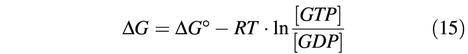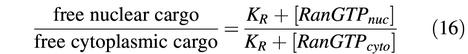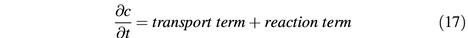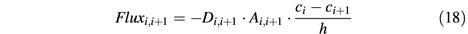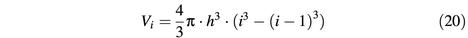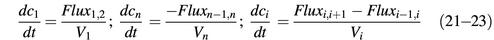Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation (original) (raw)
Abstract
Here, we analyse the RanGTPase system and its coupling to receptor-mediated nuclear transport. Our simulations predict nuclear RanGTP levels in HeLa cells to be very sensitive towards the cellular energy charge and to exceed the cytoplasmic concentration ≈1000-fold. The steepness of the RanGTP gradient appears limited by both the cytoplasmic RanGAP concentration and the imperfect retention of nuclear RanGTP by nuclear pore complexes (NPCs), but not by the nucleotide exchange activity of RCC1. Neither RanBP1 nor the NPC localization of RanGAP has a significant direct impact on the RanGTP gradient. NTF2-mediated import of Ran appears to be the bottleneck for maximal capacity of Ran-driven nuclear transport. We show that unidirectional nuclear transport can be faithfully simulated without the assumption of a vectorial NPC passage; transport receptors only need to reversibly cross NPCs and switch their affinity for cargo in response to the RanGTP gradient. A significant RanGTP gradient after nuclear envelope (NE) breakdown can apparently exist only in large cytoplasm. This indicates that RanGTP gradients can provide positional information for mitotic spindle and NE assembly in early embryonic cells, but hardly any in small somatic cells.
Keywords: importin/kinetics/nuclear transport/translocation
Introduction
Nuclear division (mitosis) as well as transport between nucleus and cytoplasm represent uniquely eukaryotic processes. The RanGTPase system plays a central role in both processes; it feeds metabolic energy into nuclear transport cycles, and guides mitotic spindle assembly at onset of mitosis as well as the reassembly of the nuclear envelope (NE) at exit from mitosis.
Nucleocytoplasmic transport (reviewed in Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Macara, 2001; Weis, 2002) is a major cellular activity. It not only includes the import of each and every nuclear protein from the cytoplasm, but also nuclear export of ribosomes, mRNAs and tRNAs to the cytoplasm. It proceeds through nuclear pore complexes (NPCs), which permit passage by passive diffusion or facilitated translocation. Passive diffusion is fast for small molecules, but becomes increasingly restricted as the mass of the transported species approaches a limit of 20–40 kDa. However, larger objects can also cross NPCs rapidly, provided appropriate nuclear transport receptors are recruited.
The members of the importin β family represent the largest class of nuclear transport receptors. They mediate facilitated NPC passage, circulate between nucleus and cytoplasm, and occur in two forms; import mediators (importins) and exportins. Their loading with cargo is controlled by a concentration gradient of RanGTP across the NE, which is sensed through RanGTP-binding domains of the transport receptors.
This gradient is generated by the asymmetric cellular distribution of regulators of the RanGTPase: RanGAP triggers hydrolysis of Ran-bound GTP (Bischoff et al., 1994), but as it is excluded from nuclei, it depletes RanGTP from the cytoplasm only. The Ran-specific nucleotide exchange factor RCC1 (Bischoff and Ponstingl, 1991a) is exclusively nuclear, chromatin bound and generates RanGTP inside the nucleus. The expected result is a steep RanGTP gradient across the NE, with high nuclear levels and low concentrations in the cytoplasm.
Importins initially recruit cargo at low RanGTP concentrations in the cytoplasm and release cargo at high RanGTP levels in the nucleus. Importin–RanGTP complexes return afterwards to the cytoplasm, where the Ran-bound GTP is finally hydrolysed and Ran dissociates from the receptor. The importin can then bind and import another cargo molecule, while NTF2 recycles RanGDP back to nucleus. The cargo binding to exportins is controlled in an opposite manner to importins; they recruit cargo at high RanGTP levels in the nucleus and release cargo at low RanGTP concentrations in the cytoplasm.
RanBP1 is a further component of the RanGTPase system with exlusively cytoplasmic localization. It binds RanGTP and facilitates the RanGAP-dependent conversion of RanGTP to RanGDP (Bischoff et al., 1995b). A related aspect of RanBP1 function appears, however, more important: RanBP1 catalyses the (cytoplasmic) disassembly of RanGTP–transport receptor complexes, which are kinetically so stable that RanGAP alone fails to trigger GTP hydrolysis (Bischoff and Görlich, 1997; Floer et al., 1997; Lounsbury and Macara, 1997). RanBP2 (Yokoyama et al., 1995) is a major constituent of the cytoplasmic filaments of NPCs and appears to fulfil similar functions as RanBP1. It contains four RanBP1 homology domains and forms a tight complex with sumoylated RanGAP (Matunis et al., 1996; Mahajan et al., 1997), thus, it is well adapted to disassemble RanGTP–transport receptor complexes that exit the nucleus.
Importin- and exportin-mediated transport cycles can accumulate cargoes against gradients of chemical activity, which is an energy-consuming task. The RanGTPase system hydrolyses one GTP molecule per transport cycle, and multiple lines of evidence suggest that this comprises the sole input of metabolic energy (Weis et al., 1996; Kose et al., 1997; Englmeier et al., 1999; Ribbeck et al., 1999). The Ran system switches transport receptors between high- and low-affinity forms for cargo binding and thereby confers directionality to the otherwise fully reversible, energy-independent and thus non-vectorial NPC passage.
Nevertheless, other models have been suggested that use the argument of asymmetry of NPCs along the nucleo–cytoplasmic axis, and describe the actual NPC passage as a vectorial process (for example, see Ben-Efraim and Gerace, 2001). To be consistent with the second law of thermodynamics, such a scenario would require an additional input of energy.
The mitotic Ran functions are probably best understood for the early embryonic cell cycles in the Xenopus system. Ran controls the assembly of the mitotic spindle by modulating the activity of certain microtubule-associated proteins, such as NUMA or TPX2 (see Kalab et al., 1999; Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001). TPX2, for example, is blocked by the Impα/ β heterodimer, and RanGTP relieves this block by dissociating Impα from Impβ. The generation of RanGTP by chromatin-bound RCC1 should therefore activate TPX2 and promote mitotic spindle formation near chromatin. Ran plays a second mitotic role in telophase, and guides the assembly of the NE such that chromatin, but no other cellular structure, becomes enclosed (Hetzer et al., 2000; Zhang and Clarke, 2000). Also, in this case, it is believed that a RanGTP gradient originating from chromatin provides positional information.
Our mechanistic insights of nuclear transport and Ran-controlled mitotic events clearly depend on a quantitative understanding of the RanGTPase system and its interplay with downstream regulatory targets. Important steps in that direction have been: enzyme kinetic characterizations of the key players (Klebe et al., 1995b; Villa Braslavsky et al., 2000); a kinetic analysis of NPC passage (Ribbeck and Görlich, 2001); a systems analysis on Ran import, which also gave a first estimate on the magnitude of the Ran gradient (Smith et al., 2002); and the development of a fluorescence-based sensor for Ran-controlled Imβ binding, which can indirectly probe the spatial distribution of RanGTP in extracts or even intact cells (Kalab et al., 2002).
