Cytokinesis in Bacteria (original) (raw)
Abstract
Work on two diverse rod-shaped bacteria, Escherichia coli and Bacillus subtilis, has defined a set of about 10 conserved proteins that are important for cell division in a wide range of eubacteria. These proteins are directed to the division site by the combination of two negative regulatory systems. Nucleoid occlusion is a poorly understood mechanism whereby the nucleoid prevents division in the cylindrical part of the cell, until chromosome segregation has occurred near midcell. The Min proteins prevent division in the nucleoid-free spaces near the cell poles in a manner that is beginning to be understood in cytological and biochemical terms. The hierarchy whereby the essential division proteins assemble at the midcell division site has been worked out for both E. coli and B. subtilis. They can be divided into essentially three classes depending on their position in the hierarchy and, to a certain extent, their subcellular localization. FtsZ is a cytosolic tubulin-like protein that polymerizes into an oligomeric structure that forms the initial ring at midcell. FtsA is another cytosolic protein that is related to actin, but its precise function is unclear. The cytoplasmic proteins are linked to the membrane by putative membrane anchor proteins, such as ZipA of E. coli and possibly EzrA of B. subtilis, which have a single membrane span but a cytoplasmic C-terminal domain. The remaining proteins are either integral membrane proteins or transmembrane proteins with their major domains outside the cell. The functions of most of these proteins are unclear with the exception of at least one penicillin-binding protein, which catalyzes a key step in cell wall synthesis in the division septum.
INTRODUCTION
Cell division in bacteria is effected by a complex macromolecular machinery containing seven or more conserved proteins, which assemble with a defined dependence hierarchy at the site of division. At the end of the process, two new cell poles have been formed, and so all of the cell envelope layers (including the membrane, wall, outer membrane, and S layer, if present) must be remodeled during division. Although the central players in division appear to be highly conserved, the diverse nature of bacterial cell envelopes clearly influences the precise pathway taken during division, so that some of the division proteins have specialized roles required only in certain groups of bacteria.
Many genes and proteins required for division have been defined by classical and molecular genetics. In the last few years, all of these proteins have been shown to be targeted to the division site and the dependence pathways for this assembly have been worked out in Escherichia coli and to a lesser extent in Bacillus subtilis (summarized in Tables 1 and 2) (25, 54). Disappointingly, despite these advances, clear biochemical functions have so far been elucidated for only two proteins. The first, FtsZ, is cytosolic and is present in virtually all eubacteria as well as in many eukaryotic organelles. It is a tubulin homologue and can polymerize, like tubulin, to form a ring-like structure (the Z ring) at the division site. The Z ring plays a key role in constriction of the cell membrane, as well as in coordination of the whole process of division. The second key biochemical activity required (for organisms with peptidoglycan [PG] cell walls), is a specialized penicillin-binding protein (PBP) required for synthesis of the wall material in the new cell poles.
TABLE 1.
Localization of cell division proteins to the division site in E. coli and their interdependences for localization
| Protein | References to work on localization to septum | References to work on dependence of localization ona: | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FtsZ | FtsA | ZipA | FtsK | FtsQ | FtsL | YgbQ | FtsW | FtsI | FtsN | |
| FtsZ | 15, 100, 113 | 1, 4, 70 | 70, 104 | 25, 167, 176 | 1 | 62 | 18, 94, 122 | 1, 140, 166, 170 | 2, 25 | |
| FtsA | 4, 113 | 4, 70, 114 | 70, 104 | 25, 167, 176 | 4 | 62 | 122 | 4, 166, 170 | 25 | |
| ZipA | 69, 104 | 70, 104, 114 | 70, 104 | 25, 167 | 26 | 62 | 122 | 104, 170 | 25 | |
| FtsK | 25, 167, 176 | 167, 176 | 167, 176 | 68 | 167 | 166, 167, 176 | ||||
| FtsQ | 21, 26 | 26 | 26 | 68 | 25 | 22, 26 | 22 | 122 | 26 | 25 |
| FtsL | 62 | 62 | 62 | 68 | 25 | 62 | 22 | 122 | 62 | 25 |
| YgbQ | 22 | 22 | 22 | 22 | 22 | |||||
| FtsW | 122, 166 | 122 | 122 | 22 | 122 | |||||
| FtsI | 166, 170, 171 | 166, 170 | 166, 170 | 68 | 25 | 170 | 170 | 22 | 122 | 25 |
| FtsN | 2 | 2 | 2 | 68 | 2 | 2, 166 |
TABLE 2.
Localization of cell division proteins to the division site in B. subtilis and their interdependences for localization
| Protein | References to work on localization to septum | References to work on dependence of localization ona: | |||||
|---|---|---|---|---|---|---|---|
| FtsZ | FtsA | DivIB | FtsL | DivIC | FtsW | FtsI | |
| FtsZ | 100, 168 | 93 | 36 | UPb | 35 | ||
| FtsA | 56 | 56 | 56 | UP | |||
| DivIB | 77 | 93 | 36 | 93 | 35 | ||
| FtsL | 34, 154 | 34 | UP | 34 | 34 | UP | 34 |
| DivIC | 92 | 93 | 93 | 36 | 35 | ||
| FtsW | UP | UP | UP | ||||
| FtsI | 35 | 35 | UP | 35 | 35 | 35 | UP |
As a result of several decades of work, the following general steps in bacterial cell division can now be recognized: (i) selection of the division site, usually at midcell between the recently replicated and segregated nucleoids; (ii) assembly of cytoplasmic apparatus, almost invariably involving FtsZ and, in most organisms, also FtsA; these proteins can bind and hydrolyze nucleoside triphosphates, which may provide the energy to remodel the cell shape, possibly counteracting forces due to turgor and/or existing wall architecture; (iii) interaction with one or more proteins that provide a linkage to the cell membrane and potentially other cell envelope layers; (iv) assembly of proteins with major extracellular domains, particularly specialized PBPs, which direct synthesis of the wall material and other cell envelope layers that will constitute the new cell poles; and constriction and possibly an additional step of septum closure.
In the last few years, substantial progress has been made in several aspects of cell division, particularly the molecular basis for division site selection by the Min system, the pathway of assembly of the division protein machinery, the biochemistry of FtsZ polymerization, and, finally, the solution of the crystal structures of several division proteins. It is also becoming clear that although some aspects of the division machinery are highly conserved, other elements, particularly the later steps, have diverged significantly. The purpose of this review is to discuss the latest developments in the field of cell division and to describe the emerging distinctions in division processes of different organisms, particularly the paradigmatic gram-negative and gram-positive species, E. coli and B. subtilis, respectively. Various aspects of division have been discussed extensively in other recent, general reviews (19, 111, 112, 116, 146).
SELECTION OF THE DIVISION SITE
Formation of the Z ring is generally accepted to be a key event in the initiation of cell division. However, little is known about how the timing of ring assembly is regulated and about how the transition from ring assembly to constriction is controlled temporally. The simplest model at present is that the timing of ring assembly is a function of the mechanism of division site placement. There is an emerging consensus for two key factors involved in selection of the correct midcell site for cell division (reviewed in references 8, 74, and 117). One involves the chromosome itself, through an effect called nucleoid occlusion (129, 172). The second, termed the Min system, is responsible for specifically blocking the unwanted potential division sites at the cell poles (Fig. 1).
FIG. 1.
Division site selection in rod-shaped bacteria. (A) Roles of nucleoid occlusion and the Min system. In cells that are about to divide, the correct midcell division site is chosen by the combined action of two negative regulatory systems. The localization of the two nucleoids blocks division in their vicinity, as shown, leaving spaces available for division at the correct midcell site and at the cell poles. The Min system acts to prevent incorrect division at the cell poles. (B) Blocking division at the cell poles. The MinCD inhibitor associates with the cytoplasmic membrane at the cell poles and prevents FtsZ polymerization. The C-terminal domain of MinC (CC) ensures MinC dimerization and interacts with MinD (D), which in turn associates with the membrane. The N-terminal domain of MinC (CN) interacts with FtsZ (Z) and prevents polymerization of FtsZ or interaction of other cell division proteins with the FtsZ ring (see the text). The MinD-MinC stoichiometry, and the presumed action of MinC, preventing the formation of correct FtsZ interactions remains hypothetical. (C and D) Contrasting functioning of the Min system in E. coli (C) and B. subtilis (D). MinD (chevrons [ATP form] and rectangles [ADP form]) is postulated to polymerize in the presence of ATP. In E. coli (A), it forms dynamic filaments emanating from one cell pole. A ring of MinE proteins associates with the ends of the filaments and moves in a poleward manner, by interaction with the MinD filaments. MinE stimulates hydrolysis of ATP by MinD, and this results in the release of monomers, probably following a conformational change (rectangles), which enter the cytoplasm. After exchange of the ADP for ATP, MinD polymers can re-form at the opposite cell pole, possibly requiring a nucleation site at the pole. In B. subtilis (B), MinD forms static filaments restricted to the polar zones as a result of nucleation by DivIVA protein targeted to the cell poles (black triangles). In both cases, MinD is associated with the division inhibitor (MinC; not shown), which blocks Z ring formation. Reproduced reference 8 with permission from the publisher.
