicistronic construct seems to be significantly higher in stationary phase when compared with yeast cells at exponential growth (J.P.G.Ballesta, personal communication). This observation supports previous findings showing that the activity of an IRES can be substantially induced when cells exit the logarithmic growth phase (Paz et al., 1999), and might indicate that yeast cells in stationary phase would be less prone to [_URE3_] propagation. The details of the mechanism of internal initiation of URE2 mRNA translation are not known, and further experiments are required to elucidate the physiological importance of expressing the truncated form of Ure2p.
We assume, however, that the balance between the full-length and truncated (94–354) Ure2p forms plays an important role in yeast cell physiology under vegetative growth conditions as well as during differentiation and their switch to invasive (filamentous) growth.
Materials and methods
Yeast strains
Wild-type CWO4 (Coppolecchia et al., 1993) and isogenic 4-2 (Table I) (Altmann et al., 1989; Altmann and Trachsel, 1997) were used in Ure2p and lacZ expression experiments and for preparation of cell-free translation extracts. The EF1(CTT) strain (Table I) was obtained as follows: ATG-Met94 of the URE2 ORF was switched to CTT-Leu by PCR (5′-GTAAAACGACGGCCAGT-3′ as upstream and 5′-GAATAC TCCACGTGACTAAGATCCGAAAATG-3′ as downstream primer), digested with _Xho_I and _Pml_I, and cloned into pBIISK+(URE2) (Komar et al., 1997). The _Pfl_MI–_Not_I fragment of _URE2__CTT was cloned into pFL39URE2 (Komar et al., 1998). The mutated URE2 copy was introduced into strain CC32 (Table I). [TRP–] colonies were selected. _URE2__CTT integration was verified by PCR of yeast genomic DNA (5′-GTGTAACTGCAGCCAAATGATGAATAAC-3′ as upstream and 5′-CCATACCATGATCGATTAAAGCTGG-3′ as downstream primer), and restriction analysis using _Hin_fI (ATG to CTT mutation results in disappearance of the _Hin_fI site). Isogenic CC30 and CC34 [_URE3_] strains (Table I) have been described previously (Fernandez-Bellot et al., 2000). eIF4E-ts mutations 4-2 and 4-4 were introduced into the haploid CC34 [_URE3_] and EF1(CTT) strains by co-transformation of a 4.4 kb cdc33::LEU2 linear DNA fragment (Altmann et al., 1987) and plasmids carrying eIF4E-ts mutations or the wild-type gene copy. Transformants, which showed a ts phenotype, were checked for homologous gene recombination by PCR (5′-CATTGCAAGAGGTAGTGTTAATTC TGG-3′ as upstream and 5′-GAGTTGTCGTTTTAACGTCATCCT CTC-3′ as downstream primer).
Cytoduction
Cytoduction was conducted essentially as described by Wickner and co-workers (Wesolowski and Wickner, 1984; Moriyama et al., 2000). The HM6B (Mata, trp1, ura2, kar1, can1 [ure-o]) strain that fails to undergo karyogamy was kindly provided by Dr Reed Wickner. The recipient cells were [_rho_O] (mitochondrial DNA was eliminated by overnight growth of the cells on YPD medium containing 1 mM ethidium bromide). Respiratory-deficient cells were screened by replica plating of clones grown on YPD medium on to YPG plates (1% yeast extract, 2% peptone, 3% glycerol). Clones unable to grow on YPG were [_rho_O]. EF1(CCT), CC34/wt, CC34/4-2 and CC34/4-4 (Matα, Table I) were donor strains. After mating recipient and donor cells on YPD plates for 6–8 h, cells with donor nuclei were counterselected by plating the mating mixture for single colonies on appropriate minimal medium; diploids were eliminated by addition to the minimal medium of l-canavanine sulfate (60 mg/ml). Respiratory-competent clones (having the recipient genotype) were screened out by replica plating on YPG plates.