We present here a comprehensive analysis by computer simulation of the RanGTPase system and its coupling to receptor-mediated cargo transport. We calculated the magnitude of mitotic and interphase RanGTP gradients and identified components and parameters that are limiting for the performance of the system. Our simulation predicts a RanGTP concentration in nuclei of interphase HeLa cells that exceeds the cytoplasmic levels ≈1000-fold. Computed nuclear RanGTP concentrations agree well with experimental measurements, where we used the endpoint of nuclear cargo accumulation as a ‘sensor’ for free RanGTP levels in the nucleus. We also studied RanGTPase-driven cargo import along the Impβ pathway, and found that the kinetics of the import process can be reproduced by a computer model based on fully reversible NPC passages and Ran-controlled cargo binding to Impβ. Importantly, the simulation predicts that NPC-associated or intranuclear activities catalyse the formation of RanGTP–Impβ complexes. A further major implication of the simulation is that a significant mitotic RanGTP gradient can apparently only be maintained in a sufficiently large cytoplasm, which is typical of early embryonic cell cycles. It thus represents an example whereby a simple physical parameter such as cell size allows embryonic cells to behave qualitatively differently from somatic cells.
Results and discussion
A computer model of the RanGTPase system
Local RanGTP concentrations are determined by local RanGTP production and consumption, as well as by Ran transport by passive diffusion and facilitated translocation. Knowing the rates of all relevant reactions would allow computation of local concentrations. To approach the problem, we first considered a simplified case and asked which nucleocytoplasmic RanGTP gradient would result if only the basic constituents of the RanGTPase system, namely Ran, RCC1, RanGAP, NTF2 and RanBP1 are present.
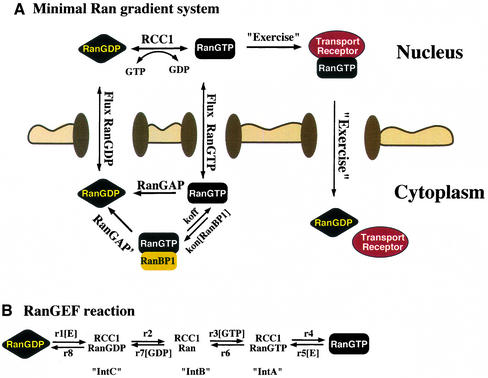
The relevant reactions are summarized in Figure 1. This scheme is the graphical presentation of an ordinary differential equation system (Figure 2) that describes nuclear and cytoplasmic consumption/generation of RanGTP and RanGDP. The differential equation system is too complex to be solved analytically; however, a numeric solution is feasible. In the following, we will first discuss each partial reaction and derive rate constants, and then combine these reactions into a mathematical model. Since most of the rate constants have been determined at 25°C, we will first discuss the simulation at this temperature, before we finally extrapolate to the physiological temperature of 37°C.
Fig. 1. Schematic overview of the RanGTPase system. (A) Ran switches between a GDP- and a GTP-bound form and circulates between nucleus and cytoplasm. Nucleotide exchange is catalysed by nuclear RCC1. Fluxes of RanGDP and RanGTP between nucleus and cytoplasm depend on their concentration differences and on the permeability of NPCs for RanGTP and RanGDP. The cytoplasmic conversion of RanGTP to RanGDP occurs either directly by RanGAP or after trapping of RanGTP by RanBP1. Receptor-mediated cargo transport occurs at the expense of the RanGTP gradient and results in a RanGTP transfer to the cytoplasm, where the bound nucleotide is hydrolysed to GDP. (B) Details of the nucleotide exchange reaction, which occurs in both directions and proceeds through three intermediates. E stands for the free RCC1 enzyme. r1-r8 are rate constants for the partial reactions (Klebe et al., 1995b).
Fig. 2. Description of the RanGTPase system by an ordinary differential equation system. Prefixes cyto and nuc stand for cytoplasmic and nuclear, respectively; fluxes of Ran between nuclear and cytoplasm are normalized to the nuclear volume. Other species and partial reactions are defined in Figure 1.
RanGAP converts RanGTP to RanGDP. Using the published kinetic constants for human RanGAP (_K_m = 0.43 µM, _k_cat = 2.1 s–1; Klebe et al., 1995a) gave rather high steady-state levels of cytoplasmic RanGTP (≈22 nM) in the simulations (see below). We therefore re-evaluated the issue, produced human RanGAP in a recombinant form, measured its kinetic constants with an optimized RanGAP assay, and found at 25°C a _k_cat of ≈10.6 s–1 and a _K_m of 0.7 µM (Table I; Supplementary data, available at The EMBO Journal Online). This number, and the cytoplasmic RanGAP concentration of 0.7 µM (see Supplementary data), were then used in subsequent computations.
Table I. Kinetic constants for the RanGAP-dependent GTPase activation on either free RanGTP or the pre-formed RanBP1–RanGTP complex.
| Substrate | Temperature (°C) | _K_cat (s–1) | _K_m (µM) |
|---|---|---|---|
| RanGTP | 25 | 10.6 | 0.7 |
| RanGTP–RanBP1 complex | 25 | 10.8 | 0.1 |
| RanGTP–RanBP1 complex | 37 | 21.2 | 0.1 |
In the second mode of the GAP reaction, RanGTP first forms a complex with RanBP1, and thereby becomes a better RanGAP substrate (Bischoff et al., 1995a). A closer inspection of the reaction (Table I; Supplementary data) revealed that RanBP1 lowers the _K_m of the reaction 7-fold, but has no effect on _k_cat.
RCC1-mediated nucleotide exchange on Ran (Bischoff and Ponstingl, 1991a) is complicated by the fact that it can occur in both directions, thus uses RanGDP as well as RanGTP as substrates, and generates both RanGDP and RanGTP. It occurs through three intermediates and involves eight partial reactions, with product formation depending on the free GDP and GTP concentrations (Klebe et al., 1995b; Figure 1B). This series of eight individual reactions cannot be easily combined into a single, simple equation and so, at this point, we make no further assumptions and describe the process as this set of differential equations.
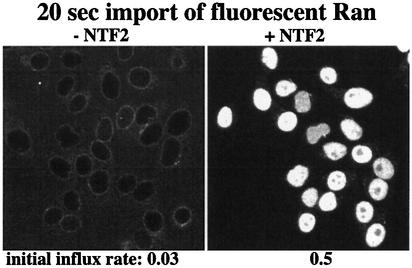
We next need to consider that NPCs permit exchange of RanGDP and RanGTP between nucleus and cytoplasm. The respective fluxes are proportional to the nucleo– cytoplasmic concentration differences and the permeabilities of NPCs (Figure 2; equations 11 and 12). In the case of RanGDP, the permeability of NPCs will not be a fixed number, but depend on the concentration of its import mediator NTF2 (Ribbeck et al., 1998; Smith et al., 1998). We therefore, determined the permeability factors (_k_Perm) for a series of RanGDP:NTF2 ratios (Figure 3; data not shown), using a kinetic nuclear transport assay described previously (Ribbeck and Görlich, 2001). The data indicate that RanGDP enters nuclei ≈16 times faster when present in a complex with NTF2. For the NPC passage of RanGTP (which cannot bind NTF2), we assume that the nucleotide-bound state has no dramatic effect on _k_Perm, and that free RanGTP traverses NPCs at the same rate as RanGDP in the absence of NTF2.