Nucleoid Occlusion
The still poorly defined mechanism of nucleoid occlusion was revealed by the observation that cell division was found to be largely inhibited in the vicinity of the nucleoid of cells in which DNA replication and/or segregation was perturbed (129). In normal (rod-shaped) cells that are about to divide, there are three sites where DNA is absent or reduced in concentration (Fig. 1A). One lies at midcell, between the replicated and segregated chromosomes; the others lie at the cell poles. In principle, the midcell site appears and disappears cyclically, in parallel with rounds of chromosome replication. If the model is correct, bacteria have developed a mechanism that senses the presence of a DNA-free zone at midcell. Importantly, because the DNA-free zone is generated only toward the end of the DNA replication cycle, it could provide not only a spatial signal but also a temporal signal for division. In its original form, this model was based on the notion that the presence of an actively replicating nucleoid inhibited cell wall synthesis in its immediate vicinity. More recently, it has generally been reinterpreted in terms of a mechanism that prevents assembly of the Z ring in the vicinity of the nucleoid. The model has been tested in both E. coli and B. subtilis. Although there are reports indicating that Z rings can form directly over the nucleoid, these rings appear to form at positions where the DNA concentration is low compared to wild-type situations (27, 66, 145, 160). This suggests that it is not the presence of DNA per se but the concentration of DNA that is important (74), which is supported by the observation that in both E. coli and B. subtilis, Z rings can assemble before chromosome segregation is complete (42, 173).
Unfortunately, the mechanistic basis for nucleoid occlusion is poorly understood, and further understanding is hampered by difficulties in defining the precise locations of the boundaries of the rather diffuse nucleoid, in identifying the moment of replication termination in individual cells, and in understanding the forces that drive segregation (i.e., separation) of sister replicated chromosomes.
The Min System
A second form of regulation of the position of the Z ring is exerted by the min system, so called because mutants give rise to minicells through inappropriate division at the cell poles. In E. coli, the min system consists of three proteins, MinC, MinD, and MinE, which prevent cell division at the poles (recently reviewed in references 74 and 117). MinC and MinD act together as a negative regulator of FtsZ ring assembly, MinCD (41). MinC is the actual inhibitor of Z ring formation and prevents the localization of FtsZ and ZipA when overproduced (16, 87, 103, 119, 137). Furthermore, E. coli MinC inhibits FtsZ polymer assembly in vitro (87).
MinC is a dimer consisting of two distinct domains connected by a flexible linker (29) (Fig. 1B). The N-terminal domain, which interacts with FtsZ (85), shows some homology to FtsA and SpoIIAA (29). It is tempting to propose that MinC and FtsA have a similar mechanism of interaction with FtsZ. The similarity to SpoIIAA is intriguing because this protein interacts with the SpoIIE protein, which in turn has a direct interaction with FtsZ (see below). The C-terminal domain of MinC mediates dimerization (29, 85, 161) as well as the interaction with MinD (85). The crystal structures of MinC (29) and MinD (30, 78) revealed no information about the MinC-MinD interaction, but mutagenesis data point to a role for residues surrounding the MinD nucleotide-binding site in MinD-MinC interaction (78). MinD is a membrane-associated ATPase (39), which sequesters MinC to the membrane (39) (Fig. 1B). The MinCD complex probably has some additional affinity for assembling septal structures (90). The crystal structures of MinD from two different archaeal species revealed a monomeric protein with a fold typical of a large family of nucleotide-binding proteins (30, 78). MinE imparts topological specificity to the MinCD inhibitor by preventing it from working at midcell (40). MinE is a dimeric protein with two separate domains: an N-terminal anti-MinCD domain (139, 178) and a C-terminal topological specificity-dimerization domain (96, 139, 177, 178). The structure of the latter domain clearly showed how MinE dimerizes, and mutagenesis identified two key residues in MinE localization, but the interaction with MinCD remains to be resolved (97).
In the past few years, fluorescence imaging has provided important new insights into the organization of the Min system. Remarkably, the MinCD inhibitor oscillates from pole to pole, with a periodicity of about 20 s (84, 142, 143) (Fig. 1C). MinE forms a band that also oscillates, but with a more restricted pattern, moving from side to side about midcell. Each sideways movement seems to be involved in sweeping MinCD from the cell pole; this is followed by reassembly of a MinCD cap at the opposite pole (59, 71). MinC seems to be a passive player, simply moving with MinD (84, 142). Experiments with spherical mutants of E. coli suggest that the Min system can help to identify the long axis of the cell, thereby promoting division parallel to this long axis (28). This could explain the presence of a Min system in various gram-negative cocci (116), which on the face of it do not have an overt polarity.
Recently, Lutkenhaus and coworkers have made considerable progress in understanding the molecular basis for oscillation (Fig. 1C). First they showed that MinD ATPase activity is strongly stimulated by MinE in the presence of phospholipid vesicles. Moreover, MinE mutant proteins affected in their stimulation of MinD ATPase showed equivalent effects on the periodicity of MinD oscillation in vivo (86). These authors went on to demonstrate that MinD binds to the phospholipid vesicles, distorting the vesicles into tubular structures coated with MinD. The MinD was probably polymerized in a helical array with a pitch of 59 Å, similar to the length of the long axis of the MinD monomer determined by X-ray crystallography (83). These results strongly suggested that the MinCD inhibitor forms a polymerized structure at the membrane with MinD in the ATP-bound form, which is displaced from the membrane by interaction with MinE. They also suggested that MinE works by stimulating ATP hydrolysis and release of MinD from the membrane. After nucleotide exchange in the cytosol, MinCD binds to the membrane in the MinE-free zone in the other cell half (86). This model is supported by computer simulations that can faithfully reproduce the oscillation patterns of the system in silico (121).
The components of the Min system are distributed unequally among bacteria. Although MinD-like proteins are present across a broad range of species, MinC is less highly conserved and MinE is even more restricted in its distribution (116). The Min system of B. subtilis has been extensively characterized. Clear homologues of MinC and MinD are present, but there is no MinE (102). The function of MinE in topological control of MinCD activity is provided by a quite different protein, DivIVA (24, 48) (Fig. 1D). DivIVA is stably associated with the cell poles, and it recruits MinCD to the cell poles, probably by a direct interaction with MinD (49, 120). The zone occupied by MinD appears to spread out from the pole more than that of DivIVA, again suggesting a cooperative self-interaction of MinD, which is perhaps nucleated at the pole by DivIVA. DivIVA-MinCD remains associated with the newly formed poles after division, thereby preventing future division at those sites (48, 119, 120). Thus, even though B. subtilis uses a homologous MinCD system to block polar division, topological control over MinCD is exerted in a strikingly different manner from that in E. coli. Various other bacterial species, particularly gram-positive cocci, lack the Min system altogether (117), and certain clostridia appear to possess both MinE and DivIVA (157), raising interesting questions about how division site selection occurs in these organisms.
ASSEMBLY OF THE FtsZ RING
FtsZ is arguably the most critical component of the division machinery for several reasons. First, it is the most widely conserved, being present in virtually all eubacteria and most archaea and in the organelles of many eukaryotes (115). Second, it is at the top of the hierarchy of assembly of division proteins — in general, all other division proteins require FtsZ for targeting to the division site (Tables 1 and 2). Third, certain ftsZ mutations that alter the form of the Z ring in the cell, for example producing helical structures, have a similar effect on the gross morphology of cell division (5).
Polymerization
The Z ring almost certainly consists of FtsZ polymers because these can readily be formed in vitro, in the presence of GTP, under a range of conditions. FtsZ polymers strongly resemble tubulin protofilaments (20, 51, 107, 108, 126). The simultaneous elucidation of the structures of FtsZ from the thermophilic archaeon Methanococcus jannaschii (105) and the αβ-tubulin heterodimer from bovine brain (133) provided conclusive evidence that FtsZ and tubulin are homologues (164). The great overall structural similarity between FtsZ and tubulin is described in detail elsewhere (131). Recent biochemical data show that not only is there great structural homology between FtsZ and tubulin but also that their polymerization mechanisms are similar. GTP hydrolysis is activated by a cation-coordinating loop (T7) that inserts into the nucleotide-binding site of an adjacent monomer (128, 150, 151). Nucleotide hydrolysis is immediate on polymerization, and the hydrolyzed phosphate is not released immediately (149). This points to an important role for phosphate release in FtsZ polymer dynamics. In tubulin, the γ-phosphate of GTP is sensed by loop T3 (132); in FtsZ, this loop is displaced depending on whether GDP or GTP is bound to the protein (45). Polymer stability is likely to be conferred by a GTP cap at one end of the FtsZ polymer, as may be inferred from the observation that FtsZ can be stabilized by the addition of nonhydrolyzable GTP-γ-S that does not exchange for most of the nucleotide that is bound to the polymer (152) and from the observation that FtsZ polymers rapidly disassemble after the addition of GDP (128, 152). Although the longitudinal interactions in the FtsZ polymer and microtubules appear very similar, the lateral interactions found in microtubules are not evident, based on the FtsZ crystal structure (106, 132). Nevertheless, this question remains to be fully resolved because FtsZ appears to be capable of forming various types of polymers, with different types of lateral arrangement, particularly in the presence of divalent cations (see, e.g., references 107 and 108).
