A Comprehensive Rice Transcript Map Containing 6591 Expressed Sequence Tag Sites (original) (raw)
Abstract
To determine the chromosomal positions of expressed rice genes, we have performed an expressed sequence tag (EST) mapping project by polymerase chain reaction–based yeast artificial chromosome (YAC) screening. Specific primers designed from 6713 unique EST sequences derived from 19 cDNA libraries were screened on 4387 YAC clones and used for map construction in combination with genetic analysis. Here, we describe the establishment of a comprehensive YAC-based rice transcript map that contains 6591 EST sites and covers 80.8% of the rice genome. Chromosomes 1, 2, and 3 have relatively high EST densities, approximately twice those of chromosomes 11 and 12, and contain 41% of the total EST sites on the map. Most of the EST-dense regions are distributed on the distal regions of each chromosome arm. Genomic regions flanking the centromeres for most of the chromosomes have lower EST density. Recombination frequency in these regions is suppressed significantly. Our EST mapping also shows that 40% of the assigned ESTs occupy only ∼21% of the entire genome. The rice transcript map has been a valuable resource for genetic study, gene isolation, and genome sequencing at the Rice Genome Research Program and should become an important tool for comparative analysis of chromosome structure and evolution among the cereals.
INTRODUCTION
cDNA sequences, known as expressed sequence tags (ESTs), can be used to analyze gene structure, expression, and function. ESTs also can be used for genome study to understand chromosomal composition, organization, and structure. With a genome size of ∼430 Mb, rice is estimated to have 30,000 to 50,000 genes. From large-scale EST sequencing at the Rice Genome Project (RGP), >29,000 cDNA clones were isolated and partially sequenced from their 5′ ends (http://rgp.dna.affrc.go.jp/Publicdata.html). The characterization of sequenced clones by similarity analysis resulted in 10,507 nonredundant sequences (Yamamoto and Sasaki, 1997). To date, ∼1450 of these cDNA clones have been mapped to chromosomes by genetic analysis (http://rgp.dna.affrc.go.jp/Publicdata). The Arabidopsis genome has a size of 125 Mb, with 25,489 predicted genes, and shows generally similar features of gene density, expression level, and repeat distribution across five chromosomes (Arabidopsis Genome Initiative, 2000). Thus, it is interesting to investigate where and how the rice genes are located and distributed along its 12 chromosomes. As ESTs are assigned to the chromosomes, they become an indispensable resource in preparing a sequence-ready physical map and in mapping sequenced DNA fragments.
We sequenced the RGP cDNA clones from their 3′ ends and classified them into different groups, each representing a unique sequence. These unique sequences were used to design clone-specific primers and for polymerase chain reaction (PCR)–based yeast artificial chromosome (YAC) screening. The rice YAC-based physical map constructed previously by the RGP was used for chromosomal assignment of ESTs (http://rgp.dna.affrc.go.jp/Publicdata/physicalmap2001/YACall2001.html). Some ESTs identified positive YAC clones that were not located on our previous YAC physical map. These ESTs were mapped by genetic analysis to improve the marker distribution of the rice linkage map and the genome coverage of the YAC-based physical map.
This article reports detailed results from our EST mapping project. A comprehensive YAC-based rice EST map was established, and its important characteristics and implications for the rice genome are discussed. Possible applications for genome analysis and plant biological research also are discussed.
RESULTS
YAC Screening
A total of 8169 primer pairs were designed using 8440 unique sequences from the 3′ ends of rice cDNA clones, and 8054 primers successfully amplified DNA fragments from rice genomic DNA under the present PCR conditions (Tables 1 and 2; see Methods). Approximately 96% of these specific primers (7718 pairs) amplified a single DNA band on 2.5% agarose gels, whereas 4% of the primers (336 pairs) amplified multiple DNA fragments, usually two to three bands. Approximately 8% (680 pairs) of the total primer pairs amplified a PCR product different from the predicted size. Most of these primers produced a larger PCR product, possibly as a result of the presence of introns in the rice genome. Among the successful primer pairs, 83.3% (6713 pairs) amplified similar-sized DNA fragments from YAC pools and rice genomic DNA. Approximately 0.6% of the primers (45 pairs) also amplified similar DNA fragments from yeast DNA. The remaining 16.1% of the primers (1296 pairs) did not amplify any DNA fragments from the YAC pools.
Table 1.
Number of cDNA Clones Used for Sequence Analysis and EST Mapping
| Library Name | Clone Identifier | Source | No. of Clones |
|---|---|---|---|
| Shoot | S1 | Green shoot from seedling | 2539 |
| Flowering panicle | E | Panicle at flowering stage | 2280 |
| Etiolated shoot | S | Etiolated shoot from seedling | 1950 |
| GA3 callusa | C5 | Scutellum | 1723 |
| BA callusb | C1 | Scutellum | 1514 |
| Mature leaf | S2 | Mature leaf | 1455 |
| Heat shock callus | C6 | Scutellum | 1438 |
| Apical meristem (S) | E6 | Immature leaf including apical meristem | 1379 |
| Root | R | Root from seedling | 1267 |
| Ripening panicle | E1 | Panicle at ripening stage | 1190 |
| Young panicle 1 | E3 | Panicle mainly at premeiotic stage | 1007 |
| Growth phase callus | C | Scutellum | 892 |
| Apical meristem (L) | E5 | Immature leaf including apical meristem | 873 |
| Chilled root | R1 | Young root | 517 |
| Young panicle 3 | E2 | Panicle mainly at meiotic stage | 429 |
| NAA callusc | C3 | Scutellum | 262 |
| C(−) callus | C2 | Scutellum | 33 |
| Root callus | C4 | Root tip | 19 |
| Young panicle 2 | E4 | Panicle mainly at early meiotic stage | 18 |
| Total | 20,785 |
Table 2.
