Expression Profile Matrix of Arabidopsis Transcription Factor Genes Suggests Their Putative Functions in Response to Environmental StressesW⃞ (original) (raw)
Abstract
Numerous studies have shown that transcription factors are important in regulating plant responses to environmental stress. However, specific functions for most of the genes encoding transcription factors are unclear. In this study, we used mRNA profiles generated from microarray experiments to deduce the functions of genes encoding known and putative Arabidopsis transcription factors. The mRNA levels of 402 distinct transcription factor genes were examined at different developmental stages and under various stress conditions. Transcription factors potentially controlling downstream gene expression in stress signal transduction pathways were identified by observed activation and repression of the genes after certain stress treatments. The mRNA levels of a number of previously characterized transcription factor genes were changed significantly in connection with other regulatory pathways, suggesting their multifunctional nature. The expression of 74 transcription factor genes responsive to bacterial pathogen infection was reduced or abolished in mutants that have defects in salicylic acid, jasmonic acid, or ethylene signaling. This observation indicates that the regulation of these genes is mediated at least partly by these plant hormones and suggests that the transcription factor genes are involved in the regulation of additional downstream responses mediated by these hormones. Among the 43 transcription factor genes that are induced during senescence, 28 of them also are induced by stress treatment, suggesting extensive overlap responses to these stresses. Statistical analysis of the promoter regions of the genes responsive to cold stress indicated unambiguous enrichment of known conserved transcription factor binding sites for the responses. A highly conserved novel promoter motif was identified in genes responding to a broad set of pathogen infection treatments. This observation strongly suggests that the corresponding transcription factors play general and crucial roles in the coordinated regulation of these specific regulons. Although further validation is needed, these correlative results provide a vast amount of information that can guide hypothesis-driven research to elucidate the molecular mechanisms involved in transcriptional regulation and signaling networks in plants.
INTRODUCTION
Plants have evolved a number of mechanisms to cope with different biotic and abiotic stresses. One important step in the control of stress responses appears to be the transcriptional activation or repression of genes. Many genes induced by stress challenges, including those encoding transcription factors, have been identified, and some of them have been shown to be essential for stress tolerance. In Arabidopsis, a number of families of transcription factors, each containing a distinct type of DNA binding domain, such as AP2/EREBP, bZIP/HD-ZIP, Myb, and several classes of zinc finger domains, have been implicated in plant stress responses because their expression is induced or repressed under different stress conditions (Shinozaki and Yamaguchi-Shinozaki, 2000). Altering the expression of certain transcription factors can greatly influence plant stress tolerance. For example, overexpression of two Arabidopsis AP2/EREBP genes, CBF1/DREB1B and DREB1A, results in enhanced tolerance to drought, salt, and freezing (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). These two transcription factors have been shown to bind to the cold-responsive cis element CRT/DRE and to activate the expression of target genes. In another example in which plant response to UV irradiation was studied, knockout mutants for the Arabidopsis AtMyb4 gene were found to produce a higher level of UV light–protective compounds, such as sinapate esters, and to be more tolerant to UV-B light, whereas transgenic plants overexpressing AtMyb4 were found to contain reduced levels of sinapate esters and to be more sensitive to UV-B light (Jin et al., 2000). The expression of AtMyb4 itself was found to be repressed by UV-B light treatment. It has been proposed that AtMyb4 functions as a negative regulator in controlling genes involved in the synthesis of protective sinapate esters.
Another well-characterized stress response is the response to various biotic stresses. The response depends on whether the interaction between the pathogen and the plant is compatible or incompatible—that is, whether the pathogen is virulent and the plant is susceptible or the pathogen is avirulent and the plant is resistant. Members of the recently identified family of WRKY transcription factors have been implicated in the control of some stress responses (Eulgem et al., 2000). The WRKY family is defined by a DNA binding domain that contains the strictly conserved amino acid sequence WRKY. Upon binding to their cognate W box binding motif (TTGACC/T), members of this family have been shown to activate transcription (Eulgem et al., 1999). WRKY genes have been found to be upregulated in response to a diverse set of stresses, including infection by pathogens and wounding, as well as during senescence. The induced accumulation of WRKY mRNA often is extremely rapid and appears not to require the de novo synthesis of regulatory factors (Eulgem et al., 2000). Recently, the expression of NPR1, which encodes a key regulator of defense responses in Arabidopsis, has been shown to be controlled by WRKY factors (Yu et al., 2001).