In vivo, Z rings can both form and disassemble rapidly, on a timescale of 1 to 3 min for assembly and 1 min for disassembly (3, 159). Recently, Stricker et al. (158) used fluorescence recovery after photobleaching to show that the Z ring is highly dynamic throughout its existence. They also showed that about 30% of cellular FtsZ is present in the Z ring and that the FtsZ in the Z ring readily exchanges with FtsZ in the cytosol. The speed of exchange of protein subunits in the ring and the rate of GTP hydrolysis showed a good correlation (158). Combined with the in vitro data on FtsZ polymerization (described above), a model can be envisaged in which the Z ring is made up of a meshwork of filaments that undergoes continuous rapid exchange with a pool of short FtsZ polymers formed in the cytosol (158).
Mechanism of Constriction
As mentioned above, the orientation of the Z ring seems to determine the location of the division plane. The results with both the ftsZ26 mutant, which makes helical Z rings and septa, and of rodA mutants, which grow as a sphere instead of a rod, show that constriction does not require closure of a complete FtsZ ring (5). “Purse string” type models for constriction can therefore probably be excluded. Several models for Z ring constriction have been described (reviewed in reference 19). (All of these models require the presence of an anchor to connect FtsZ to the membrane [discussed below].) In the first model, short filaments of FtsZ slide relative to one another, aided by an as yet unidentified motor protein, to reduce the circumference of the ring. In the second model, FtsZ filaments lose subunits through depolymerization at an anchor site (which must somehow remain attached to the filament end), again resulting in ring constriction. The observation that overproduction of FtsZ inhibits constriction of the FtsZ ring (169), which can be explained by the inhibition of depolymerization through the concentration increase, lends support to this model. In a third model, constriction is effected by bending of FtsZ filaments, resulting from the conformational change that occurs on hydrolysis of GTP to GDP (108) or release of Pi. More sophisticated in vitro reconstitution systems are probably needed to test these models. Moreover, although FtsZ is a ubiquitous, key player in division, models for constriction will ultimately have to incorporate the functions of the various other division proteins that have been identified.
Asymmetric Division during Sporulation
A key event during sporulation in B. subtilis is the formation of an asymmetric division septum at one of the cell poles. This asymmetric septum formation is preceded by a change of localization of the Z ring from midcell to the cell poles (100) followed by localization of other division proteins and the sporulation-specific FtsZ-associated protein SpoIIE (recently reviewed in reference 54 (see below). The switch in FtsZ localization was previously thought to be effected by a mechanism which blocked formation of the Z ring at midcell and released the inhibition of polar septation (9, 100). However, recently Ben-Yehuda and Losick (14) showed that a medial Z ring can be converted into, or replaced by, a helical FtsZ filament that grows outward toward the cell poles, where it is then converted into bipolar Z rings. Both FtsA and EzrA were shown to colocalize with the FtsZ helical structures. The switch from midcell to polar Z rings was shown to be driven by a combination of increased cellular concentration of FtsZ and the presence of sporulation protein SpoIIE (14).
Helical FtsZ structures have previously been observed in misshapen vegetative cells of B. subtilis (91) and in E. coli cells that overproduce FtsZ (113, 159) or that lack the membrane phospholipid phosphatidylethanolamine (123). This suggests that the ability to form such structures is a general property of FtsZ proteins and even raises the possibility that the general form of the Z ring at division sites may actually comprise a helix of very short pitch (14).
FtsA
FtsA is the only known cell division protein, other than FtsZ, that lacks a clear membrane-spanning sequence. The FtsA primary sequence places it in the actin/Hsp70/sugar kinase ATPase superfamily (17). The crystal structure of FtsA from Thermotoga maritima was recently solved (165) and confirmed the similarity to actin, except for the absence of the 1B domain of actin and the presence of a novel domain dubbed 1C. Generally, however, FtsA is typical of the actin superfamily (17), possessing two domains with a common core and an interdomain cleft harboring the nucleotide-binding site. The significance of the structural similarity to actin is not clear. Although FtsA does not appear to polymerize in vitro, B. subtilis FtsA has recently been shown to dimerize and to bind and hydrolyze ATP (56). There is evidence that E. coli FtsA can be phosphorylated (148), but phosphorylation has not been observed with B. subtilis FtsA (A. Feucht, personal communication), and the molecular consequences of phosphorylation of E. coli FtsA are not clear.
As shown in Table 1, FtsA assembles at the Z ring early. This is due to a direct interaction between the two proteins, as demonstrated by yeast two-hybrid interaction studies. The extreme C terminus of FtsZ seems to be the site of interaction, as for other interactions (see below) (Fig. 1B) (46, 114, 174). Two particular residues in the E. coli FtsZ C terminus, L372 and the highly conserved P375 (50), are critical for its interaction with FtsA. Interestingly, although there is a clear overall homology between FtsZ and tubulin, the C termini of both proteins are quite different (although both are highly acidic). The C-terminal tail of FtsZ is highly conserved in bacteria, with a central Pro-X-(Phe/hydrophobic residue) motif (50), which mediates the interaction with both FtsA and ZipA.
In E. coli, the cellular ratio of FtsA to FtsZ is 1:100, this ratio is important for correct division (31, 44). This has important implications for the nature of the FtsZ-FtsA interaction in vivo because there is not sufficient FtsA to form a complete ring and it presumably makes only widely interspersed contacts with FtsZ filaments. FtsA dimerization could allow it could cross-link adjacent FtsZ filaments. Interestingly, the level of FtsA appears to be much higher in B. subtilis; with an intracellular FtsA-to-FtsZ ratio of 1:5 (56). Given the distribution of both FtsZ and ZipA in ring-associated and non-ring-associated fractions and their dynamics (158; see below), the actual stoichiometry in the division complex may differ significantly from the ratios measured by quantitative immunoblotting. Ultimately, one specific role for FtsA is recruitment of other cell division proteins to the division site (Tables 1 and 2). In principle, it is possible that FtsA uses the energy of ATP hydrolysis to drive assembly. Alternatively, the ATP hydrolysis could in some way help to power cell constriction.
MEMBRANE ANCHOR
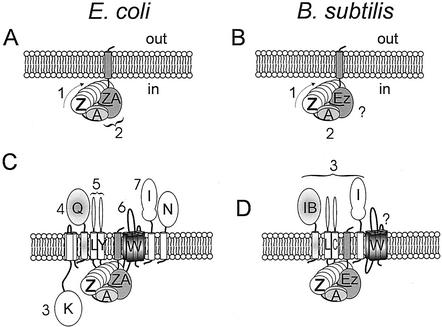
As mentioned above, all current models for FtsZ ring constriction require the ring to be tightly bound to the membrane. Moreover, membrane association is clearly required to coordinate membrane invagination, with synthesis of septal PG. Several candidates for the putative membrane anchor of the Z ring exist. One set of candidates would be the PBPs or proteins involved in recruiting the PBP complex to the septum (see below). However, these proteins are recruited to the division complex at a relatively late step in the hierarchy (see below). The best current candidate is a protein that assembles early in the division cycle, ZipA (Fig. 1B).
ZipA
ZipA was identified in E. coli by affinity blotting for FtsZ-interacting proteins (69). It is a membrane protein with an unusual membrane topology in which the N-terminal membrane anchor is followed by a large cytoplasmic portion that consists of two clear domains connected by a Pro-Gln rich flexible tether (134) (Fig. 2A). Although ZipA is essential for division in E. coli, its primary sequence is poorly conserved, and obvious ZipA homologues have been reported only for Haemophilus influenzae (144). ZipA localizes to the Z ring in an FtsZ-dependent, FtsA-independent manner, and overexpression of zipA abolishes cell division (69, 70, 104) (Table 1). Interestingly, FtsZ rings are formed in the presence of either ZipA or FtsA but not in the absence of both proteins (138), so that these proteins play a partially redundant role in Z ring assembly, although both are needed for septal constriction (Fig. 2A; Table 1). Increased zipA expression suppresses the thermosensitivity of the ftsZ84(Ts) mutant through FtsZ ring stabilization, and purified ZipA stabilizes both FtsZ and FtsZ84 polymers in vitro (144). Polymers formed in the presence of ZipA show enhanced bundling (72, 144), similar to FtsZ polymers that have been stabilized with divalent cations (127). The bundling of FtsZ protofilaments could be important in formation of the macroscopic Z ring. Additionally, or alternatively, bundling could enhance the stability of individual FtsZ protofilaments through a reduction of GTPase activity (P. A. J. de Boer, personal communication).
FIG. 2.