Summary of EST Primer Designing and PCR Screening
| Contents | No. |
|---|---|
| Unique EST sequences | 8440 |
| Designed EST primers | 8169 |
| EST primers amplifying DNA bands from rice genomic DNA | 8054 |
| EST primers amplifying DNA bands from YAC DNA pools | 6713 |
| Total selected YAC clones by EST primers | 4387 |
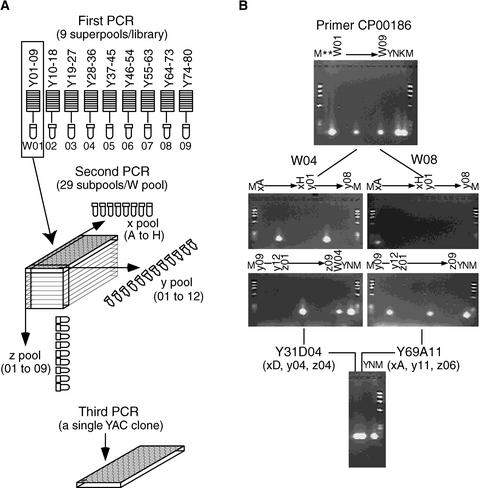
To perform the PCR-based YAC screening, 6713 specific primers, including 1290 primers designed from the cDNA sequences corresponding to genetic markers on our rice genetic map, were used. For example, Figure 1B shows the screening result of primer pair CP00186 designed from the 3′ end sequence of cDNA clone R3001. In all, ∼756,400 PCRs against the YAC library were performed (60,400 at the first PCR, 643,500 at the second PCR, and 52,500 at the third PCR). The average number of PCRs against the YAC DNA for the second and third PCRs was 95.9 (3.3 positive superpools) and 7.8 per primer pair, respectively. As a result, 4387 individual YAC clones were selected from our YAC library. The number of positive YACs selected by one EST primer pair varied from one to 27, with an average of 4.3 YACs per EST. Conversely, one YAC clone was selected by an average of 5.9 EST primers, with the maximum of 40.
Figure 1.
Construction of YAC DNA Pools for the Three-Step PCR and YAC Screening Using the EST Primer.
(A) YAC DNA pools.
(B) Two positive YAC clones, Y31D04 and Y69A11, were screened by the primer pair CP00186 designed from the 3′ end sequences of cDNA clone R3001. On the basis of the chromosomal positions of the two positive YACs, cDNA clone R3001 was assigned to a site ∼200 kb from the genetic marker of C69 (117.0 cM) on chromosome 6. Asterisks indicate special DNA templates pooled from the YAC clones mapped on chromosomes 1 and 6.
K, Kasalath rice; M, a size marker (ΦX174 HaeIII digest); N, Nipponbare rice; Y, yeast.
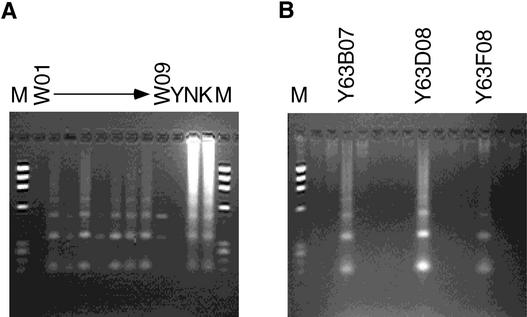
To investigate the position of rice centromeric regions on the YAC-based physical map, YAC screening using the centromere-specific primer RCS2CP02 also was performed, as shown in Figure 2. A total of 105 YAC clones were identified from the entire YAC library, including 67 YAC clones also selected by the EST primers.
Figure 2.
YAC Screening Using the Centromere-Specific Primer RCS2CP02.
(A) Screening of YAC superpools (W01 to W09).
(B) Confirmation of candidate YACs. Three positive YAC clones, Y63B07, Y63D08, and Y63F08, are shown. These clones were not mapped to the rice physical map because of the absence of associated genetic markers or EST sites.
K, Kasalath rice; M, a size marker (ΦX174 HaeIII digest); N, Nipponbare rice; Y, yeast.
YAC Analysis by SEGMAP
Among the 4425 YACs selected by the EST and centromere-specific primers, 1846 clones were mapped previously on our YAC-based physical map (Saji et al., 2001). Approximately 700 additional YAC clones were localized onto the physical map through SEGMAP analysis (Green and Green, 1991). These YAC clones, selected only by EST primers, mapped as bridge clones to close the gaps between two mapped YAC contigs or mapped redundantly to the regions of mapped contigs. Approximately 1870 YAC clones selected by ∼1500 ESTs were not present on our previous YAC-based physical map, remaining as chromosome unmapped.