Plasmids
For pET3a_URE2(94–354), DNA encoding Ure2p (amino acids 94–354) was amplified by PCR (5′-ATGAGTCACGTGGAGTATTCC-3′ as upstream and 5′-CCGCGGTGGCGGCCGCT-3′ as downstream primer), digested with _Bam_HI and cloned into pET3a (Stratagene) in which the _Nde_I site was filled in with Klenow fragment prior to vector digestion with _Bam_HI. For pYeF2L_URE2(94–354), DNA encoding Ure2p (amino acids 94–354) was amplified by PCR (5′-CGGGATCCATGAGTCA CGTGGAG-3′ as upstream and 5′-GCGCCTTAGGTCATTCACCACG CAATGCCTTGATG-3′ as downstream primer) and subcloned as a _Bam_HI–_BsU_36I fragment into pYeF2L under the control of a hybrid GAL10-CYC1 promoter. pBIISK+(URE2) has been described previously (Komar et al., 1997). For the production of pFL39URE2ΔΔAUG1,2 (lacking both AUG codons in the URE2 ORF), DNA encoding Ure2p was amplified by PCR [5′-GCAAATTAAGTTGTACACCAAGGAT CCAATAACAACGGCAACCAAGTGTCG-3′ as upstream primer, which replaces AUGAUG by GGATCC (_Bam_HI site) and 5′-GCT TCGTCCATTTGTAAACTTCTGG-3′ as downstream primer]. The PCR fragment was digested with _Bsr_GI and _Nco_I, and ligated into pFL39URE2. The TGA81-Stop of the URE2 ORF was introduced by double PCR (Landt et al., 1990) (5′-GAATACCTGAGAACAAC-3′ as upstream and 5′-GGAAACAGCTATGACCATG-3′ as downstream primer in the first PCR, 5′-GTAAAACGACGGCCAGT-3′ as upstream primer and the first PCR product as downstream primer in the second PCR). The PCR product was digested with _Xho_I and _Eco_RI, and cloned into pBIISK+.
pSP64(polyA) derivatives carrying wild-type or mutant URE2 were obtained by PCR (5′-GTGTAACTGCAGCCAAATGATGAATAAC-3′ as upstream and 5′-GTGAGGATCCTTTGCACTTAATTTTCC-3′ as downstream primer), digested with _Bam_HI and _Pst_I, and cloned into pSP64poly(A) vector (Promega). For p281-4-URE2 and p281-4-URE2_CTT plasmids, DNA encoding Ure2p (amino acids 3–353) was amplified by PCR (5′-AAAAAACTCGAGAATAACAACGGCAACC-3′ as upstream and 5′-TTCCGGAATTCTACCACGCAATGCCTTG-3′ as downstream primer), digested with _Xho_I and _Eco_RI, and cloned into p281-4 vector under the control of the GAL1/10 promoter (Altmann et al., 1993). The reporter plasmid p281-IRES_URE2 bearing a 165 bp fragment of the URE2 gene (bp 203–368 of the URE2 ORF; amino acids 67–122) was constructed by PCR amplification of pBluescriptIISK+(URE2) (linearized with _Xmn_I) (5′-CCAGATCTG CCAAAATAATGATAACGAG-3′ as upstream and 5′-CCGGATCC GGCGCAGACCTGTGAGAGAA-3′ as downstream primer). The PCR fragment was digested with _Bgl_II and Bam_HI, and ligated into plasmid p281 (Mueller et al., 1987). A bicistronic CAT (chloramphenicol acetyltransferase)–_lacZ (β-galactosidase) construct bearing the above-mentioned URE2 gene region was produced by subcloning the CAT ORF as a _Bam_HI fragment into the unique _Bgl_II site of p281-IRES_URE2 plasmid, resulting in p281-CAT_IRES_URE2.
Miscellaneous
Standard yeast media, cultivation procedures and genetic methods were used (Sherman et al., 1986; Rose et al., 1990). Yeast were transformed using the lithium acetate method (Ito et al., 1983). Molecular cloning and sequencing were performed following general procedures described in Sambrook et al. (1989). SDS–PAGE was performed according to Schagger and von Jagow (1987). ‘Rainbow [14C]methylated colored proteins’ (mol. wts 2350–46 000 Da, Amersham) were used as molecular weight markers. In vitro transcription and translation were performed as described previously (Altmann et al., 1989; Altmann and Trachsel, 1997; Komar et al., 1997). Western blotting was done following standard procedures (Towbin et al., 1979). Extracts for western blotting were prepared by glass-bead cell disruption in electrophoresis sample buffer [50 mM Tris–HCl pH 6.8, 4% SDS, 2% mercaptoethanol (v/v), 12% glycerol (w/v), 0.01% Serva Blue G]. Western blots were decorated with polyclonal anti-Ure2p rabbit antibodies, diluted 1:5000 in TBS buffer (10 mM Tris–HCl pH 7.0, 150 mM NaCl) containing 0.5% bovine serum albumin (BSA) for 1.5 h, decorated with rabbit anti-IgG antibody (Sigma A-8275) coupled to horseradish peroxidase (1:2500 dilution in TBS, 0.5% BSA) and stained with ECLplus (Amersham) reagent. Northern blotting was done as described (Alwine et al., 1977). Yeast genomic DNA was isolated using the DNA-Pure™ Yeast Genomic Kit from CPG, Inc. following the manufacturer’s recommendations. Total RNA was prepared following the method of Chomczynski and Sacchi (1987). The PCR-amplified 3 kb lacZ fragment (5′-CGCCTTGCAGCACATCC-3′ as upstream and 5′-GGTAGCGACCGGCGC-3′ as downstream primer) and 1 kb URE2 fragment (5′-CGGCAACCAAGTGTCGAATC-3′ as upstream and 5′-CCTTGATGACCGCGGGTC-3′ as downstream primer) were used for random primed labeling. Ambion Random Primed Strip Able DNA probe synthesis and removal kit were used for random primed labeling. RNA electrophoresis was done using a glyoxal-based system (Ambion kit no. 1946). Ambion RNA Millennium Markers™ were used as molecular size standards. LacZ activity was measured following the protocol described in the Clontech Yeast Protocols Handbook using _O_-nitrophenyl-β-d-galactopyranoside as a substrate. Cell extracts were prepared by subsequent cycles of cell freezing in liquid nitrogen and thawing at 37°C. CAT activity was measured using the FAST CAT Green (deoxy) Chloramphenicol Acetyltransferase Assay Kit from Molecular Probes following the manufacturer’s recommendations.