Fig. 3. Panels compare nuclear influx of RanGDP in the absence or presence of NTF2 (one homodimer per two RanGDP molecules). Quantitation of initial rates at 25°C revealed a permeability factor of NPCs towards free RanGDP of 0.03 s–1, i.e. for a nucleocytoplasmic concentration difference of 1 µM, the nuclear RanGDP concentration changes at 0.03 µM/s. The permeability factor for the NTF2–RanGDP complex is 0.48 s–1.
Having obtained the kinetic constants required to describe the minimal Ran system, we next computed the dynamic behaviour of the whole system, whereby the dimensions of nucleus and cytoplasm, as well as the concentrations of RanGAP, RanBP1, RCC1 and NTF2 were set to the numbers found for HeLa cells (see Supplementary tables). The free Ran concentration was set to 3 µM, i.e. we assume that half of the 6 µM Ran in HeLa cells (Bischoff and Ponstingl, 1991b; our unpublished data) is bound to transport receptors and thus is not available for either nucleotide exchange or GTP hydrolysis. This set of parameters will be, unless noted otherwise, the standard conditions for all further computations.
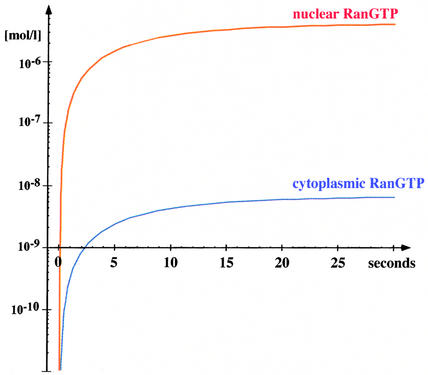
In the simulation, all Ran was initially cytoplasmic and in the GDP-bound form. From that, a nuclear RanGTP pool is rapidly generated (relaxation time ≈7 s). It reaches a steady-state concentration of 4.3 µM, while the cytoplasmic RanGTP concentration remains low at ≈8 nM (Figure 4; Table II). Thus, a steep RanGTP gradient across the NE builds up, reaching a nuclear:cytoplasmic RanGTP ratio of >550.
Fig. 4. Simulated time course for generation of the RanGTP gradient. At time zero of the simulation, all Ran was cytoplasmic and GDP bound. With these starting conditions, a nucleocytoplasmic RanGTP gradient builds up (half time ≈7 s). The figure shows a log linear plot for nuclear and cytoplasmic RanGTP concentrations. The following steady-state concentrations result: 4.3 µM nuclear RanGTP, 7.7 nM cytoplasmic RanGTP, 0.4 µM nuclear RanGDP and 1.5 µM cytoplasmic RanGDP. Parameters used for the simulation were: 1.2 pl nuclear volume, 1.8 pl cytoplasmic volume, 0.7 µM cytoplasmic RanGAP, 2 µM cytoplasmic RanBP1, 0.7 µM nuclear RCC1, 0.3 µM cellular NTF2 concentration (homodimers) and 3 µM cellular Ran concentration. For kinetic constants, see Supplementary tables and main text.
Table II. Effect of the single parameters on the steady state of the RanGTP gradient.
| Difference to standard conditions | Nuclear RanGTP (steady state; µM) | Cytoplasmic RanGTP (steady state; nM) | Dynamic capacity of the system for Ran-driven transport (µM/s) |
|---|---|---|---|
| Standard | 4.3 | 7.7 | 0.60 |
| Omission of RanBP1 | 4.3 | 8.1 | 0.60 |
| 200% RCC1 | 4.0 | 7.1 | 0.60 |
| 50% RCC1 | 4.3 | 7.7 | 0.60 |
| 10% RCC1 | 3.6 | 6.4 | 0.48 |
| 1% RCC1 | 1.4 | 2.5 | 0.08 |
| GTP:GDP = 500:0 | 4.8 | 8.6 | 0.60 |
| GTP:GDP = 500:50 | 0.8 | 1.5 | 0.58 |
| GTP:GDP = 500:500 | 0.12 | 0.21 | 0.34 |
| Saturating NTF2 | 5.1 | 9.2 | 2.2 |
| Omission of NTF2 | 2.5 | 4.5 | 0.16 |
| 200% RanGAP | 4.3 | 3.9 | 0.60 |
| 50% RanGAP | 4.3 | 14 | 0.60 |
| Extrapolated from 25 to 37°C | 4.9 | 4.3 | 1.2 |
These numbers refer to a temperature of 25°C, but they can be extrapolated to the physiological temperature of 37°C as follows. A shift to 37°C roughly doubles the rate of facilitated translocation, while the rate of passive NPC passage remains unaffected (data not shown). Likewise, the activity of enzymes from mesophilic organisms doubles per 10°C temperature increase, and this also appears to apply to RanGAP (Table I). A simulation with these modified numbers indicates that a shift from 25°C to the physiological temperature of 37°C would steepen the RanGTP gradient to a nuclear:cytoplasmic ratio of >1000:1 (Table II).
To address the question as to which components are limiting for the performance of the RanGTPase system, we altered the individual parameters in the simulations and compared the effects with the just mentioned standard conditions (at 25°C). The effects were assessed not only by the criteria of nuclear and cytoplasmic steady-state levels of RanGTP; we also considered that transport at the expense of the Ran gradient depletes the nuclear RanGTP pool. Therefore, it is an important criterion which level of work load or ‘exercise’ the system can sustain. This number reflects the maximum rate of nuclear RanGTP production (in µM/sec), and we refer to it here as the dynamic capacity for Ran-driven transport.
The simulation showed that GAP-independent (intrinsic) GTP hydrolysis or RCC1-independent (intrinsic) nucleotide exchange are fully negligible for the RanGTPase system (data not shown). The gradient is dominated by RanGAP and RCC1; omission of either component abolishes the gradient.
RanBP1 has only a negligible effect on the gradient of free RanGTP
The omission of RanBP1 has no effect on the nuclear RanGTP levels and only a minimal effect on the cytoplasmic RanGTP concentration (5% increase, see Table II). The smallness of the effect might seem a contradiction of the fact that RanBP1 stimulates the GAP reaction at subsaturating RanGTP concentrations 7-fold (Table I). This stimulation, however, applies to a pre-formed RanGTP–RanBP1 complex, and the apparent discrepancy is resolved by the fact that formation of the RanBP1– RanGTP complex (Villa Braslavsky et al., 2000) is ≈20 times slower than the direct turnover of RanGTP by RanGAP (calculated for the physiological RanBP1 and RanGAP concentrations). This finding supports the assumption that the primary role of RanBP1 is the disassembly of transport receptor–RanGTP complexes and not just the co-stimulation of the GTPase in free RanGTP.