Genetic Mapping of EST Clones Associated with Unmapped YACs
To improve the genome coverage of the YAC-based physical map, genetic analysis of 431 EST clones assigned to unmapped YAC contigs was performed with the same mapping population (186 F2 plants) from a cross of rice cvs Nipponbare and Kasalath using methods described previously (Harushima et al., 1998). The short DNA fragments amplified from the 3′ end regions were used as probes to detect the restriction fragment length polymorphism. Most of the ESTs showed a single hybridized DNA band with rice genomic DNA and mapped to the gap regions on our previous rice genetic linkage map. Fifty-eight loci mapped to chromosome 1, 45 to chromosome 2, 37 to chromosome 3, 39 to chromosome 4, 37 to chromosome 5, 23 to chromosome 6, 36 to chromosome 7, 38 to chromosome 8, 29 to chromosome 9, 35 to chromosome 10, 44 to chromosome 11, and 42 to chromosome 12. These additional genetic markers were used as anchors for the chromosomal assignment of new YAC clones. Through DNA gel blot hybridization of YAC DNA with the genetically mapped EST clones, ∼1400 new YACs were confirmed and mapped to the gap regions on our previous physical map, leading to an increase of >75 Mb of YAC contigs as well as successful chromosomal assignment of >1300 EST sites.
YAC Chimerism and EST Duplication
A number of EST primer pairs identified multiple YAC clones that mapped to more than one chromosomal position on the physical map. Because each EST primer pair amplifies an identical DNA fragment for all positive YACs, two possibilities, YAC chimerism and EST duplication, must be considered. DNA gel blot analysis of YAC DNA with genetic markers or ESTs indicated that ∼1200 YAC clones (27.1% of the total screened YACs) were chimeric because they contained the same-sized DNA fragments hybridizing to probes on different chromosomal regions. Approximately 200 ESTs were confirmed to have multiple sites on the YAC-based physical map as a result of gene duplication. Chimeric YAC clones could seriously affect the construction of a physical map and the correct assignment of EST location and order. Because most of these chimeric clones were associated with YAC contigs containing multiple YACs, they could be ignored for the construction of the YAC-based physical map and EST assignment.
Comprehensive YAC-Based Rice EST Map
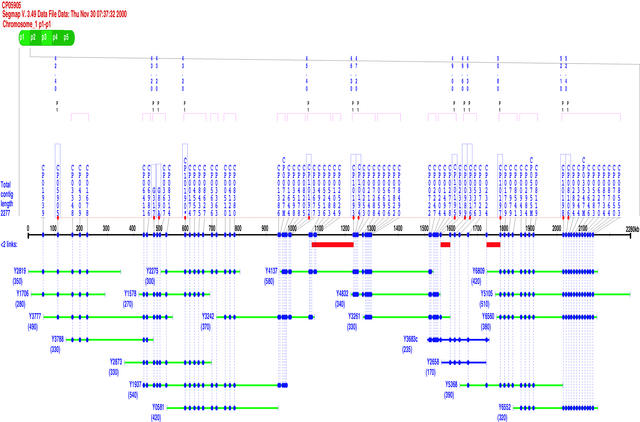
YACs and YAC contigs with known chromosome locations served as the framework for us to assign EST sites directly to specific genomic regions of rice chromosomes. After adding the newly mapped YAC clones and removing the chimeric YAC clones, a comprehensive YAC-based rice EST map was established using SEGMAP run under the travelling salesman problem (TSP) algorithm. Figure 3 shows an example of SEGMAP analysis for the construction of YAC contigs and the assignment of EST sites. Total results of EST mapping and important features of the rice EST map are summarized in Table 3. The map is composed of 2782 YAC clones containing 364 YAC contigs and 6591 putatively assigned EST sites from 6421 unique sequences. Seventy chimeric YAC clones were used because they were bridge clones for linking two neighboring YAC contigs on the final physical map.
Figure 3.
Construction of YAC Contigs and Assignment of EST Sites by SEGMAP Analysis.
The map includes the contig name, data file date, and chromosome name at top. Beneath that is a horizontal line representing the chromosome region spanned by the contig. Dots on the line represent the EST sites shown vertically with their primer names (CP). Genetically mapped EST sites are shown with their linkage distances (centimorgans [cM]; the asterisk indicates the floating marker) and chromosomal locations above the names. ESTs assigned to multiple sites on different chromosome regions are indicated by the letter M after the name. YAC clones are displayed under the line with their insert sizes (kb) shown in parentheses under the names. The closed circles on the YAC line indicate the sites corresponding to ESTs. The red lines above some YACs immediately below the scale line indicate that two sites are linked by one YAC. YACs shown in blue represent the new clones mapped in the present study. Chimeric YACs are shown with the letter c after the name. The total physical length is given at left of the scale line. This contig, covering the genomic region of chromosome 1 from 42.4 to 52.4 cM with an estimated physical length of 2277 kb, was made of 20 YAC clones and contained 74 putatively assigned EST sites. The relative order and positions of ESTs that selected the common YACs could not be determined. Two genomic clones (G317 and G89) also were used as framework markers in this region.
Table 3.