The Arabidopsis genome encodes >1500 transcription factors (Riechmann et al., 2000), the majority of which are members of large families. It is a challenge to understand and discriminate the function of closely related members within each family. To meet this challenge, we took a genomics approach and monitored their regulation at the mRNA level in 81 developmental stages, genetic backgrounds, and environmental conditions using the Arabidopsis GeneChip system (Zhu and Wang, 2000). Here, we report the expression profiles and the potential functions of 402 genes coding for known and putative transcription factors based on our observations.
RESULTS
Building an Expression Profile Matrix for Genes Encoding Known and Putative Transcription Factors
A total of 402 potential stress-related genes that encode known or putative transcription factors were selected for this study from ∼8300 genes (corresponding to approximately one-third of the genome) covered by the Arabidopsis GeneChip (see supplemental data). The detailed selection criteria for these genes are described in Methods. These genes include 63 AP2/EREBP genes, 121 AtMyb genes, 34 bZIP genes, 152 members of the diverse zinc finger gene classes, 12 AtHD-ZIP genes, and 21 IAA/AXR genes. The complex zinc finger gene classes can be divided further into distinct zinc finger gene families based on their structural features. These families include plant-specific WRKY (Eulgem et al., 2000) and Dof proteins (Yanagisawa and Schmidt, 1999), GATA-type zinc finger proteins, and RING zinc finger proteins (Jensen et al., 1998). Although some zinc finger proteins, such as the RING zinc finger proteins, might be involved in protein–protein interactions rather than direct DNA binding, they were included in this study because some studies have shown that they too can be involved in the transcriptional regulation of gene expression (Borden, 2000; Capili et al., 2001).
Expression levels of these 402 transcription factor genes were monitored in various organs, at different developmental stages, and under various biotic and abiotic stresses. A two-dimensional transcription matrix (genes versus treatments or developmental stages/tissues) describing the changes in the mRNA levels of the 402 transcription factor genes was constructed for these experiments. The data represent 19 independent experiments, with samples derived from different organs such as roots, leaves, inflorescence stems, flowers, and siliques and at different developmental stages (Zhu et al., 2001a) and >80 experiments representing 57 independent treatments with cold, salt, osmoticum, wounding, jasmonic acid, and different types of pathogens at different time points (see supplemental data for a detailed sample description). To make results comparable across all of the experiments in the study of stress response, transcript levels of the stress-treated samples were compared with those of the corresponding mock-treated samples, and fold change values were used for further clustering analysis.
Transcription Factor Genes Apparently Involved in Stress Responses Exhibit Nonspecific and Specific Alterations in Expression Profiles
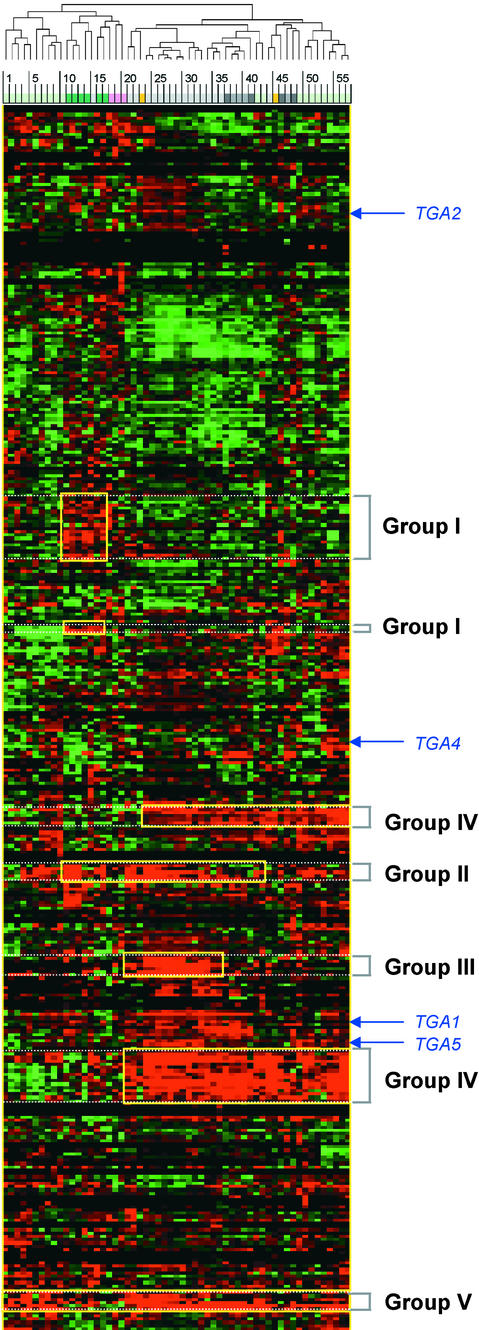
The majority of the transcription factor genes analyzed are expressed differentially after various stress treatments (Figure 1). With the intention of identifying transcription factor genes that are regulated by specific types of stress treatments and/or coregulated by combinations of different types of stress treatments, five groups (groups I to V) with distinct expression patterns were created based on the results of cluster analysis; these are highlighted in Figure 1. The members of each group are listed in the supplemental data.