Organization of the cell division apparatus during various steps in the division process. A comparison of the E. coli system (A and C) with that of B. subtilis (B and D) is shown. Numbers beside proteins indicate the hierarchy of assembly; a question mark indicates that this is not yet clear. (A and B) Formation of the Z ring and anchoring of the Z ring to the membrane. FtsZ (Z) interacts with the cytoplasmic cell division protein FtsA (A) via the C-terminal tail of Z, which is flexible. FtsA (at least of B. subtilis [56]) is a dimer. In E. coli, the FtsZ ring is anchored to the membrane, at least partly, by ZipA (ZA), which also binds to the flexible C-terminal FtsZ tail. In most bacteria, functional equivalents of ZipA have not yet been found. In B. subtilis, EzrA protein has a similar topology to ZipA, although it is not essential for division. (C and D) Assembly of the remainder of the cell division machinery. In E. coli (C), the hierarchy for assembly of the remaining proteins is essentially linear, as numbered, except for FtsL (L) and YgbQ (Y). The other proteins are labeled according to their fts suffix. In B. subtilis (D), there is no apparent homologue of FtsN and the FtsK homologue (SpoIIIE) is not required for division. The DivIB (IB), DivIC (C), FtsL (L), and PBP-2B (I) proteins assemble in a concerted manner. The order of assembly of FtsW (W) is not known.
Various lines of evidence have shown that it is the C-terminal domain of ZipA that interacts with FtsZ and is responsible for the enhanced bundling observed in vitro (72, 104, 124, 125). As with FtsA, ZipA binds to the extreme C terminus of FtsZ (72, 73). The structure of the C-terminal domain of ZipA containing the C-terminal 17 residues of FtsZ revealed a split β-α-β motif with the FtsZ peptide bound in a shallow, hydrophobic, solvent-exposed cavity that runs the width of the ZipA C-terminal domain (124, 125). A major portion of the FtsZ fragment is in contact with ZipA residues, mainly through hydrophobic interactions but also via two hydrogen bonds (124). Interestingly, the highly conserved Pro residue of the C-terminal tail of FtsZ does not contact ZipA. Various other methods have been applied to characterize FtsZ residues implicated in the ZipA interaction, yielding unequivocal results. A surface plasmon resonance analysis of a peptide comprising the C-terminal 17 FtsZ residues and the C-terminal ZipA domain identified D373, I374, F377 and L378 as the most critical FtsZ residues (124). In an in vivo assay, an L378A mutant interacted with ZipA whereas D373A, I374A, and F377A mutants did not (114). Finally, in a yeast two-hybrid assay, a D373G mutant did not bind ZipA (73).
The nature of the interaction between FtsZ and ZipA is clearly well established in vitro. In vivo, ZipA is 10- to 100-fold less abundant than FtsZ (69), and only about 30% of total ZipA localizes to the division site (158). Strikingly, ZipA displays a dynamic behavior similar to that of FtsZ, with a constant exchange of protein between the Z ring and an external pool, in this case located in the membrane rather than the cytosol, as analyzed by fluorescence recovery after photobleaching (158) (H. P. Erickson, personal communication). An intriguing question is whether the observed dynamics of FtsZ and ZipA (158) reflects the movement of both proteins individually or of FtsZ-ZipA complexes.
As with FtsA (see above), the low abundance of ZipA means that it can interact with only a small proportion of FtsZ subunits. Moreover, even though FtsA and ZipA both interact with the C-terminal tail of FtsZ, they probably do not compete for binding sites on the same FtsZ molecules.
Equivalents of ZipA in Other Organisms?
In B. subtilis, EzrA was identified as a negative regulator of FtsZ polymerization (98). In an ezrA null mutant, the critical concentration for formation of the Z ring was lowered, with polar ring formation that could overcome the effects of minCD overexpression, suggesting that EzrA acts to destabilize FtsZ polymers (103). The gene is conserved in several gram-positive bacteria. Although the phenotype of an ezrA mutant is, on the face of it, the opposite of the zipA phenotype, the two proteins share the unusual membrane topology mentioned above for ZipA. Thus, the predicted EzrA protein also has a single N-terminal membrane span and a large cytoplasmic domain (98) (Fig. 2B). Since ZipA and EzrA have similar topologies (although there is no significant primary sequence similarity) and interact with FtsZ to promote or destabilize polymers, it seems possible that the proteins are functionally homologous; however, more work is needed to substantiate this idea.
Also in B. subtilis, as mentioned above, a membrane-bound protein, SpoIIE, is implicated in the shift of cell division to polar sites that occurs when this organism initiates sporulation (52, 100). SpoIIE is made specifically during sporulation and is targeted to the division site in an FtsZ-dependent manner (6, 100, 101). Targeting does not require later division proteins encoded by divIB, divIC, or ftsL (55, 101). Although polar Z rings and septa are made in the absence of SpoIIE, they are delayed and substantially reduced in frequency (57, 95). SpoIIE has 10 transmembrane spans at its N terminus (domain 1), a linker domain of about 250 residues (domain 2), and a C-terminal domain involved in a quite separate regulatory function (52). SpoIIE interacts directly with FtsZ, probably through domain 2 (109), but the nature of this interaction and its function in vivo have not yet been elucidated. In principle, however, its role could be similar to that of ZipA in medial division of E. coli.
In addition to these proteins, it is conceivable that one or more of the transmembrane proteins involved in the synthesis of cell wall peptidoglycan could contribute to anchoring the Z ring to the membrane. So far, single mutations affecting these genes do not seem to prevent formation of the Z ring, although, as described below, several more transmembrane proteins are required for constriction of the ring.
SEPTAL PEPTIDOGLYCAN MACHINERY AND ASSOCIATED PROTEINS
Most of the remaining proteins involved in cell division have at least one membrane span and a substantial extracellular domain. Surprisingly, the only protein with a defined biochemical function is FtsI, which is required for septal peptidoglycan synthesis. In principle, the other division proteins could simply be involved in directing the PG-synthesizing machinery (FtsI plus various other proteins) to the division site, or they could have other functions, such as an active involvement in PG synthesis or a role in the membrane dynamics associated with invagination.
Complex and Diverse Assembly Pathways for Late Division Proteins?
The assembly pathways for the late, membrane-bound division proteins have been extensively analyzed in both E. coli and B. subtilis. These experiments have revealed surprising differences between the two organisms. In E. coli, the dependence pathway for assembly is, with one recent exception (22), completely linear (Table 1; Fig. 2C), with the sequence FtsK → FtsQ → [FtsL YgbQ] → FtsW → FtsI → FtsN. Thus, FtsK requires none of the other proteins to assemble at the Z ring, whereas FtsN depends on all of the other proteins. Brackets indicate that localization of FtsL and localization of YgbQ are codependent. In contrast, the equivalent proteins of B. subtilis appear to be recruited in a much more concerted manner (Table 2; Fig. 2D) (54). DivIB, DivIC, FtsL, PBP-2B, and probably FtsW (R. A. Daniel and L. J. Wu, unpublished data) are all completely interdependent for assembly at the division site; mutation or depletion of any of the proteins prevents all of the others from assembling. Although it is satisfying to have established the details of these dependence pathways, they shed little light on the mechanisms of division; however, in E. coli at least, they provide indications of where to look for specific protein-protein interactions.
The next sections summarize what is known about the functions and interactions of these proteins in E. coli and B. subtilis.
The Penicillin-Binding Protein FtsI
As mentioned above, septation requires a dedicated PBP for synthesis of the PG forming the new poles of the daughter cells. An extensive and diverse family of proteins involved in the final stages of PG synthesis have been identified on the basis of their ability to bind penicillins (reviewed recently in reference 65). Many bacteria have multiple PBPs, and there appears to be considerable redundancy in the way they function (43, 141). The high-molecular-weight (class A) PBPs (e.g., PBP-1a of E. coli and PBP-1 of B. subtilis) typically possess both transglycosylase and transpeptidase activity and seem to be involved in general PG synthesis (65). Class B PBPs are somewhat smaller and have only transpeptidase activity (65). In E. coli, two members of this class of protein appear to play important, spatially specialized roles in PG synthesis. Mutations in the mrdA gene (encoding PBP-2) give a characteristic round-cell phenotype, apparently as a result of a loss of ability to synthesize PG in the cylindrical part of the cell (155). In contrast, mutation of the ftsI or pbpB gene (encoding PBP-3) abolishes PG synthesis during division but still support cell elongation (156).
Functional homologues of FtsI (PBP-3) are recognizable both by their amino acid sequence and by the conserved position of their gene within a cluster of genes involved in division and cell wall synthesis, in many diverse bacteria. In B. subtilis, the equivalent gene (also called pbpB and encoding PBP-2B) is required for septation (35, 175).