Summary of Rice EST Mapping Results
| Chromosome No. | YAC Clones | YAC Contigs | Total Length ofYAC Contigs (Mb) | Coverage (%)a | EST Sites | Average EST Density(EST/100 kb)b |
|---|---|---|---|---|---|---|
| 1 | 303 | 36 | 39.09 | 76.5 | 977 | 2.50 |
| 2 | 293 | 32 | 34.65 | 78.1 | 781 | 2.25 |
| 3 | 296 | 28 | 34.36 | 73.5 | 963 | 2.80 |
| 4 | 261 | 31 | 30.29 | 83.2 | 476 | 1.57 |
| 5 | 199 | 25 | 26.45 | 77.0 | 555 | 2.10 |
| 6 | 206 | 25 | 27.87 | 78.5 | 522 | 1.87 |
| 7 | 238 | 30 | 27.80 | 83.4 | 503 | 1.81 |
| 8 | 213 | 29 | 27.64 | 81.2 | 446 | 1.61 |
| 9 | 168 | 19 | 18.58 | 70.7 | 296 | 1.59 |
| 10 | 191 | 28 | 22.83 | 97.0 | 369 | 1.62 |
| 11 | 197 | 34 | 25.79 | 76.8 | 328 | 1.27 |
| 12 | 193 | 34 | 26.46 | 86.0 | 322 | 1.22 |
| Unknown | 24 | 13 | 5.46 | 1.3 | 53 | 0.97 |
| Total | 2782 | 364 | 347.27 | 80.8 | 6591 | 1.90 |
Locations of rice centromeres for all chromosomes except chromosome 11 were mapped on the physical map with YAC clones that contain the centromere-specific repetitive sequence (Figure 2). The estimated physical length of the map, including ∼5.5 Mb from 13 unmapped YAC contigs (because ESTs on these YACs did not reveal polymorphism during genetic analysis), was 347.3 Mb, which corresponds to ∼80.8% of the rice genome. Both single and multiple gene duplications were observed. A large, previously known, duplicated region was observed on the distal ends of the short arms of chromosomes 11 and 12 (Wu et al., 1998). Duplication of 46 ESTs in these two chromosomal regions was detected by their 3′ primers. The order of these duplicated genes was highly conserved.
Map Accuracy
To assess the accuracy of the rice EST map, EST sites and physical positions were examined by matching them to 40 Mb of chromosome 1 genome sequence released by RGP to DDBJ as of May 2001 (http://rgp.dna.affrc.go.jp/cgi-bin/giot/ine.pl). Only five of 961 ESTs mapped on the sequenced regions of this chromosome were not present on the genomic sequence, corresponding to a misassignment error rate of 0.5%. The placement of 18 EST sites disagreed with the genomic sequence by <200 kb. The quality of the EST map is attributable primarily to our attention to ambiguous or inconsistent data indicated by SEGMAP during the construction of the YAC contigs.
DISCUSSION
Learning about the physical organization of genes can be greatly helpful for understanding genome organization and evolution in plants. Such studies also may provide the basis for some important discoveries in biology. In this article, we report the successful construction of a comprehensive rice transcript map. A total of 6591 rice EST sites were assigned to specific chromosomal regions, and the genetic marker distribution on the rice linkage map as well as the resolution and genome coverage of the YAC-based physical maps were greatly improved. These results will aid future research into plant structural and functional genomics.
A total of 463 new genetic loci derived from 431 EST clones were added to the rice genetic linkage map. These loci increased the coverage of the marker-dense region (marker interval <2 centimorgan [cM]) from 46 to 60% and decreased the number of gaps (marker interval >5 cM) from 39 to 24. As a result, 13% of the genomic regions on the latest genetic map are now composed of marker-rare regions (interval >5 cM). The physical distance between 50 markers from the two distal ends of 25 YAC contigs (27.8 Mb) in chromosome 1 was determined by integrating the YAC physical map to the P1-derived artificial chromosome (PAC)/bacterial artificial chromosome (BAC) physical map (our unpublished data) and their genomic sequences. Statistical analysis of the difference of the marker distance from the two types of data indicates that the standard error is only ±9.0%. Thus, it is possible to explore extensively the variation in the frequency of recombination across the rice chromosomes of Nipponbare rice by comparing the genetic and physical distances between the two maps.
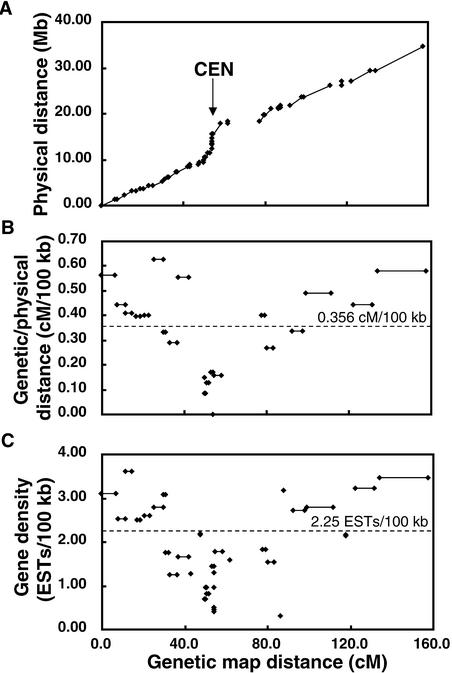
The average ratio of physical to genetic length of each YAC contig varied significantly along the length of the rice chromosomes. Figures 4A and 4B show an example of the relationship between the genetic and physical distance of YAC contigs in chromosome 2. This is particularly important and useful for gene isolation through map-based cloning. As in other plants, the recombination frequency in the centromere regions of each rice chromosome was suppressed significantly (>2740 kb/cM or <0.037 cM/100 kb), as shown in Figures 4B and 5, possibly as a result of the formation of dense heterochromatin made of numerous repetitive elements. Some regions of low recombination frequency (500 to 1000 kb/cM or 0.100 to 0.200 cM/100 kb) also were present on the chromosomal arms. Chromosomal regions with a relatively high frequency of recombination (70 to 180 kb/cM or 0.556 to 1.429 cM/100 kb) were found throughout the arms of each chromosome. It is interesting that the genomic regions with greater recombination frequencies usually had greater EST densities (Figure 4C).