RCC1 is essential, but not limiting for the RanGTP gradient
Surprisingly, and in contrast to the assumption of others (Smith et al., 2002), the simulation clearly shows that the amount of nuclear RCC1 is not limiting for the magnitude of the gradient (Table II). An increase above the physiological concentration even reduces available nuclear RanGTP, because more Ran stays complexed with RCC1. A reduction of RCC1 to 50% of the physiological level has no effect. To become limiting for the dynamic capacity of the system, it has to be reduced by 90%. A significant decrease in the steady-state levels of nuclear RanGTP in the absence of exercise even requires a further reduction.
We can see two explanations as to why interphase cells are equipped with such excess of RCC1. First, high amounts of RCC1 keep the transporter system operational at low GTP:GDP ratios also (see next section). Secondly, during mitosis, high RanGTP levels must be maintained around chromatin in the absence of an NE, which stretches the limits of RanGTP production more than an interphase situation.
The cellular phosphorylation potential has a large impact on the availability of nuclear RanGTP
With 500 µM GTP, but no GDP present in the system, the simulations predict 4.8 µM nuclear RanGTP at steady state. This number was reduced to 4.3 µM when a phosphorylation potential as measured by NMR in a resting muscle was assumed (NTP:NDP ≈300:1; Stryer, 1995). At a GTP:GDP ratio of 50:50, it even dropped to 0.1 µM. This is still >100-fold above the dissociation constants for typical importin–RanGTP interactions, however, for import and export reactions where the on-rate of RanGTP binding to transport receptors is rate limiting, this could mean an up to 40-fold drop in the rates. The explanation for this sensitivity is that Ran binds GDP 22-fold better than GTP. Free GDP not only competes RanGTP formation strongly, it also indirectly favours the decay of RanGTP.
A drop in GTP:GDP ratio strongly affects the nuclear steady-state level of RanGTP, but only has a moderate effect (40% reduction at a 50:50 ratio) on the dynamic capacity of RanGTP production (Table II). This is not a contradiction to the just drawn conclusions, instead, it simply emphasizes the fact that the nucleotide exchange capacity is not limiting.
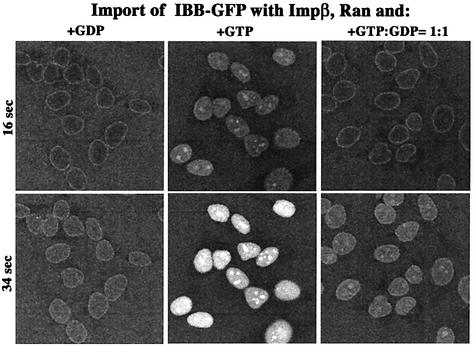
The simulation thus predicted a great impact of the phosphorylation potential on import and export reactions, where RanGTP binding to the transport receptors is rate limiting. To test this, we performed the following experiment: import of an IBB–GFP fusion was performed with Impβ and a limiting concentration of Ran (Figure 5). With GDP alone, nucleocytoplasmic equilibration and an even distribution of the substrate within the nuclei was observed. In the presence of a GTP-regenerating system, rapid influx occurred and some cargo appeared in the nucleoli, indicating its displacement from the receptor. With the combination of 500 µM GTP and 500 µM GDP, almost no import stimulation over the GDP control was observed. The only clear indication for the generation of nuclear RanGTP was the (weak) concentration of substrate at the nucleoli. Ran-driven influx over equilibration reached a mere 10% of the energy-mix control, which indicates that nuclear RanGTP became limiting. It should be noted that the lower GTP:GDP ratio not only slowed down cargo influx at early time points, but also caused a 4-fold lower endpoint of nuclear cargo accumulation (data not shown), which is approximately the effect predicted from simulating the complete RanGTPase/Impβ system (see below).
Fig. 5. Effect of GTP:GDP ratio on the rate of receptor-mediated nuclear accumulation. IBB–GFP (2 µM) fusion was complexed stoichiometrically with Impβ and imported into nuclei of permeabilized cells in the presence of a GTP-regenerating system, which keeps the GTP:GDP ratio >200, or with GDP, or with 500 µM GTP plus 500 µM GDP. The figure depicts, at indicated time points, the nuclear:cytoplasmic distribution of the import cargo, as visualized by confocal laser scanning microscopy.
At first, it appears very odd that Ran binds GDP so much better than GTP. We suggest that it relates to the fact that the released free energy per hydrolysed GTP molecule increases with the GTP:GDP ratio. Assuming constant water and inorganic phosphate concentrations, this change in free energy equals:
At a GTP:GDP ratio of 1:1, ≈30 kJ/mol hydrolysed GTP are released, whereas at the physiological ratio of ≈300:1, ≈45 kJ/mol are released. To exploit this higher energy charge, the GTPase has to bind GTP weaker than GDP; the difference in binding energies (7.8 kJ/mol in the case of Ran) then adds to the free energy released by the actual hydrolysis of the phosphodiester bond. Ran, therefore, yields more energy from GTP hydrolysis than a GTPase with no preference for GDP. The drawback is that the Ran system requires a very high GTP:GDP ratio for effective operation and so Ran appears optimized for most efficient operation at high energy charge. However, this drawback is well compensated for by the large excess of RCC1 in the system.
Effect of NTF2 on the Ran gradient
NTF2 is the import receptor for Ran and increases the flux of RanGDP through NPCs (Ribbeck et al., 1998; Smith et al., 1998). When the system is not under a work load, the presence or absence of NTF2 has a modest effect on the RanGTP gradient. Increase of NTF2 to saturating concentrations makes 15% more nuclear RanGTP available, while its complete omission reduces this number by no more than 30%. A far greater effect is evident when looking at another criterion, namely the dynamic capacity of the system, i.e. the maximum rate by which the system can produce RanGTP and support RanGTPase-driven cargo transport. The dynamic capacity is indeed limited by the supply of nuclei with RanGDP. This is because the RCC1 capacity exceeds, under standard conditions, the RanGDP transport capacity 12-fold. The RCC1 capacity becomes limiting only at very low GTP:GDP ratios (≈50:50 or less).
Effect of RanGAP concentration and RanGAP distribution on the RanGTP gradient
Assuming a physiological RanGAP concentration and a homogenous distribution in the cytoplasm, a steady state with 8 nM cytoplasmic RanGTP is reached in the simulation (Table II). This is 15 times above the _K_D of Impβ for RanGTP binding (Bischoff and Görlich, 1997) and thus still inhibitory for cargo binding to this receptor (∼30% inhibition for an IBB-fusion protein at 1 µM free concentration). This suggests that RanGAP is limiting for the performance of the system. Indeed, doubling the cytosolic RanGAP concentration halved the cytoplasmic RanGTP levels (Table II). Interestingly, the same effect occurs when the cytoplasmic volume is doubled at a constant RanGAP concentration (data not shown). In fact, as long as homogeneous mixing in the cytoplasm occurs, it is the total amount of cytoplasmic RanGAP (in ratio to nuclei) that determines the cytoplasmic RanGTP levels. This is an important consideration for the in vitro transport reactions using permeabilized cells, where the ‘cytoplasmic’ volume exceeds that of the nuclei by three orders of magnitude, and where rather low cytoplasmic RanGAP concentrations are optimal for nuclear import.