The FtsI proteins have, in their N-terminal 50 or so residues, a noncleavable transmembrane span. This is followed by a domain of about 200 amino acids (aa) of poorly understood function (the non-penicillin-binding [nPB] domain) and then by a C-terminal domain of about 300 aa, which contains the characteristic sequence motifs representing the catalytic residues of PBPs (the penicillin-binding [PB] domain) (65). In an attractive current model for PG synthesis, in gram-negative bacteria at least, FtsI acts in a multiprotein complex that introduces three new glycan strands in parallel with hydrolysis of an existing docking or template strand (82). Full details of how FtsI fits into this complex remain to be determined. The crystal structure of the extracellular part of the equivalent protein from Streptococcus (PBP-2X) was determined several years ago (135). The PB domain shows the expected fold for the catalytic portion of a transpeptidase. The nPB domain has an interesting “sugar tongs” structure, with the legs of the tongs projecting away from the PB domain and the head of the tongs embedded into the side of the PB domain. Site-directed mutagenesis of the nPB domain resulted in the isolation of some mutations that destabilize the protein, suggesting that the nPB domain may behave as an intramolecular chaperone, required for proper folding of the PB domain (64, 118). However, other explanations are possible, including a site for interaction with other division proteins or proteins of the general PG synthetic machinery. It is even possible that this domain plays a direct role in PG synthesis: some of the mutants do not significantly affect catalysis, as judged by penicillin binding, but it is possible that they are unable to act on more physiologically relevant substrates.
It is generally thought that the class A PBPs of E. coli are nonspecialized, being equally involved in both cylindrical-wall and septal PG synthesis. However, in B. subtilis the major class A PBP, PBP-1, encoded by the ponA gene, is partially targeted to septal sites, and on depletion of the protein, the most prominent effect is on cell division (136). This again might again reflect differences in the way PG synthesis is organized in gram-positive and -negative organisms.
Nanninga's group has obtained evidence for the existence of a penicillin-insensitive enzymatic step in PG synthesis at the onset of septal constriction (130). However, so far, the putative enzyme responsible for this function has not been identified.
FtsW
FtsW is a member of the SEDS family of proteins (for “shape, elongation, division, and sporulation”) (81, 88). These proteins appear to have 10 transmembrane spans (60) and are frequently encoded by a gene that lies close to the gene for a class B PBP. Thus, in E. coli, ftsW lies in the same operon as ftsI (18, 89) and mrdB (encoding SEDS protein RodA) lies adjacent to mrdA (encoding PBP-2) (162). In both cases, the phenotype obtained when the SEDS-encoding gene is knocked out is similar to that obtained when its cognate PBP-encoding gene is knocked out. In B. subtilis, the gene in the equivalent position to ftsW of E. coli, spoVE, is specialized for sporulation (80). However, a functional homologue of ftsW (ylaO) has been found elsewhere in the genome (81) (Daniel and Wu, unpublished). According to a survey of sequenced bacterial genomes (116), there is a perfect presence-absence correlation between FtsW and FtsI, suggesting that their functions are intimately connected. The most likely function for the SEDS proteins, apart from targeting their cognate PBPs, would seem to be translocation of the lipid-linked precursor for PG synthesis from the inside to the outside of the membrane and delivery to its cognate PG-synthesizing complex (82). Such a role would be consistent with the multiple transmembrane structure of these proteins. However, the class B PBP has no transglycosylase activity and so is unlikely to be the immediate recipient for the precursor. Presumably, at least one class A PBP needs to be involved. Why should different substrate translocation proteins be required? There is some evidence that the transpeptidase activities of PBP-2 and FtsI have different substrate preferences, with the former preferring a pentapeptide side chain and the latter preferring a tripeptide side chain (13). Perhaps, RodA and FtsW have different precursor affinities and channel the appropriate precursor to differentiated PG-synthesizing machineries containing their cognate class B PBPs (82).
Although a role for FtsW in stabilization of FtsZ has been suggested for E. coli, no further evidence for a direct FtsZ-FtsW interaction has been reported. Interestingly, Datta et al. (38) have recently reported an FtsZ-FtsW interaction in Mycobacterium tuberculosis, mediated by a prominent C-terminal extension to FtsW, which is absent from the homologues of most other bacteria.
FtsK
FtsK is a large protein with several distinct roles in the late stages of cell division. It has a large soluble cytoplasmic domain that is required for chromosome segregation (53) and is highly conserved almost throughout the eubacteria. This is separated, by an extremely variable linker, from an N-terminal membrane domain containing four membrane spans. In E. coli (but not B. subtilis), the N-terminal domain is essential for division. The original temperature-sensitive mutations affecting the N-terminal domain of FtsK caused a late block in division (12), but protein depletion experiments have subsequently demonstrated an early block (25, 47, 167). Interestingly, the late-blocking ftsK(Ts) mutations (but not the depletion mutations) could be suppressed by deletion of dacA, which codes for carboxypeptidase PBP5 (47). Draper et al. (47) speculated that the late stages of septation could require a PBP with a preference for pentapeptide side chains, which should be enhanced in the absence of PBP5. In contrast, all of the ftsK mutations were suppressed by overexpression of ftsN (47; see below).
The C-terminal domain of FtsK is capable of ATP-dependent translocation along DNA, consistent with a role in pumping DNA through a closing septum (7). This behavior is similar to the DNA translocation activity displayed by the B. subtilis FtsK homologue SpoIIIE, which ensures proper chromosome translocation into the prespore compartment of sporulating cells (10). Moreover, the C-terminal domain of FtsK promotes Xer recombination reactions, required to resolve chromosome dimers before chromosome segregation (7). Given these functions of the FtsK C-terminal domain during the final steps of DNA replication and segregation, it is conceivable that FtsK helps to ensure that division occurs only in the region between newly replicated chromosomes.
FtsN
The ftsN gene was isolated as a multicopy suppressor of a temperature-sensitive ftsA mutation (32). Surprisingly, it also suppressed, to a greater or lesser extent, temperate-sensitive mutations in the ftsI, ftsQ, and ftsK genes (32). The protein has an N-terminal transmembrane segment that directs the major C-terminal domain to the periplasmic side of the membrane, but this segment can be replaced by heterologous sequences, even a cleavable signal peptide, suggesting that its division-specific function is contained in the C-terminal domain (33). The protein is poorly conserved, with clear homologues present only in enteric bacteria and Haemophilus spp. (33). However, FtsN shows weak sequence similarity to cell wall amidases (for example, Cw1C and SpoIIB in B. subtilis), suggesting a possible role in wall hydrolysis. Conceivably, FtsN could be required to cleave certain bonds in the wall to allow initiation of constriction. The suppressor activity of FtsN overproduction could work by further weakening the wall in such a way that a partially nonfunctional division machinery (e.g., one containing thermosensitive mutations affecting FtsA, FtsQ, or FtsI [32]) can operate more normally.
FtsL/DivIC
FtsL of E. coli is a small (121-aa) transmembrane protein with poorly conserved primary sequence (67, 163). The only potentially informative sequence feature is a coiled-coil motif in its major extracellular (C-terminal) domain (67). Such domains are typically involved in protein-protein interactions (110). Little is known about the function of FtsL in division, except that there is biochemical support for self-interaction through the detection of protein dimers (61). Originally, two temperature-sensitive alleles of ftsL were isolated: one produced filaments at the nonpermissive temperature, whereas the other caused lysis (163). The significance of this is not clear. B. subtilis appears to have a functional homologue of FtsL. Although the primary sequence similarity is negligible, the protein is a likely homologue on the grounds of similar size and predicted transmembrane topology, division phenotype, and the location of its gene immediately upstream of pbpB (36, 37). Mutagenesis of the B. subtilis ftsL gene lent support to the notion that few if any of its residues are critical for function (153) and therefore that its function is more likely to be structural than catalytic.
B. subtilis has a second _ftsL_-like gene, divIC, which is also required for division (99). It appears that DivIC and FtsL interact to form a heterodimer or -oligomer, on the basis of both yeast two-hybrid experiments and native gel electrophoresis (154). Furthermore, depletion of FtsL results in degradation of DivIC, indicating that formation of an FtsL-DivIC complex could stabilize DivIC (36). Interestingly, FtsL (of B. subtilis) is itself an intrinsically unstable protein (34). It is possible that FtsL plays an important regulatory role in division, because septation is shut down very rapidly if the gene is turned off (36). One important factor controlling FtsL turnover in B. subtilis is DivIB protein (34), as described below. Recently, a new essential cell division gene, ygbQ, was identified in Vibrio cholerae and E. coli (22). Depletion of the protein gave a filamentous phenotype typical of other cell division mutants. The YgbQ protein is a small transmembrane protein with a predicted structure similar to that of FtsL and DivIC. The protein localized to the division site, and this localization was codependent with FtsL. Although the protein was not significantly similar to any protein of B. subtilis, it did show weak but significant similarity to the DivIC protein of Bacillus halodurans. Taken together, these properties strongly point to YgbQ being homologous to DivIC.