Figure 4.
Relationships of Genetic Distance, Physical Distance, and Average EST Density of 32 YAC Contigs on Rice Chromosome 2.
The map position of each genetic marker on the rice linkage map is indicated by a diamond. YAC contigs are shown as solid lines linking two markers on two distal ends of each contig.
(A) The physical distance of each YAC contig is plotted against the position of each marker on the rice linkage map. The YAC contigs cover 78.1% of the entire chromosome. The physical distance for gaps (indicated by spaces between contigs) is unknown. CEN, rice centromere region.
(B) The ratio of genetic to physical distance between marker pairs of each YAC contig is plotted against the map positions of markers on the linkage map. The standard error for the ratio is estimated to be ±9.0%, based on the results of statistical analysis of chromosome 1. The ratio of genetic to physical distance for seven contigs could not be determined, either because markers could not be separated (a YAC clone spanning two markers) or because of the presence of only one marker on a contig. The dotted line indicates the average ratio (0.356 cM/100 kb) of genetic to physical distance calculated from the total genetic distance and the estimated size of the entire rice genome (1530.4 cM/430 Mb).
(C) The average gene density of each contig is plotted against the map position of markers on the linkage map. The dotted line indicates the average value (2.25 ESTs/100 kb) of gene density for the entire YAC physical map of this chromosome.
Figure 5.
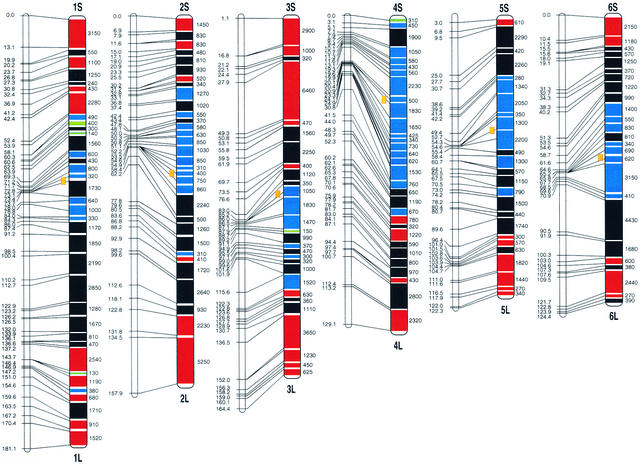
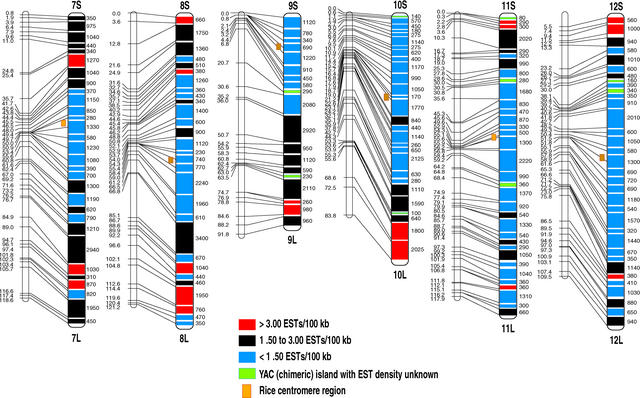
Chromosomal Distribution of the 6591 Rice EST Sites.
Genetic (left; cM) and YAC-based physical (right; kb) maps are shown for each chromosome. The solid lines across each chromosome show the positions and regions of YAC contigs on the genetic map. Different colors represent different EST densities observed on the individual contigs.
Compared with the previous rice YAC-based physical map, the number of YAC contigs in the present EST map decreased from 440 to 364 and the genome coverage increased from 62.8 to 80.8% (including 5.5 Mb of unmapped YAC contigs containing 53 EST sites). This was obtained through gap closure of previous YAC contigs and chromosomal mapping of new YAC clones by genetic analysis of associated ESTs. The largest YAC contig was on rice chromosome 3, containing 64 YACs with an estimated physical length of 6460 kb, that completely covered the region from 27.9 to 49.3 cM on our rice genetic map (Figure 5). Approximately 20% of the rice genome remains uncovered. The uncovered regions include probably centromeres, pericentromeres (surrounding the centromeres), and nucleolar organizers as well as telomeres, in which construction of the physical map by the methods we used is difficult because of highly repeated DNA sequences. There exist, of course, some other genomic regions in which DNA sequences are not cloned in our YAC library, as indicated by those EST primer pairs (16.1% of the total screened) that did not amplify DNA fragments from the YAC pools but did amplify fragments from rice genomic DNA. Further efforts will be made to perform physical analysis of these regions using BAC or PAC clones as well as chromosome in situ hybridization.