In higher eukaryotes, a fraction of RanGAP is immobilized to the cytoplasmic fibrils of the NPCs (Matunis et al., 1996; Mahajan et al., 1997), i.e. in proximity to where the outflow of RanGTP occurs. This poses the questions as to whether this strategic RanGAP location would steepen the Ran gradient. The attachment of eight copies of RanGAP per pore through the 50 nm long cytoplasmic filaments will result in a local concentration of ≈35 µM within a 50 nm range around the pore. Diffusion at this scale, however, is so fast that a molecule of Ran’s size will stay on average for only 60 µsec within this zone, and this time suffices only to convert ≈3% of the out-flowing RanGTP to RanGDP (for details, see Supplementary data). This makes it highly unlikely that the primary ‘purpose’ of the NPC localization of RanGAP is to steepen the gradient of free RanGTP. Rather, it is consistent with the recent observation that RanBP2 is dispensable for simple Ran-driven import processes (Walther et al., 2002) and with the fact that, for example, yeasts lack a RanBP2 equivalent altogether.
What about the disassembly of transport receptor– RanGTP complexes? The concentration of RanGAP close to NPCs would facilitate the process probably only if additional trapping mechanisms would keep the complexes in the zone for longer. To be efficient, such trapping devices should have a very large target area. The extended FG-repeat domains at cytoplasmically exposed Nups (such as CAN or RanBP2) could well fulfil this function by binding the RanGTP complexes via the transport receptors.
Extrapolation to cellular conditions
Our mathematical model of the Ran system is based on rate constants obtained in vitro, which poses the question in how far an extrapolation to cellular conditions is justified. Of course, we have to assume that the model includes all relevant reactions. In addition, we need to consider that living cells are not just filled with the dilute buffers used for determining kinetic constants. Instead, they represent a very crowded environment with content of ≈200–300 mg/ml macromolecules. This crowding causes two opposing effects on enzymatic reactions (Ellis, 2001): first, it increases viscosity 3- to 10-fold and thereby decreases the rate of bimolecular collisions by the same factor. Secondly, there is a volume exclusion effect, which favours association between macromolecules and thus accelerates bimolecular reactions. The magnitude of the latter effect depends on the size of the reacting molecules and in the case of the RanGAP reaction (the most critical one for the interphase Ran gradient), the two effects are likely to just compensate each other (see Ellis, 2001). However, this issue clearly deserves further, thorough investigation.
Coupling of Impβ-mediated cargo transport to the RanGTP gradient
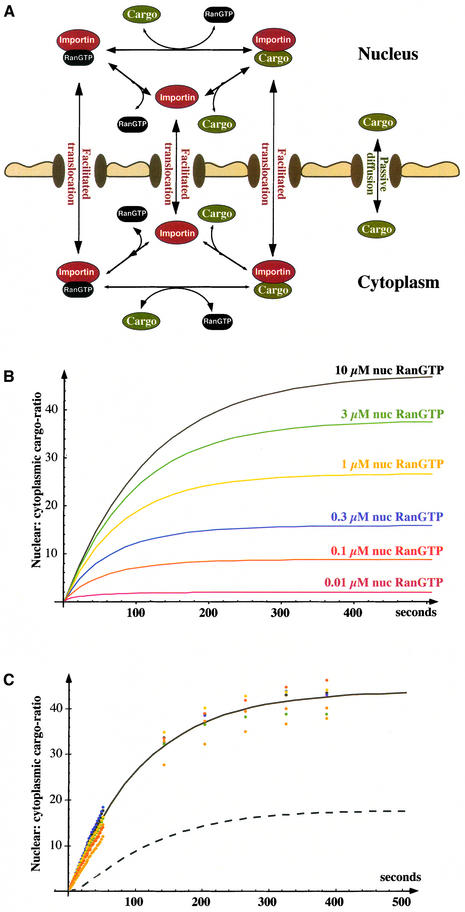
The RanGTPase system drives a large number of nuclear transport pathways by switching transport receptors between a low- and a high-affinity state for cargo binding. In the following, we will attempt to simulate such a coupled system and derive the relationship between the magnitude of the primary RanGTP gradient and extent of possible cargo accumulation. For that, we will consider (as a typical example) Impβ-mediated import of a cargo that binds the receptor directly (i.e. no adapter involved), contains a single import signal and is displaced by RanGTP from Impβ.
Impβ circulates between nucleus and cytoplasm, and exhibits its greatest possible import efficiency when its cargo load is 100% upon nuclear entry and zero during exit. In reality, however, this maximum efficiency cannot be reached. The steeper the cargo concentration gradient against which the transporter system operates, the lower the import efficiency will be; an increase in nuclear concentration of free cargo will raise the cargo load of those Impβ molecules that exit the nucleus and thereby enhance receptor-mediated re-export of the import cargo. Eventually, a steady state will be reached, where the same fraction of entering and exiting Impβ molecules are loaded with cargo. With the assumption that no passive diffusion of cargo occurs, this endpoint only depends on the nuclear and cytoplasmic RanGTP concentrations and on the dissociation constant (_K_R) for RanGTP–Impβ interaction (for details see Supplementary data):
It may be a surprise that the cargo ratio here is not identical to the ratios of RanGTP concentrations. However, this simply indicates that receptor-dependent RanGTP efflux is not rigidly coupled to cargo import. A rigid coupling would require that only the RanGTP– importin and cargo–importin complexes, but not the free importin, were able to pass the NPCs.
The non-rigid coupling lowers the performance of the system, not only in terms of maximum degree of cargo accumulation, but also in the efficiency by which the energy of the (primary) Ran gradient is used. Nevertheless, for our estimate of nuclear and cytoplasmic RanGTP concentrations (at 37°C) in a living HeLa cell (Table II), Impβ could achieve a very respectable ≈1000-fold nuclear accumulation of the cargo.
In reality, however, such a high level of accumulation cannot be reached, because ‘real’ import substrates traverse NPCs not only as a transport–receptor complex, but also by passive diffusion (Figure 6A; Table IV). Such passive diffusion is normally very slow compared with the receptor-mediated passage, but it becomes sub stantial once a significant nucleocytoplasmic gradient of free-cargo concentration has built up, and in fact it is one of the limiting parameters for cargo accumulation (Table IV).