FtsQ/DivIB
FtsQ of E. coli is a 276-aa protein of low abundance (∼22 copies per cell [23]) with a similar transmembrane topology to FtsL, FtsN, and FtsI (23, 75). The gene appears essential for division in E. coli, but its precise function is unclear (26). The ftsQ gene lies immediately upstream of ftsA and ftsZ in a range of organisms, but the overall sequence conservation is again poor. B. subtilis has a probable homologue of ftsQ called divIB (11, 76), but some of the properties of DivIB highlight possible differences in function. First, as with FtsA (see above), DivIB appears to be much more abundant (about 100-fold more) than its E. coli counterpart (147). Second, mutants with null mutations of divIB are viable, although they are temperature sensitive and fail to divide at higher temperatures (11). The likely basis for temperature-sensitive division was recently illuminated through the finding that the division defect can be largely overcome by overexpression of ftsL (34). Therefore, the main division function of DivIB could be to protect FtsL from degradation at higher temperatures. Interestingly, the dependence of DivIC stability on FtsL appears to be mediated in some way by DivIB, because in the absence of DivIB, DivIC stability is no longer dependent on FtsL (R. A. Daniel, unpublished data). All of these effects point to the formation of one or more complexes between these proteins. Recent results from this laboratory suggest that the target for these effects could be the PG-synthesizing machinery, because divIB null mutants can be suppressed by mutations affecting the nPB module of FtsI (Daniel, unpublished). This would support the notion that one important role of the nPB domain of FtsI is assembly of the septum-specific PG-synthesizing machinery at the division site.
CONSTRICTION AND CLOSURE OF THE DIVISION SEPTUM
The final steps of cell division pose a number of interesting questions, most of which remain to be resolved. During constriction, the assembled proteins described in the previous sections coordinately organize the annular constriction of the cytoplasmic membrane, cell wall, and any other cell envelope layers present, such as the outer membrane of gram-negative bacteria. E. coli and B. subtilis appear to be strikingly different in the mechanics of this process. In E. coli, division proceeds by gradual constriction of the cylindrical part of the cell, whereas in B. subtilis, a new cross-wall is formed with no concomitant constriction of the existing cylinder. One important consequence of this latter scheme is that the initial product of division is a pair of sister cells that are sealed off from each other by a double membrane but joined by a layer of wall material. Cell separation occurs later by the action of various cell wall autolysins (58). The distinction between these processes was recently blurred by the finding that a combination of mutations inactivating two autolytic enzymes results in E. coli cells that divide via a septum-like structure, with delayed cell separation (79).
Constriction is presumably an energy-dependent process. There are at least two potential drivers. One is constriction of the cytosolic Z ring, with associated factors and membrane anchor (see above). The other is inward growth of an annulus of PG. Indirect support for the first mechanism comes from the similarity of FtsZ and FtsA to eukaryotic cytoskeletal proteins (tubulin and actin). Moreover, the ability of certain wall-less organisms to divide suggests that PG synthesis is not essential for division, although there could be an equivalent extracellular mechanism substituting for PG synthesis in these organisms. Interestingly, the FtsZ ring is missing from chlamydiae. Although these organisms do not have a full PG wall, they retain the proteins needed for most steps in PG synthesis (63). Intriguingly, they do not appear to have a PBP with transglycosylase activity, but they retain several class B PBPs, including one closely related to FtsI (63). In principle, this could allow them to make a peptide-based polymer that substitutes in part for PG (63).
Insights into the possible interplay between the FtsZ and PG components of the division machinery of B. subtilis came from the effects of depletion of PBP 2B (FtsI) (35). As expected, inhibition of cell division occurs, giving rise to filamentous cells. The filaments can assemble Z rings at regularly spaced intervals, but none of the extracellular proteins (FtsL, DivIB, and DivIC) are recruited. However, at about the midpoint of many of the filaments, a complete assembly of division proteins is formed. These assemblies probably depend on residual PBP-2B activity giving rise to some septal PG synthesis. Interestingly, complete invagination of a pair of septal membranes occurs at many of these sites with little or no concomitant septal PG synthesis. This shows that septal membrane invagination probably needs a little PG synthesis to initiate but that, once started, it can largely be disengaged from PG synthesis (35). Conceivably, the major distortion of membrane conformation that is needed when ingrowth of the septum starts is an energetic barrier to initiation. A small annulus of newly synthesized PG at the division site could be sufficient to stabilize the distortion in the membrane, after which membrane invagination can readily proceed to completion.
During division, most of the proteins involved appear to be gradually lost from the division site as this process progresses and the diameter of the septal annulus decreases. Very little is known about how the complex division apparatus accommodates changes in the diameter of the annulus.
Finally, a specialized form of PG synthesis might be required to insert a small circular disk of wall material at the centre of the septum. Certain temperature-sensitive mutations, notably those of the ftsK gene, appear to result in arrest of division with deep invaginations of the septum, consistent with this idea (12).
THE FUTURE
Cell division is one of the most central processes in biology. For no cellular system do we have a clear idea of the molecular mechanisms underlying this process. However, for bacteria, particularly E. coli and B. subtilis, we have reached the point where most, if not all, of the essential players have been identified and the hierarchy with which they are assembled at the Z ring has been worked out. Furthermore, the crystal structures of most of the players will soon be available and the sites of some protein-protein interactions have been characterized. Of the five steps described in the Introduction, significant progress has been made in understanding the first four, which really comprise the assembly of the machinery. The major challenges now lie in understanding how the assembled machinery is triggered into action and how it shrinks and disassembles as the annulus closes, as discussed in the preceding section. Some progress is being made toward understanding the regulation of FtsZ polymerization in vitro. Ultimately, however, progress in understanding the molecular details of division may be limited by the availability of new experimental methods that allow in vitro reconstruction of more complete versions of the division apparatus.
Acknowledgments
This work was supported by a grant from the BBSRC. J.E. gratefully acknowledges receipt of a BBSRC Senior Research Fellowship. D-J.S. is supported by a Marie-Curie postdoctoral fellowship (HPMF-CT-2001-01421) of the European Union.
We are grateful to Harold P. Erickson and Piet A. J. de Boer for helpful comments and communication of unpublished data.
REFERENCES
- 1.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178**:**3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25**:**303-309. [DOI] [PubMed] [Google Scholar]
- 3.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 179**:**4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178**:**7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22**:**231-237. [DOI] [PubMed] [Google Scholar]
- 6.Arigoni, F., K. Pogliano, C. D. Webb, P. Stragier, and R. Losick. 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270**:**637-640. [DOI] [PubMed] [Google Scholar]
- 7.Aussel, L., F. X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108**:**195-205. [DOI] [PubMed] [Google Scholar]
- 8.Autret, S., and J. Errington. 2001. Dynamic proteins in bacteria. Dev. Cell 1**:**10-11. [DOI] [PubMed] [Google Scholar]
- 9.Barák, I., P. Prepiak, and F. Schmeisser. 1998. MinCD proteins control the septation process during sporulation of Bacillus subtilis. J. Bacteriol. 180**:**5327-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290**:**995-997. [DOI] [PubMed] [Google Scholar]
- 11.Beall, B., and J. Lutkenhaus. 1989. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J. Bacteriol. 171**:**6821-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177**:**6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Begg, K. J., A. Takasuga, D. H. Edwards, S. J. Dewar, B. G. Spratt, H. Adachi, T. Ohta, H. Matsuzawa, and W. D. Donachie. 1990. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J. Bacteriol. 172**:**6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109**:**257-266. [DOI] [PubMed] [Google Scholar]
- 15.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354**:**161-164. [DOI] [PubMed] [Google Scholar]
- 16.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175**:**1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89**:**7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24**:**1263-1273. [DOI] [PubMed] [Google Scholar]
- 19.Bramhill, D. 1997. Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13**:**395-424. [DOI] [PubMed] [Google Scholar]
- 20.Bramhill, D., and C. M. Thompson. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91**:**5813-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 180**:**6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 99**:**6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carson, M. J., J. Barondess, and J. Beckwith. 1991. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J. Bacteriol. 173**:**2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha, J.-H., and G. C. Stewart. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179**:**1671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol 42**:**395-413. [DOI] [PubMed] [Google Scholar]
- 26.Chen, J. C., D. S. Weiss, J.-M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181**:**521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook, W. R., and L. I. Rothfield. 1999. Nucleoid-independent identification of cell division sites in Escherichia coli. J. Bacteriol. 181**:**1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbin, B. D., X. C. Yu, and W. Margolin. 2002. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J 21**:**1998-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordell, S. C., R. E. Anderson, and J. Lowe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J 20**:**2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordell, S. C., and J. Löwe. 2001. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492**:**160-165. [DOI] [PubMed] [Google Scholar]