With 6591 EST sites assigned, the map resolution now stands at an average of 53 kb per marker. The RGP now is sequencing rice chromosomes 1, 2, 6, 7, and 8. The YAC-based physical maps for these five chromosomes have an estimated coverage ranging from 76.5 to 83.4%. We selected and used some EST primer pairs assigned on these chromosomes to identify specific clones from the RGP PAC library for the effective construction of a sequence-ready physical map. On chromosome 1, for example, the use of EST primers permitted the construction of a 1.9-Mb PAC contig, covering the genomic region from 43.2 to 52.4 cM, without chromosomal walking (http://rgp.dna.affrc.go.jp/cgi-bin/giot/ine.pl). Although the completion of a sequence-ready map for the genomic regions with a low EST density might be difficult, the EST primer pairs should be a valuable resource as anchor markers for selecting seed clones in these regions.
The cDNA clones used in the present study derived from 19 cDNA libraries that are made from various tissues with different treatments (Table 1). Although the number of assigned EST sites might correspond to only 10 to 20% of the total genes estimated in rice, the EST map is the densest one constructed so far except for the human map (Deloukas et al., 1998) and should yield insights into the distribution pattern and expression levels of genes along the 12 rice chromosomes. The average EST density calculated by comparing the physical length and mapped EST sites for the entire YAC-based physical map was 1.90 ESTs per 100 kb (Table 3). YAC contigs flanking the centromeric regions for most of the chromosomes had a lower EST density (0 to 1.00 EST/100 kb). The average EST density for each rice chromosome, however, varies significantly, with a range from 1.22 to 2.80 ESTs per 100 kb, differing by a factor of 2.30. The highest EST density of YAC contigs was observed from a region on chromosome 8 with 5.39 ESTs per 100 kb (Figure 5).
It is surprising to find that up to 41% of the total mapped EST sites are distributed on the physical maps of rice chromosomes 1, 2, and 3, although the total physical length from all YAC contigs on these three chromosomes is 108.0 Mb, or only ∼25% of the entire genome of rice. Such an uneven chromosomal distribution of genes is not observed in Arabidopsis (Arabidopsis Genome Initiative, 2000). This finding should be useful for analysis of the structure and evolution of rice chromosomes as well as its genome variation with the other plants. A total of 59 gene-rich regions (>3.00 ESTs/100 kb) are observed on the rice EST map (Figure 5). These regions, more frequent on chromosomes 1, 2, and 3, were distributed mainly on the distal regions of the chromosomes and had a total physical length of 74.6 Mb with 2642 assigned EST sites (average density of 3.54 ESTs/100 kb), as shown in Table 4. This finding indicates that 21% of the rice genome could contain 40% of the rice genes. On the other hand, 190 gene-poor regions (<1.50 ESTs/100 kb) are seen on the map. These regions have a total physical length of 148.2 Mb, corresponding to 43% of the rice genome, but only 1270 EST sites. These observations may be informative in analyzing the chromosomal regions relating to the gene-dense or gene-poor compartments of the rice genome (Barakat et al., 1997; Schmidt and Heslop-Harrison, 1998).
Table 4.
Genome Compartments of Rice Inferred by Gene Density
| Range of Gene Density (ESTs/100 kb) | YAC Contigs | Length of YAC Contigs (Mb) | Mapped ESTs | Average Gene Density (ESTs/100 kb)a |
|---|---|---|---|---|
| >3.00 | 59 | 74.60 | 2642 | 3.54 |
| 1.50 to 3.00 | 115 | 124.46 | 2679 | 2.15 |
| <1.50 | 190 | 148.20 | 1270 | 0.87 |
Use of this map information and the estimated functions of the assigned EST clones through homology search should have the practical value of accelerating the discovery of rice genes by positional and positional candidate cloning. On the conventional rice map, ∼200 gene loci were mapped and an integrated map of the phenotypes with DNA molecular markers has been constructed (Yoshimura et al., 1997). At the RGP, the use of EST markers on the map has resulted in the effective and successful isolation of several important rice genes (Ashikari et al., 1999; Yano et al., 2000). The rice EST map also contains sites for ∼110 cDNA sequences with putative proteins that carry nucleotide binding sites, Leu-rich repeats, or other conserved domains showing high similarities to plant resistance genes. Many of these ESTs map to regions corresponding to the map locations of rice resistance genes. To further rice functional genomics, clone-specific primers of all of the mapped ESTs have been used for populating gene expression arrays to monitor gene expression profiles (Yazaki et al., 2000).
The ultimate gene map for an organism is the complete sequence of its genome, annotated with the beginning and ending coordinates of every gene. To estimate the total number of rice genes at this time, we used the EST map to compare EST density with the annotation result of 1941 kb of completed genomic DNA sequence on chromosome 1, between 43.2 and 52.4 cM. The average EST density observed on the EST map from the YAC contig covering the corresponding region was 3.25 ESTs/100 kb. On the other hand, 345 genes were predicted from the genomic DNA sequence, revealing an average density of 17.78 predicted genes per 100 kb in this region (http://rgp.dna.affrc.go.jp/cgi-bin/giot/ine.pl). There were 5.47 times as many predicted genes as ESTs in this region of the map. Applying this ratio to the entire genome based on the average EST density, ∼44,700 predicted genes could be estimated for rice.
We have constructed a comprehensive rice transcript map containing 6591 EST sites. This map will facilitate annotation of the rice genome sequence, map-based cloning and other efforts in genetic research in rice. This reference resource represents a substantial contribution to the advancement of structural and functional genomics, to biological and physiological research of rice genes, and to comparative analysis of chromosome structure and evolution among the cereal plants.