Acknowledgments
Acknowledgements
We thank Drs F.Lacroute and C.Reiss for their support and discussions, Dr R.Wickner for the gift of HM6B yeast strain and helpful advice, Drs R.Melki and C.Thual for the gift of anti-Ure2p antibodies and Dr J.P.G.Ballesta for sharing his results prior to publication. This research was supported by grants from Swiss National Science Foundation (grant 31-45528.95 to H.T. and M.A.) and from the National Institute for General Medical Science, NIH (grant GM26796 to W.C.M.).
References
- Altmann M and Trachsel,H. (1997) Translation initiation factor-dependent extracts from yeast Saccharomyces cerevisiae. Methods, 11, 343–352. [DOI] [PubMed] [Google Scholar]
- Altmann M., Handschin,C. and Trachsel,H. (1987) mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Sonenberg,N. and Trachsel,H. (1989) Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol. Cell. Biol., 9, 4467–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Blum,S., Pelletier,J., Sonenberg,N., Wilson,T.M. and Trachsel,H. (1990) Translation initiation factor-dependent extracts from Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1050, 155–159. [DOI] [PubMed] [Google Scholar]
- Altmann M., Muller,P.P., Wittmer,B., Ruchti,F., Lanker,S. and Trachsel,H. (1993) A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J., 12, 3997–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J.C., Kemp,D.J. and Stark,G.R. (1977) Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl Acad. Sci. USA, 74, 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., De Long,S.K. and Sachs,A.B. (2000) Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell, 11, 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., Slaven,J.W., De Long,S.K., Ibrahimo,S. and Sachs,A.B. (2001) A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J., 20, 6464–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G.J. and Jackson,R.J. (2000) Translation initiation on picornovirus RNA. In Sonenberg,N., Hershey,J.W.B. and Matthews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 869–900.
- Belsham G.J. and Sonenberg,N. (1996) RNA–protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev., 60, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A.M. and Kean,K.M. (1997) Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology, 237, 129–136. [DOI] [PubMed] [Google Scholar]
- Cardenas M.E., Cutler,N.S., Lorenz,M.C., Di Como,C.J. and Heitman,J. (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev., 13, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.S., Kuhn,K.M. and Sarnow,P. (2000) Cellular internal ribosome entry site elements and the use of cDNA microarrays in their investigation. In Sonenberg,N., Hershey,J.W.B. and Matthews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 615–635.
- Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Coppolecchia R., Buser,P., Stotz,A. and Linder,P. (1993) A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J., 12, 4005–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S., Bruynooghe,Y., Denecker,G., Van Huffel,S., Tinton,S. and Beyaert,R. (2000) Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell, 5, 597–605. [DOI] [PubMed] [Google Scholar]
- Coschigano P.W. and Magasanik,B. (1991) The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione _S_-transferases. Mol. Cell. Biol., 11, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K.H., Rai,R., Distler,M., Daugherty,J.R., Coffman,J.A. and Cooper,T.G. (2000) Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J. Biol. Chem., 275, 17611–17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov Y.L., Skulachev,M.V., Ivanov,P.A., Zvereva,S.D., Tjulkina,L.G., Merits,A., Gleba,Y.Y., Hohn,T. and Atabekov,J.G. (2002) Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl Acad. Sci. USA, 99, 5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H.K., Gray,V.T. and Wickner,R.B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl Acad. Sci. USA, 96, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet,E. and Cullin,C. (2000) The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J., 19, 3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet,E., Ness,F., Baudin-Baillieu,A., Ripaud,L., Tuite,M. and Cullin,C. (2002) The [URE3] phenotype: evidence for a soluble prion in yeast. EMBO rep., 3, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Hardwick J.S., Kuruvilla,F.G. Tong,J.K. Shamji,A.F. and Schreiber,S.L. (1999) Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl Acad. Sci. USA, 96, 14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C.U. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick,W.C. (2000) Pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Matthews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 615–635.