- 31.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol 174**:**6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai, K., Y. Xu, and J. Lutkenhaus. 1993. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J. Bacteriol. 175**:**3790-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai, K., Y. Xu, and J. Lutkenhaus. 1996. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol 178**:**1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel, R. A., and J. Errington. 2000. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36**:**278-289. [DOI] [PubMed] [Google Scholar]
- 35.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35**:**299-311. [DOI] [PubMed] [Google Scholar]
- 36.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29**:**593-604. [DOI] [PubMed] [Google Scholar]
- 37.Daniel, R. A., A. M. Williams, and J. Errington. 1996. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J. Bacteriol. 178**:**2343-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta, P., A. Dasgupta, S. Bhakta, and J. Basu. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J Biol Chem 277**:**24983-24987. [DOI] [PubMed] [Google Scholar]
- 39.de Boer, P. A. J., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10**:**4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56**:**641-649. [DOI] [PubMed] [Google Scholar]
- 41.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174**:**63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Den Blaauwen, T., N. Buddelmeijer, M. E. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181**:**5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181**:**3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dewar, S. J., K. J. Begg, and W. D. Donachie. 1992. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J. Bacteriol. 174**:**6314-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz, J. F., A. Kralicek, J. Mingorance, J. M. Palacios, M. Vicente, and J. M. Andreu. 2001. Activation of cell division protein FtsZ. Control of switch loop T3 conformation by the nucleotide γ-phosphate. J. Biol. Chem. 276**:**17307-17315. [DOI] [PubMed] [Google Scholar]
- 46.Din, N., E. M. Quardokus, M. J. Sackett, and Y. V. Brun. 1998. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 27**:**1051-1063. [DOI] [PubMed] [Google Scholar]
- 47.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180**:**4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24**:**905-915. [DOI] [PubMed] [Google Scholar]
- 49.El Karoui, M., and J. Errington. 2001. Isolation and characterization of topological specificity mutants of minD in Bacillus subtilis. Mol. Microbiol. 42**:**1211-1221. [DOI] [PubMed] [Google Scholar]
- 50.Erickson, H. P. 2001. The FtsZ protofilament and attachment of ZipA—structural constraints on the FtsZ power stroke. Curr. Opin. Cell Biol. 13**:**55-60. [DOI] [PubMed] [Google Scholar]
- 51.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologues of tubulin polymers. Proc. Natl. Acad. Sci. USA 93**:**519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Errington, J. 2001. Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr. Opin. Microbiol. 4**:**660-666. [DOI] [PubMed] [Google Scholar]
- 53.Errington, J., J. Bath, and L. J. Wu. 2001. Bacterial DNA transport. Nat. Rev. Mol. Cell. Biol. 2**:**538-545. [DOI] [PubMed] [Google Scholar]
- 54.Errington, J., and R. A. Daniel. 2001. Cell division during growth and sporulation, p. 97-109. In L. Sonenshein, R. Losick, J. A. Hoch, (ed.), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 55.Feucht, A., R. A. Daniel, and J. Errington. 1999. Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis. Mol. Microbiol. 33**:**1015-1026. [DOI] [PubMed] [Google Scholar]
- 56.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40**:**115-125. [DOI] [PubMed] [Google Scholar]
- 57.Feucht, A., T. Magnin, M. D. Yudkin, and J. Errington. 1996. Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis. Genes Dev. 10**:**794-803. [DOI] [PubMed] [Google Scholar]
- 58.Foster, S. J., and D. L. Popham. 2001. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 59.Fu, X., Y.-L., Shih, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98**:**980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerard, P., T. Vernet, and A. Zapun. 2002. Membrane topology of the Streptococcus pneumoniae FtsW division protein. J Bacteriol 184**:**1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghigo, J.-M., and J. Beckwith. 2000. Cell division in Escherichia coli: role of FtsL domains in septal localization, function, and oligomerization. J. Bacteriol. 182**:**116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghigo, J.-M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31**:**725-737. [DOI] [PubMed] [Google Scholar]
- 63.Ghuysen, J.-M., and C. Goffin. 1999. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob. Agenets Chemother. 43**:**2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goffin, C., C. Fraipont, J. Ayala, M. Terrak, M. Nguyen-Distèche, and J. M. Ghuysen. 1996. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J. Bacteriol. 178**:**5402-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62**:**1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gullbrand, B., and K. Nordström. 2000. FtsZ ring formation without subsequent cell division after replication runout in Escherichia coli. Mol. Microbiol. 36**:**1349-1359. [DOI] [PubMed] [Google Scholar]
- 67.Guzman, L.-M., J. J. Barondess, and J. Beckwith. 1992. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J. Bacteriol. 174**:**7716-7728. [PMC free article] [PubMed] [Google Scholar]
- 68.Hale, C. A., and P. A. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184**:**2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hale, C. A., and P. A. J. De Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88**:**175-185. [DOI] [PubMed] [Google Scholar]
- 70.Hale, C. A., and P. A. J. De Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181**:**167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hale, C. A., H. Meinhardt, and P. A. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20**:**1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hale, C. A., A. C. Rhee, and P. A. J. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182**:**5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. de Boer. 2001. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276**:**11980-11987. [DOI] [PubMed] [Google Scholar]
- 74.Harry, E. J. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40**:**795-803. [DOI] [PubMed] [Google Scholar]
- 75.Harry, E. J., B. J. Stewart, and R. G. Wake. 1993. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol. Microbiol. 7**:**611-621. [DOI] [PubMed] [Google Scholar]
- 76.Harry, E. J., and R. G. Wake. 1989. Cloning and expression of a Bacillus subtilis division initiation gene for which a homolog has not been identified in another organism. J. Bacteriol. 171**:**6835-6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harry, E. J., and R. G. Wake. 1997. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol. Microbiol. 25**:**275-283. [DOI] [PubMed] [Google Scholar]
- 78.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20**:**1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Holtje. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41**:**167-178. [DOI] [PubMed] [Google Scholar]
- 80.Henriques, A. O., H. De Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74**:**735-748. [DOI] [PubMed] [Google Scholar]
- 81.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28**:**235-247. [DOI] [PubMed] [Google Scholar]
- 82.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62**:**181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99**:**6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34**:**82-90. [DOI] [PubMed] [Google Scholar]
- 85.Hu Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182**:**3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7**:**1337-1343. [DOI] [PubMed] [Google Scholar]
- 87.Hu Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96**:**14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ikeda M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171**:**6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ishino, F., H. K. Jung, M. Ikeda, M. Doi, M. Wachi, and M. Matsuhashi. 1989. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J. Bacteriol. 171**:**5523-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184**:**2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones, L. J. F., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104**:**913-922. [DOI] [PubMed] [Google Scholar]
- 92.Katis, V. L., E. J. Harry, and R. G. Wake. 1997. The Bacillus subtilis division protein DivIC is a highly abundant membrane-bound protein that localizes to the division site. Mol. Microbiol. 26**:**1047-1055. [DOI] [PubMed] [Google Scholar]
- 93.Katis, V. L., R. G. Wake, and E. J. Harry. 2000. Septal localization of the membrane-bound division proteins of Bacillus subtilis DivIB and DivIC is codependent only at high temperatures and requires FtsZ. J. Bacteriol. 182**:**3607-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khattar, M. M., S. G. Addinall, K. H. Stedul, D. S. Boyle, J. Lutkenhaus, and W. D. Donachie. 1997. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J. Bacteriol. 179**:**784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khvorova, A., L. Zhang, M. L. Higgins, and P. J. Piggot. 1998. The spollE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J. Bacteriol. 180**:**1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.King, G. F., S. L. Rowland, B. Pan, J. P. Mackay, G. P. Mullen, and L. I. Rothfield. 1999. The dimerization and topological specificity functions of MinE reside in a structurally autonomous C-terminal domain. Mol. Microbiol. 31**:**1161-1169. [DOI] [PubMed] [Google Scholar]
- 97.King, G. F., Y. L. Shih, M. W. Maciejewski, N. P. Bains, B. Pan, S. L. Rowland, G. P. Mullen, and L. I. Rothfield. 2000. Structural basis for the topological specificity function of MinE. Nat. Struct. Biol. 7**:**1013-1017. [DOI] [PubMed] [Google Scholar]
- 98.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96**:**9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Levin, P. A., and R. Losick. 1994. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J. Bacteriol. 176**:**1451-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Levin, P. A., and R. Losick. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Devol. 10**:**478-488. [DOI] [PubMed] [Google Scholar]
- 101.Levin, P. A., and R. Losick, P. Stragier, and F. Arigoni. 1997. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol. Microbiol. 25**:**839-846. [DOI] [PubMed] [Google Scholar]
- 102.Levin, P. A., and P. S. Margolis, P. Setlow, R. Losick, and D. Sun. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174**:**6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levin, P. A., and R. L. Schwartz, and A. D. Grossman. 2001. Polymer stability plays an important role in the positional regulation of FtsZ. J. Bacteriol. 183**:** 5449-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31**:**1853-1861. [DOI] [PubMed] [Google Scholar]
- 105.Löwe J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391**:**203-206. [DOI] [PubMed] [Google Scholar]