METHODS
Sequence Analysis of Rice ESTs
Rice (Oryza sativa cv Nipponbare) cDNA clones from 19 different libraries were used as materials (Table 1). Partial sequences from their 3′ end regions were obtained by the same method described previously (Yamamoto and Sasaki, 1997). After cutting off vector sequences and deleting clones containing organelle sequences, all of the sequences were determined to have a length > 150 bases with <5% unidentified bases. In all, 20,785 sequences with an average length of 453 bases were obtained and used for identification of unique rice expressed sequence tags (ESTs). Similarity analysis of each clone was performed with BLASTN2.0 (Altschul et al., 1997). Using the threshold limit values of (1) P < 10−30 and (2) identity of >95% <100 bp or >90% >100 bp, 8440 unique sequences were identified, including 3504 groups and 4936 singletons. The average value of clone redundancy was 2.46. The maximum redundancy was 103.
Designing EST Primers
The 8440 unique 3′ end sequences were used for EST mapping. Primer pairs (20 bp long) were designed using Oligo 4.s (National Biosciences, Plymouth, MN). The 3′ primer was always made from sequences within the expected 3′ untranslated region, usually <50 bp from the poly(A) site, except for cases in which the sequence was not suitable for primer design. The 5′ primer was designed from sequences ∼200 bp from the 3′ primer. The 3′ end sequences were used for primer design because of clone specificity as well as the convenience of polymerase chain reaction (PCR) screening (http://rgp.dna.affrc.go.jp/rgp/ricegenomenewslet/nl11/02.html).
Centromere-Specific Primers
Rice genomic clone RCS2 carrying centromere-specific sequences was used. The 639-bp insert of this clone was isolated from an Indica rice line (IR-BB21) known to contain four copies of a 168-bp tandemly arranged repeat (Dong et al., 1998). Fiber fluorescence in situ hybridization analysis demonstrated that the RCS2 family is organized into various sizes of uninterrupted arrays in the rice genome with an estimated 6200 copies of the 168-bp monomer. Using Oligo 4.s, four sets of primers were designed and tested with rice genomic DNA and yeast artificial chromosome (YAC) DNA templates under different PCR conditions to show a PCR band pattern known to derive from the sequence of tandem repeats. Using an annealing temperature of 55°C, one primer pair, RCS2CP02, with sequences of 5′-gagtgtattgggtgcgttcg-3′ (5′ primer) and 5′-gtgagtttttcccacgaacg-3′ (3′ primer), generated multiple DNA fragments from both rice genomic and YAC DNA templates and appeared on the 2.5% agarose gels as a ladder pattern (Figure 2).
YAC Library Screening
A YAC library constructed from cultured cells of rice that contained 7680 clones with an average clone insert size of 350 kb (covering more than six equivalents of the rice genome) was used (Umehara et al., 1995). Total DNA of each YAC clone was isolated by an automatic plasmid isolation system (Kurabo, Osaka, Japan) with a modified protocol after YAC culture in AHC solution (6.7 g of yeast nitrogen base without amino acids, 10 g of casein hydrolysate acid, 25 mg of adenine hemisulphate, and 20 g of glucose per L, pH 5.8). The DNA concentration of the YAC clones was adjusted after checking samples on 0.8% gels with a standard DNA sample. YAC DNA pools were constructed using the same purified and concentration-adjusted DNA solution for the three-step PCR amplification. As shown in Figure 1A, the first PCR contained 12 kinds of DNA pools. Nine come from the rice YAC library (W01 to W09 superpools, each containing the DNA of 882 YAC clones from nine titer plates except W08 and W09, which contain YAC clones from 10 and seven titer plates, respectively). One comes from yeast (Y) cells as the negative control, and two come from the rice cultivars Nipponbare (N) and Kasalath (K; an Indica variety) as the positive controls. The second PCR is called three-dimensional PCR, containing 29 (28 to 30) subpools of YAC DNA prepared from each W pool (xA to xH, y01 to y12, and z01 to z09) and the controls. The third PCR is run for the confirmation of YAC candidates using DNA from a single, corresponding YAC clone.
Each PCR (20 μL) contained 2 μL of 10 × buffer, 2 μL of MgCl2 (25 mM), 2 μL of deoxynucleotide triphosphate (2 mM), 0.2 μL of Taq polymerase (5 units/μL), 0.3 μL of primer DNA (a mixture of upper and lower primers with a concentration of 10 μM for each), 4 μL of 50% glycerol, 5 μL of template DNA (5 ng/μL), and 4.5 μL of water. PCR amplification was performed in a MJ Research (San Francisco, CA) PCR machine (PTC-225 DNA Engine Tetrad) with cycling conditions of 94°C for 30 sec, a standard annealing temperature of 60°C for 60 sec, and 72°C for 60 sec over 35 cycles. A specific annealing temperature of 55°C also was used for a very small number of primers because they did not amplify DNA fragments at the standard temperature. PCR products run on 2.5% agarose gels were visualized by ethidium bromide staining and recorded by gel analysis (Densitograph; ATTO Ltd., Tokyo, Japan).