Fig. 6. Dependence of substrate accumulation from the Ran gradient. (A) Scheme of the Impβ import system, representing the differential equation system used for import simulation. (B) Simulated import of an IBB–MBP fusion protein (1 µM) mediated by Impβ (0.5 µM). Parameters for the simulation were equivalent to experimental conditions of import into nuclei of permeabilized cells [i.e. the volume outside of the nuclei is very large compared with the nuclear volume; see also (C)]. Cytoplasmic RanGTP was fixed to 1 nM and nuclear RanGTP varied from 10 nM to 10 µM. For binding constants, see Supplementary table; see also Table IV. (C) Import of an IBB–MBP fusion into nuclei of permeabilized cells was performed at 37°C with the following components: 1 µM fluorescent IBB–MBP, 0.5 µM Impβ, 3 µM Ran, 0.4 µM NTF2 (dimers), 0.05 µM RanGAP, 0.1 µM RanBP1 and a GTP-regenerating system. The nuclear:cytoplasmic distribution of the import substrate is plotted for early time points in 2 s intervals and for later time points in 90 s intervals. Different colours indicate behaviour of individual nuclei in a typical experiment. Curves show simulated time course of nuclear IBB–MBP accumulation, whereby the mathematical model linked the generation of the RanGTP gradient (Figure 1) with transport at the expense of the gradient (Figure 6A). The simulation predicts 6 µM nuclear RanGTP at steady state. It reproduces time course and endpoint of nuclear cargo accumulation, provided the on-rate for RanGTP binding to Impβ is assumed not to be limiting (continuous curve). For that, RanGTP binding to Impβ must occur at least one to two orders of magnitude faster in the context of intact nuclei than observed with pure components. The dashed curve shows simulation using the on-rate of the RanGTP–Impβ interaction as measured with pure components (8·104 M–1s–1; Villa Braslavsky et al., 2000). The rates, by which the binding equilibria between cargo, RanGTP and Impβ are adjusted within the cytoplasm, are also not known. This uncertainty is, however, irrelevant for the cytoplasmic side of this simulation because the reaction volume outside nuclei is >1000-fold larger than the nuclear volume.
Table IV. Numbers to the simulation of nuclear cargo accumulation as depicted in Figure 6B.
| Cytoplasmic RanGTP (nM) | Nuclear RanGTP (µM) | Endpoint of cargo accumulation (nuclear:cytoplasmic ratio) | Endpoint w/o passive NPC passage of cargo |
|---|---|---|---|
| 100 | 10 | 11 | 88 |
| 10 | 10 | 34 | 560 |
| 1 | 10 | 48 | 1400 |
| 1 | 6 | 44 | 1020 |
| 1 | 3 | 38 | 640 |
| 1 | 1 | 27 | 280 |
| 1 | 0.3 | 16 | 94 |
| 1 | 0.1 | 8.9 | 32 |
| 1 | 0.01 | 2.0 | 2.9 |
| 1 | 0.001 | 1.0 | 1.0 |
Passive cargo diffusion complicates the mathematical model of receptor-mediated cargo accumulation considerably, and the system can only be analysed by computer simulation. We also had to determine the permeability of NPCs for our free model substrate (a fusion between the IBB domain and the maltose binding protein MBP) and for the cargo–Impβ complex. For the Impβ–IBB–MBP complex, we found a _k_Perm of 0.95 s–1 and for free IBB–MBP a _k_Perm of 0.008 s–1 (data not shown; see below), i.e. the transport receptor facilitates the NPC passage ≈120-fold.
We then used the numbers to simulate the transport process under conditions that apply to in vitro import into nuclei of permeabilized cells, simply because the simulation could then be directly compared with experimental data (for details see legend to Figure 6). We fixed the cytoplasmic RanGTP concentration to 1 nM and simulated IBB–MBP import for nuclear RanGTP concentrations between 1 nM and 10 µM. If nuclear and cytoplasmic RanGTP concentration are identical, only equilibration of cargo occurs. At 10 µM nuclear RanGTP, a 47-fold nuclear cargo accumulation can be reached. The simulation shows a strict dependence of the endpoint on nuclear RanGTP concentration (Figure 6B; Table IV) and thus offers an excellent opportunity to use an experimental endpoint determination as a probe for the free RanGTP concentration in the nucleus. Experimentally, we observed a 37- to 46-fold nuclear accumulation, whereby the range reflects variability between individual nuclei (Figure 6C). This points to nuclear RanGTP levels between 3 and 10 µM. A simulation predicts steady-state levels of 6 µM nuclear RanGTP under these conditions, and we can thus conclude that simulation and experimental data are in excellent agreement.
Finally, we combined the generation of the RanGTP gradient with transport at the expense of the RanGTP gradient into a single mathematical model and simulated the complete system. Figure 6C documents the good agreement between the simulation and experimental import data. This excellent agreement only required one additional assumption that concerns the kinetics of RanGTP binding to Impβ. When measured with pure components, the on-rate for the reaction is rather slow (<105 M–1s–1; see Bischoff and Görlich, 1997; Villa Braslavsky et al., 2000). This rate is far too slow to explain the rate of IBB import (Figure 6C, dashed curve). The RanGTP binding to Impβ must therefore occur at least one to two orders of magnitude faster in the context of intact nuclei than with purified components. Thus, intranuclear or NPC-associated components might catalyse the formation of RanGTP–Impβ complexes. The Ran-binding zinc fingers in Nup153 (Nakielny et al., 1999) or other nucleoporins in the nuclear basket would be excellent candidates for such a function (for example, see Gilchrist et al., 2002)
We can thus conclude that the simulation faithfully reproduces the transport process. The mathematical model clearly shows that the transporter system can efficiently work without any vectorial cargo/receptor transfer through NPCs. It is fully sufficient to let the receptor circulate and regulate its cargo loading by the RanGTPase system and no further assumptions (such as vectorial NPC-passage via affinity gradients) need to be made.
Effect of NE permeability on the RanGTP gradient
Nuclear pore complexes are not absolute barriers, so RanGTP can flow out of nuclei, which will diminish the gradient. The simulation clearly shows (Table III) that the magnitude of the gradient is limited by the restriction of RanGTP outflow and is inversely proportional to the permeability of the NE towards RanGTP: if the permeability is reduced to 50%, a slight increase in the nuclear RanGTP concentration and a nearly 50% reduction in the cytoplasmic levels occurs; conversely, if the permeability for RanGTP is doubled, then the magnitude of the gradient is halved. This is in line with the conclusion drawn above about the receptor-mediated cargo accumulation in nuclei, which is also extremely sensitive to the permeability of NPCs for the cargo.
Table III. Influence of the permeability of NPCs on the RanGTP gradient.
| Difference to standard conditions | Nuclear RanGTP (steady state; µM) | Cytoplasmic RanGTP (steady state; nM) |
|---|---|---|
| Standard | 4.3 | 7.7 |
| 50% NPC permeability towards RanGTP | 4.9 | 4.4 |
| 200% NPC permeability towards RanGTP | 3.4 | 12.3 |
| 400% NPC permeability for RanGTP | 2.5 | 17.8 |
| No nuclear envelope | 0.83 | 480 |
| No nuclear envelope and RanGAP evenly distributed between nucleus and cytoplasm | 0.72 | 520 |
To build up a substantial gradient across the NE, the gradient-forming agent must be sufficiently retained. As NPCs restrict passage of inert objects according to size, it is plausible why the primary gradient that drives nuclear transport can only be formed by a sizeable protein (Ran) and not by a small molecule, such as a nucleotide.
Constrains for mitotic RanGTP gradients
In interphase, the RanGTP gradient orients the direction of nuclear transport and feeds metabolic energy into transport cycles, while the NE protects the nuclear RanGTP pool from being dissipated into the cytoplasm. At the onset of mitosis, the permeability of NPCs suddenly increases, before the entire NE eventually disintegrates (Terasaki et al., 2001). However, also during mitosis, it is believed that a Ran gradient originates from chromatin and orients not only the mitotic spindle, but also guides NE reassembly during anaphase/telophase. We therefore also looked into the mitotic situation.