- 106.Löwe J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18**:**2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Löwe, J., and L. A. Amos. 2000. Helical tubes of FtsZ from Methanococcus jannaschii. Biol. Chem. 381**:**993-999. [DOI] [PubMed] [Google Scholar]
- 108.Lu, C., M. Reedy, and H. P. Erickson. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182**:**164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lucet, I., A. Feucht, M. D. Yudkin, and J. Errington. 2000. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 19**:**1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem Sci. 21**:**375-382. [PubMed] [Google Scholar]
- 111.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66**:**93-116. [DOI] [PubMed] [Google Scholar]
- 112.Lutkenhaus, J., and E. Bi. 2001. Cell morphogenesis and division. Curr. Opin. Microbiol. 4**:**621-624. [Google Scholar]
- 113.Ma, X., D. W. Ehrhardt, and W. Margolin. 1997. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93**:**12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181**:**7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Margolin, W. 2000. Organelle division: self-assembling GTPases caught in the middle. Curr. Biol. 10**:**328-330. [DOI] [PubMed] [Google Scholar]
- 116.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24**:**531-548. [DOI] [PubMed] [Google Scholar]
- 117.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4**:**647-652. [DOI] [PubMed] [Google Scholar]
- 118.Marrec-Fairley, M., A. Piette, X. Gallet, R. Brasseur, H. Hara, C. Fraipont, J. M. Ghuysen, and M. Nguyen-Distèche. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 37**:**1019-1031. [DOI] [PubMed] [Google Scholar]
- 119.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33**:**84-96. [DOI] [PubMed] [Google Scholar]
- 120.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Devl. 12**:**3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meinhardt, H., and P. A. de Boer. 2001. Pattern formation in Escherichia coli: A model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98**:**14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184**:**904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mileykovskaya, E., Q. Sun, W. Margolin, and W. Dowhan. 1998. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol. 180**:**4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19**:**3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moy, F. J., E. Glasfeld, L. Mosyak, and R. Powers. 2000. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry 39**:**9146-9156. [DOI] [PubMed] [Google Scholar]
- 126.Mukherjee, A., and J. Lutkenhaus. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176**:**2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mukherjee, A., and J. Lutkenhaus. 1999. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181**:**823-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mukherjee, A., C. Saez, and J. Lutkenhaus. 2001. Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J. Bacteriol. 183**:**7190-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mulder, E., and C. L. Woldringh. 1989. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J. Bacteriol. 171**:**4303-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nanninga, N. 1991. Cell division and peptidoglycan assembly in Escherichia coli. Mol. Microbiol. 5**:**791-795. [DOI] [PubMed] [Google Scholar]
- 131.Nogales, E., K. H. Downing, L. A. Amos, and J. Löwe. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5**:**451-458. [DOI] [PubMed] [Google Scholar]
- 132.Nogales, E., M. Whittaker, R. A. Milligan, and K. H. Downing. 1999. High-resolution model of the microtubule. Cell 96**:**79-88. [DOI] [PubMed] [Google Scholar]
- 133.Nogales, E., S. G. Wolf, and K. H. Downing. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391**:**199-203. [DOI] [PubMed] [Google Scholar]
- 134.Ohashi, T., C. A. Hale, P. A. de Boer, and H. P. Erickson. 2002. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J. Bacteriol. 184**:**4313-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3**:**284-289. [DOI] [PubMed] [Google Scholar]
- 136.Pedersen, L. B., E. R. Angert, and P. Setlow. 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181**:**3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183**:**6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21**:**685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pichoff, S., B. Vollrath, C. Touriol, and J-P. Bouché. 1995. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 18**:**321-329. [DOI] [PubMed] [Google Scholar]
- 140.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94**:**559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class a high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178**:**2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Raskin, D. M., and P. A. J. De Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181**:**6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Raskin, D. M., and P. A. J. De Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96**:** 4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18**:**2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Regamy, A., E. J. Harry, and R. G. Wake. 2000. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: co-ordinating chromosome replication with cell division. Mol. Microbiol. 38**:**423-434. [DOI] [PubMed] [Google Scholar]
- 146.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33**:**423-448. [DOI] [PubMed] [Google Scholar]
- 147.Rowland, S. L., V. L. Katis, S. R. Partridge, and R. G. Wake. 1997. DivIB, FtsZ and cell division in Bacillus subtilis. Mol. Microbiol. 23**:**295-302. [DOI] [PubMed] [Google Scholar]
- 148.Sanchez, M., A. Valencia, M. J. Ferrandiz, C. Sander, and M. Vicente. 1994. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 13**:**4919-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Scheffers, D., and A. J. Driessen. 2002. Immediate GTP hydrolysis upon FtsZ polymerization. Mol. Microbiol. 43**:**1517-1521. [DOI] [PubMed] [Google Scholar]
- 150.Scheffers, D. J., J. G. de Wit, T. den Blaauwen, and A. J. Driessen. 2001. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 494**:**34-37. [DOI] [PubMed] [Google Scholar]
- 151.Scheffers, D. J., J. G. de Wit, T. den Blaauwen, and A. J. M. Driessen. 2002. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41**:**521-529. [DOI] [PubMed] [Google Scholar]
- 152.Scheffers, D-J., T. den Blaauwen, and A. J. M. Driessen. 2000. Non-hydrolysable GTP-γ-S stabilizes the FtsZ polymer in a GDP-bound state. Mol. Microbiol. 35**:**1211-1219. [DOI] [PubMed] [Google Scholar]
- 153.Sievers, J., and J. Errington. 2000. Analysis of the essential cell division gene ftsL of Bacillus subtilis by mutagenesis and heterologous complementation. J. Bacteriol. 182**:**5572-5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sievers, J., and J. Errington. 2000. The Bacillus subtilis cell division protein FtsL localizes to sites of septation and interacts with DivIC. Mol. Microbiol. 36**:**846-855. [DOI] [PubMed] [Google Scholar]
- 155.Spratt, B. G. 1975. Distinct penicillin-binding proteins involved in the division, elongation and shape of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 72**:**2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Spratt, B. G. 1977. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J. Bacteriol. 131**:**293-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stragier, P. 2000. A gene odyssey: exploring the genomes of endospore-forming bacteria, p. 519-525. In Bacillus subtilis and its relatives: from genes to cells. L. Sonenshein, R. Losick, and J. A. Hoch American Society for Microbiology, Washington, D. C.
- 158.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99**:**3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sun, Q., and W. Margolin. 1998. FtsZ dynamics during the division cycle of live Escherichia coli cells. J. Bacteriol. 180**:**2050-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sun, Q., X-C. Yu, and W. Margolin. 1998. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol. 29**:**491-503. [DOI] [PubMed] [Google Scholar]
- 161.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183**:**6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tamaki, S., H. Matsuzawa, and M. Matsuhashi. 1980. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J. Bacteriol. 141**:**52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ueki, M., M. Wachi, H. K. Jung, F. Ishino, and M. Matsuhashi. 1992. Escherichia coli mraR gene involved in cell growth and division. J. Bacteriol. 174**:**7841-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van den Ent F., L. Amos, and J. Löwe. 2001. Bacterial ancestry of actin and tubulin. Curr. Opin. Microbiol. 4**:**634-638. [DOI] [PubMed] [Google Scholar]
- 165.van den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19**:**5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang, L., M. K. Khattar, W. D. Donachie, and J. Lutkenhaus. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180**:**2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang, L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29**:**731-740. [DOI] [PubMed] [Google Scholar]
- 168.Wang, X., and J. Lutkenhaus. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9**:**435-442. [DOI] [PubMed] [Google Scholar]
- 169.Ward, J. E., Jr., and J. Lutkenhaus. 1985. Overproduction of FtsZ induces minicell formation in E. coli. Cell 42**:**941-949. [DOI] [PubMed] [Google Scholar]
- 170.Weiss D. S., J. C. Chen, J-M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181**:**508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weiss, D. S., K. Pogliano, M. Carson, L-M, Guzman, C. Fraipont, M. Nguyen-Distèche, R. Losick, and J. Beckwith. 1997. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol. Microbiol. 25**:**671-681. [DOI] [PubMed] [Google Scholar]
- 172.Woldringh, C. L., E. Mulder, J. A. Valkenburg, F. B. Wientjes, A. Zaritsky, and N. Nanninga. 1990. Role of the nucleoid in the toporegulation of division. Res. Microbiol. 141**:**39-49. [DOI] [PubMed] [Google Scholar]
- 173.Wu, L. J., A. H. Franks, and R. G. Wake. 1995. Replication through the terminus region of the Bacillus subtilis chromosome is not essential for the formation of a division septum that partitions the DNA. J. Bacteriol. 177**:**5711-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yan, K., K. H. Pearce, and D. J. Payne. 2000. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 270**:**387-392. [DOI] [PubMed] [Google Scholar]
- 175.Yanouri, A., R. A. Daniel, J. Errington, and C. E. Buchanan. 1993. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J. Bacteriol. 175**:**7604-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu, X-C., A. H. Tran, Q. Sun, and W. Margolin. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol. 180**:**1296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang, Y., S. Rowland, G. King, E. Braswell, and L. Rothfield. 1998. The relationship between hetero-oligomer formation and function of the topological specificity domain of the Escherichia coli MinE protein. Mol. Microbiol. 30**:**265-273. [DOI] [PubMed] [Google Scholar]
- 178.Zhao, C-R, P. A. J. De Boer, and L. I. Rothfield. 1995. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. USA 92**:**4313-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]