Data Analysis
SEGMAP (version 3.48), an interactive graphical tool for analyzing and displaying physical mapping data developed by C. Magness and P. Green (http://www.genome.washington.edu/UWGC/analysistools/segmap.htm), was used for the analysis of YAC screening data and the establishment of the EST map. Our YAC-based physical map, made previously using 1439 genetic DNA markers from the rice linkage map with an estimated coverage of 63% of the rice genome, was used as the framework for the analysis of EST sites (Saji et al., 2001). SEGMAP automatically grouped the YACs and ESTs into contigs and determined their order and relative distances. When YAC contigs contained ambiguous or inconsistent data, they were checked by supplemental PCR to avoid false-negative or false-positive screening results, inaccurate data entry, and incomplete screening. To confirm the presence of chimeric YACs and gene duplication on these contigs, DNA gel blot hybridization of YAC DNA was performed with genetic or EST markers. When contigs were formed by SEGMAP without chromosomal location, the associated EST clones were subjected to genetic analysis to obtain chromosomal locations for the selected YACs.
DNA Gel Blot Hybridization and Genetic Analysis
DNA gel blot hybridization of YAC or rice genomic DNA with genetic markers or EST clones was conducted using the enhanced chemiluminescence system (Amersham, Buckinghamshire, UK). Genetic analysis was performed with the same F2 mapping population described previously (Harushima et al., 1998) using short DNA fragments amplified from the 3′ end regions as probes.
Data Access and Clone Availability
All of the data obtained in the present study are available at http://rgp.dna.affrc.go.jp/publicdata/estmap2001. YAC screening data with EST primer pairs used for the construction of the EST map contain the anchors (genetic markers) and anchor positions, cDNA clones and positions, DDBJ accession numbers, selected YAC clones, amplified PCR band sizes, annealing temperatures, and EST primer sequences. The rice EST map redrawn from SEGMAP contains the anchors and anchor positions, YAC clones and YAC contigs, cDNA clones, and their putative positions. The latest rice high-density genetic linkage map can be viewed at http://rgp.dna.affrc.go.jp/publicdata/geneticmap2000/index.html. All of the DNA clones used in the present study are available for research purposes. For EST clones, contact the DNA bank at the Ministry of Agriculture, Forestry, and Fisheries of Japan (http://rgp.dna.affrc.go.jp/Cloneaccess.html). For YAC clones, contact the Rice Genome Research Program at tsasaki@nias.affrc.go.jp.
Accession Number
The accession number for rice genomic clone RCS2 is AF058902.
Acknowledgments
We thank Drs. Chuck Magness and Philip Green for providing SEGMAP, Dr. Benjamin Burr for his critical reading of the manuscript and useful comments, Drs. Masahiro Nakagahra and Kyozo Eguchi for advice and encouragement, and all Rice Genome Research Program members for their kind cooperation in this project. This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project No. GS-1102).
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**,** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408**,** 796–815. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Wu, J., Yano, M., Sasaki, T., and Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-banding protein. Proc. Natl. Acad. Sci. USA 96**,** 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat, A., Chrels, N., and Bernardi, G. (1997). The distribution of genes in the genomes of Gramineae. Proc. Natl. Acad. Sci. USA 94**,** 6857–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas, P., et al. (1998). A physical map of 30,000 human genes. Science 282**,** 744–746. [DOI] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.-L., Ronald, P.C., and Jiang, J. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95**,** 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E.D., and Green, P. (1991). Sequence-tagged site (STS) content mapping of human chromosomes: Theoretical considerations and early experiences. PCR Methods Appl. 1**,** 77–90. [DOI] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2,275 markers using a single F2 population. Genetics 148**,** 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saji, S., Umehara, Y., Antonio, B.A., Yamane, H., Tanoue, H., Baba, T., Aoki, H., Ishige, N., Wu, J., Koike, K., Matsumoto, T., and Sasaki, T. (2001). A physical map with yeast artificial chromosome (YAC) clones covering 63% of the 12 rice chromosomes. Genome 44**,** 32–37. [DOI] [PubMed] [Google Scholar]
- Schmidt, T., and Heslop-Harrison, J.S. (1998). Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends Plant Sci. 3**,** 195–199. [Google Scholar]
- Umehara, Y., Inagaki, A., Tanoue, H., Yasukochi, Y., Nagamura, Y., Saji, S., Otsuki, Y., Fujimura, T., Kurata, N., and Minobe, Y. (1995). Construction and characterization of a rice YAC library for physical mapping. Mol. Breed. 1**,** 79–89. [Google Scholar]
- Wu, J., Kurata, N., Tanoue, H., Shimokawa, T., Umehara, Y., Yano, M., and Sasaki, T. (1998). Physical mapping of duplicated genomic regions of two chromosome ends in rice. Genetics 150**,** 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K., and Sasaki, T. (1997). Larger-scale EST sequencing in rice. Plant Mol. Biol. 35**,** 135–144. [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd-1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12**,** 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki, J., Kishimoto, N., Nakamura, K., Fujii, F., Shimbo, K., Otsuka, Y., Wu, J., Yamamoto, K., Sakata, K., Sasaki, T., and Kikuchi, S. (2000). Embarking on rice functional genomics via cDNA microarray: Use of 3′UTR probes for specific gene expression analysis. DNA Res. 7**,** 367–370. [DOI] [PubMed] [Google Scholar]
- Yoshimura, A., Ideta, O., and Iwata, N. (1997). Linkage map of phenotype and RFLP markers in rice. Plant Mol. Biol. 35**,** 49–60. [PubMed] [Google Scholar]