We could so far assume that the diffusion of RanGDP and RanGTP within nucleus and cytoplasm is fast compared with transport between the two compartments. This assumption is not fulfilled without an NE, and so modelling a mitotic situation requires a more sophisticated mathematical model that also considers diffusion of RanGDP and RanGTP within the two compartments (for details, see Supplementary data). Applying this model to our standard interphase situation gave the same average nuclear and cytoplasmic RanGTP concentrations as the simpler model outlined in Figure 2. In addition, it can also now predict concentration profiles of RanGTP within nucleus and cytoplasm. While RanGTP is very homogeneously distributed within the nucleus (4.3 µM), there is a shallow gradient in the cytoplasm with ≈10 nM in immediate vicinity of NPCs and ≈6 nM at a distance of 2 µm (Figure 7; Smith et al., 2002).
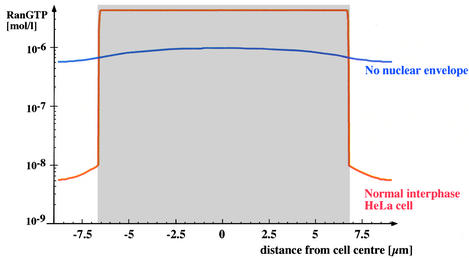
Fig. 7. Effect of the NE on the RanGTP gradient. Figure shows profile of simulated RanGTP levels within a HeLa cell. The nuclear area is represented in grey. The red curve shows interphase cell with intact NE and steep nucleocytoplasmic RanGTP gradient. The blue curve shows a mitotic cell lacking a NE; here, the RanGTP gradient has fully collapsed. For the mitotic situation, it was assumed that RCC1 was still chromatin bound, while RanGAP was evenly distributed between chromatin and cytoplasm.
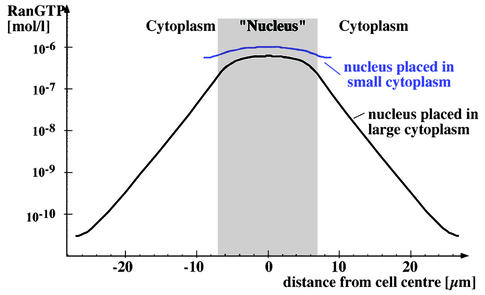
Without a nuclear membrane, the simulation predicts that RanGTP levels decrease to ≈0.8 µM in the centre of the nucleus, while 0.5 µM are reached at the cell periphery (Figure 7). Thus, the RanGTP gradient collapses and RCC1 dominates the nucleotide-bound state of Ran. This result appears in striking contrast to the proposed role of RanGTP gradients in mitosis (Hetzer et al., 2002) and the recent indirect detection of a mitotic RanGTP gradient using an IBB-based sensor (Kalab et al., 2002). We therefore asked wether conditions could be found that permit a mitotic gradient. It turned out that the size of the cytoplasm is the key parameter. Figure 8 compares simulated mitotic Ran gradients in a HeLa cell with a situation where the mitotic nucleus is placed into a cell three times larger. Within this larger cell, a respectable gradient is generated with a RanGTP concentration of ≈0.5 µM in the centre of the nucleus and ≈10 pM at the cell periphery. This illustrates the extreme scale-dependence of diffusion, and indicates that 10 µm of cytoplasm are as good a diffusion barrier as the NE.
Fig. 8. Effect of cytoplasm size on a mitotic RanGTP gradient. The RanGTP profile was simulated for a mitotic HeLa cell (blue curve) and for a cell with a larger cytoplasm (black curve). Note, only the larger cytoplasm allows the formation of a significant mitotic RanGTP gradient.
This also suggests that mitotic RanGTP gradients may be particularly relevant in the early embryonic cell cycles of organism in which initially only a very small number of nuclei multiply in a huge cytoplasm. Indeed, it appears far more difficult for a mitotic spindle to capture mitotic chromatin in a large cytoplasm than in a small one, and this consideration gives a plausible explanation as to why the guidance of the Ran gradient must be employed early in development.
The mitotic RanGTP gradient can be considered as a long-range cellular navigation system that reports the position of chromatin with an accuracy of a few micrometres. RanGTP transforms the microtubule dynamics in the mitotic cytoplasm such that mitotic asters can eventually be established in the vicinity of chromatin (Karsenti and Vernos, 2001). This transformation would affect the entire volume of a (small) HeLa cell; during early embryonic cell cycles, however, it would be restricted to a radius of only a few micrometres around mitotic chromatin.
The spatial resolution of the mitotic RanGTP gradient appears sufficient to polarize mitotic asters towards chromatin, however, it cannot be precise enough for an accurate attachment of microtubules to kinetochores and chromatin, which requires a resolution of at least 100 nm. The final, high-resolution targeting could be accomplished by the microtubules themselves, e.g. according to the search and capture model (Holy and Leibler, 1994). Kinetically instable microtubules alternate here between growth and shrinking, but become protected from catastrophic shrinking if an attachment to chromosomes was successful. This random search ultimately selects for chromosome-attached microtubules. The RanGTP gradient would aid this process by stabilizing microtubules in the vicinity of chromatin and thus restricting the ‘search’ to this area.
Materials and methods
Kinetic measurements of Ran- and cargo-flux through NPCs
Nuclear transport assays with permeabilized HeLa cells and their quantitation was essentially performed as described previously (Ribbeck and Görlich, 2001); however, two improvements were implemented: ovalbumin and maltose binding protein (20 mg/ml each) were added as crowding reagents, which improves the stability of nuclei, and the energy mix was replaced by a GTP-regenerating system, containing ATP, GTP, creatine phosphate, creatine kinase and nucleoside diphosphate kinase.
Simulation of the RanGTPase system
Ordinary differential equation systems for two-compartment models (Figures 1, 2, 4 and 6) were solved with the NDSolve function of Mathematica 4.1.
Two-compartment models are valid when mixing within nucleus and cytoplasm is fast compared with exchange between nucleus and cytoplasm, i.e. when the NE is intact. In a mitotic situation or for predicting concentration profiles within cytoplasm and nucleus, however, not only reaction terms, but also diffusion must be considered. The two terms are additive and so the local concentration c(x,y,z,t) of a diffusing species (RanGDP or RanGTP) changes over time according to:
The reaction terms are analogous to the RanGAP and RCC1 reactions of the two-compartment model (Figure 2, equations 1–14). The cellular distributions of RCC1 and RanGAP were kept constant in the simulation.
To compute diffusion terms, the cell was divided into n spatial segments. The flux between adjacent segments (i and i + 1) follows Ficks’ first law:
whereby _D_i,i+1 is the local diffusion constant, _A_i,i+1 the contact area between the segments, _c_i and _c_i + 1 the concentrations in segments i and i + 1, and h the distance between the segments.