- Hofman-Bang J. (1999) Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol., 12, 35–73. [DOI] [PubMed] [Google Scholar]
- Iizuka N., Najita,L., Franzusoff,A. and Sarnow,P. (1994) Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. (2000) Comparative view of initiation site selection mechanisms. In Sonenberg,N., Hershey,J.W.B. and Matthews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 127–183.
- Jackson R.J. and Kaminski,A. (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- Johannes G., Carter,M.S., Eisen,M.B., Brown,P.O. and Sarnow,P. (1999) Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA, 96, 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A., Lesnik,T., Cullin,C., Guillemet,E., Ehrlich,R. and Reiss,C. (1997) Differential resistance to proteinase K digestion of the yeast prion-like (Ure2p) protein synthesized in vitro in wheat germ extract and rabbit reticulocyte lysate cell-free translation systems. FEBS Lett., 415, 6–10. [DOI] [PubMed] [Google Scholar]
- Komar A.A., Guillemet,E., Reiss,C. and Cullin,C. (1998) Enhanced expression of the yeast Ure2 protein in Escherichia coli: the effect of synonymous codon substitutions at a selected place in the gene. Biol. Chem., 379, 1295–1300. [PubMed] [Google Scholar]
- Lacroute F. (1971) Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol., 106, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt O., Grunert,H.P. and Hahn,U. (1990) A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene, 96, 125–128. [DOI] [PubMed] [Google Scholar]
- Lauring A.S. and Overbaugh,J. (2000) Evidence that an IRES within the Notch2 coding region can direct expression of a nuclear form of the protein. Mol. Cell, 6, 939–945. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Cutler,N.S. and Heitman,J. (2000) Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 183–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D., Nagel,A.C. and Preiss,A. (2002) Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation. Proc. Natl Acad. Sci. USA, 99, 15480–15485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison D.C. and Wickner,R.B. (1995) Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science, 270, 93–95. [DOI] [PubMed] [Google Scholar]
- Masison D.C., Maddelein,M.L. and Wickner,R.B. (1997) The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl Acad. Sci. USA, 94, 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Edskes,H.K. and Wickner,R.B. (2000) [_URE3_] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol., 20, 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P.P., Harashima,S. and Hinnebusch,A.G. (1987) A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc. Natl Acad. Sci. USA, 84, 2863–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I., Abramovitz,L. and Choder,M. (1999) Starved Saccharomyces cerevisiae cells have the capacity to support internal initiation of translation. J. Biol. Chem., 274, 21741–21745. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky,I.N., Fletcher,S.P., Jackson,R.J. and Hellen,C.U. (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev., 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D. Winston,F. and Heiter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sachs A.B. (2000) Cell cycle-dependent translation initiation: IRES elements prevail. Cell, 101, 243–245. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Sagliocco F.A. Vega Laso,M.R., Zhu,D., Tuite,M.F., McCarthy,J.E. and Brown,A.J. (1993) The influence of 5′-secondary structures upon ribosome binding to mRNA during translation in yeast. J. Biol. Chem., 268, 26522–26530. [PubMed] [Google Scholar]
- Sambrook J. Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schagger H. and von Jagow,G. (1987) Tricine-sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Thompson S.R., Gulyas,K.D. and Sarnow,P. (2001) Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl Acad. Sci. USA, 98, 12972–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thual C., Komar,A.A., Bousset,L., Fernandez-Bellot,E., Cullin,C. and Melki,R. (1999) Structural characterization of Saccharomyces cerevisiae prion-like protein Ure2. J. Biol. Chem., 274, 13666–13674. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin,T. and Gordon,J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Velden A.W. and Thomas,A.A. (1999) The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell Biol., 31, 87–106. [DOI] [PubMed] [Google Scholar]
- Wesolowski M. and Wickner,R.B. (1984) Two new double-stranded RNA molecules showing non-mendelian inheritance and heat inducibility in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [_URE3_] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 528–529. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Taylor,K.L., Edskes,H.K., Maddelein,M.L., Moriyama,H. and Roberts,B.T. (2000) Prions of yeast as heritable amyloidoses. J. Struct. Biol., 130, 310–322. [DOI] [PubMed] [Google Scholar]
- Wilson J.E., Pestova,T.V., Hellen,C.U. and Sarnow,P. (2000) Initiation of protein synthesis from the A site of the ribosome. Cell, 102, 511–520. [DOI] [PubMed] [Google Scholar]
- Zhou W., Edelman,G.M. and Mauro,V.P. (2001) Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl Acad. Sci. USA, 98, 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]