To limit the complexity of the mathematical model, we assumed rotational symmetry of the cell and chose spatial segments in the form of concentric spherical layers with a thickness of h. The contact area between two adjacent layers (i and i + 1) equals, therefore, the surface area of a sphere with radius = h_·_i:
_A_i,i+1 = 4π·_h_2·_i_2(19)
The volume of such layer equals the difference between the volumes of two spheres with radii of h_·_i and _h_·(i – 1):
The change in concentration over time within a given layer depends on the flux to and from neighbouring layers and on the volume of the layer. For the innermost (first) layer, for the outermost (_n_th) layer and for all other layers (i = 2…_n_–1), the following transport terms apply:
Ran (Stokes radius 2.2 nm) was assumed to diffuse in a medium of approximately cellular viscosity (η = 0.05 _g_cm–1s–1), corresponding to a diffusion constant of 20 µm2/s. To introduce a barrier (i.e. a NE), the diffusion constant was adjusted locally to the barrier-specific value. The differential equation system comprising equations 17–23 and the reaction terms were solved with the NDSolve function of Mathematica. h was set to 81 nm.
The correctness of the simulations was verified by formal criteria (correct mass balance and grid convergence) by modelling special cases that could also be solved analytically and by comparing the simulations with actual nuclear import reactions (Ran-import, IBB–GFP and IBB–MBP import).
Please note that the programmes used for the simulations are available on request (dg@zmbh.uni-heidelberg.de).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank U.Jäkle and P.Rübmann for excellent technical help, I.Vetter and A.Wittinghofer for support of the RanGAP project, M.Kirkilionis and T.Fischer for advice and stimulating discussions, and the DFG for financial support.
References
- Ben-Efraim I. and Gerace,L. (2001) Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J. Cell Biol., 152, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R. and Ponstingl,H. (1991a) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature, 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. and Ponstingl,H. (1991b) Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc. Natl Acad. Sci. USA, 88, 10830–10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R. and Görlich,D. (1997) RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett., 419, 249–254. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe,C., Kretschmer,J., Wittinghofer,A. and Ponstingl,H. (1994) RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc. Natl Acad. Sci. USA, 91, 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Krebber,H., Kempf,T., Hermes,I. and Ponstingl,H. (1995a) Human RanGTPase activating protein RanGAP1 is a homolog of yeast RNA1p involved in messenger RNA processing and transport. Proc. Natl Acad. Sci. USA., 92, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Krebber,H., Smirnova,E., Dong,W.H. and Ponstingl,H. (1995b) Coactivation of RanGTPase and inhibition of GTP dissociation by Ran GTP binding protein RanBP1. EMBO J., 14, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J. (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol., 11, 114–119. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo,J.C. and Mattaj,I.W. (1999) Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol., 9, 30–41. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel,G. and Rexach,M. (1997) Disassembly of RanGTP–karyopherin β complex, an intermediate in nuclear protein import. J. Biol. Chem., 272, 19538–19546. [DOI] [PubMed] [Google Scholar]
- Gilchrist D., Mykytka,B. and Rexach,M. (2002) Accelerating the rate of disassembly of karyopherin cargo complexes. J. Biol. Chem., 277, 18161–18172. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Gruss O.J. et al. (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell, 104, 83–93. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Bilbao-Cortes,D., Walther,T.C., Gruss,O.J. and Mattaj,I.W. (2000) GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell, 5, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Gruss,O.J. and Mattaj,I.W. (2002) The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol., 4, E177–E184. [DOI] [PubMed] [Google Scholar]
- Holy E.T. and Leibler,S. (1994). Dynamic instability of microtubules as an efficient way to search in space. Proc. Natl Acad. Sci. USA, 91, 5682–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu,R.T. and Dasso,M. (1999) The ran GTPase regulates mitotic spindle assembly. Curr. Biol., 9, 481–484. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis,K. and Heald,R. (2002) Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science, 295, 2452–2456. [DOI] [PubMed] [Google Scholar]
- Karsenti E. and Vernos,I. (2001) The mitotic spindle: a self-made machine. Science, 294, 543–547. [DOI] [PubMed] [Google Scholar]
- Klebe C., Bischoff,F.R., Ponstingl,H. and Wittinghofer,A. (1995a) Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry, 34, 639–647. [DOI] [PubMed] [Google Scholar]
- Klebe C., Prinz,H., Wittinghofer,A. and Goody,R.S. (1995b) The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry, 34, 12543–12552. [DOI] [PubMed] [Google Scholar]
- Kose S., Imamoto,N., Tachibana,T., Shimamoto,T. and Yoneda,Y. (1997) Ran-unassisted nuclear migration of a 97 kD component of nuclear pore- targeting complex. J. Cell Biol., 139, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury K.M. and Macara,I.G. (1997) Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J. Biol. Chem., 272, 551–555. [DOI] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin,C., Guan,T., Gerace,L. and Melchior,F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell, 88, 97–107. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas,E. and Blobel,G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol., 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Maresca,T.J., Salmon,W.C., Waterman-Storer,C.M., Heald,R. and Weis,K. (2001) Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell, 104, 95–106. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K. and Görlich,D. (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J., 20, 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky,G., Kent,H.M., Stewart,M. and Görlich,D. (1998) NTF2 mediates nuclear import of Ran. EMBO J., 17, 6587–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Kutay,U., Paraskeva,E. and Görlich,D. (1999) The translocation of transportin–cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol., 9, 47–50. [DOI] [PubMed] [Google Scholar]
- Smith A., Brownawell,A. and Macara,I.G. (1998) Nuclear import of ran is mediated by the transport factor NTF2. Curr. Biol., 8, 1403–1406. [DOI] [PubMed] [Google Scholar]
- Smith A.E., Slepchenko,B.M., Schaff,J.C., Loew,L.M. and Macara,I.G. (2002) Systems analysis of Ran transport. Science, 295, 488–491. [DOI] [PubMed] [Google Scholar]
- Stryer L. (1995) Biochemistry. 4th Edn. Freemann and Company, New York, NY, pp. 448.
- Terasaki M., Campagnola,P., Rolls,M.M., Stein,P.A., Ellenberg,J., Hinkle,B. and Slepchenko,B. (2001) A new model for nuclear envelope breakdown. Mol. Biol. Cell., 12, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa Braslavsky C.I., Nowak,C., Görlich,D., Wittinghofer,A. and Kuhlmann,J. (2000) Different structural and kinetic requirements for the interaction of ran with the ran-binding domains from RanBP2 and importin-β. Biochemistry, 39, 11629–11639. [DOI] [PubMed] [Google Scholar]
- Walther T.C., Pickersgill,H.S., Cordes,V.C., Goldberg,M.W., Allen,T.D., Mattaj,I.W. and Fornerod,M. (2002) The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol., 158, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Wilde,A., Moore,M.S., Adam,S.A., Merdes,A. and Zheng,Y. (2001) Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science, 291, 653–656. [DOI] [PubMed] [Google Scholar]
- Weis K. (2002) Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol., 14, 328–335. [DOI] [PubMed] [Google Scholar]
- Weis K., Dingwall,C. and Lamond,A.I. (1996) Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J., 15, 7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N. et al. (1995) A giant nucleopore protein that binds Ran/TC4. Nature, 376, 184–188. [DOI] [PubMed] [Google Scholar]
- Zhang C. and Clarke,P.R. (2000) Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science, 288, 1429–1432. [DOI] [PubMed] [Google Scholar]