Figure 1.
Expression Profiles of the Arabidopsis Transcription Factor Genes under Different Stress Conditions.
The fold change values for each sample, relative to untreated control samples, were log2 transformed and subjected to complete linkage hierarchical clustering, as described in Methods. Expression values higher and lower than those of the control are shown in red and green, respectively. The higher the absolute value of a fold difference, the brighter the color. The yellow rectangles indicate five potentially interesting groups as discussed in the text. The arrows indicate four genes that belong to the Arabidopsis TGA subfamily. The numbered color bar at the top indicates the type of stress applied for each experiment: light gray indicates bacterial; medium gray indicates fungal; dark gray indicates oomycetic; light green indicates viral; dark green indicates abiotic; pink indicates chemical; and light orange indicates wounding (see supplemental data for details). The horizontal dendrogram (top) indicates the relationship among experiments across all of the genes included in the cluster analysis.
Group I contains 21 genes that are induced preferentially by abiotic stress—that is, 3 and 27 hr after treatment with cold and/or high salt and/or osmoticum, and 2 and 12 hr after jasmonic acid treatment. This group includes the genes encoding the DRE/CRT binding factors DREB1B/CBF1, DREB1C/CBF2, and DREB1A/CBF3 that were shown previously to be activated transcriptionally by cold stresses (Liu et al., 1998; Medina et al., 1999). Other known genes in this group are CCA1 and Athb-8, which are regulated by the circadian clock and the hormone auxin, respectively (Wang and Tobin, 1998; Baima et al., 2001). In addition, a number of genes encoding putative zinc finger proteins, Myb proteins, bZIP/HD-ZIPs, and AP2/EREBP domain-containing proteins, also were found in this group.
Group II contains five genes that are activated preferentially by both abiotic stress and bacterial infection. Several isogenic pairs of strains of virulent or avirulent bacteria were included in this study, such as Pseudomonas syringae pv tomato DC3000 with (avirulent) or without (virulent) different avr genes and P. syringae pv maculicola ES4326 with (avirulent) or without (virulent) avrRpt2, and mRNA level changes were monitored over time. Genes encoding different types of leucine zipper DNA binding proteins, the bZIPs, such as GBF3, and HD-ZIPs, such as Athb-7 and Athb-12, belong to this group. GBF3 is thought to play a role in light regulation and in plant response to abscisic acid (Lu et al., 1996). The expression of both Athb-7 and Athb-12 is induced by abscisic acid and water stress (Soderman et al., 1996; Lee and Chun, 1998). The other two genes included in this group encode putative bZIP and Myb transcription factors.
Group III contains six genes that are activated mainly by bacterial infection. This group contains genes that belong to various types of transcription factor superfamilies, including AP2/EREBP, Myb proteins, WRKY type, and other types of zinc finger proteins. We were surprised to find that none of the genes in this group has been characterized with regard to plant stress responses.
Group IV contains 20 genes that are activated by infection with different types of pathogens, including bacteria, fungi, oomycetes, and viruses. Genes such as AtERF1, AtERF2, and ERF1 are in this group. Although their functions in other pathways are known, none of these transcription factor genes has been shown to be activated by pathogen infections. ERF1 plays a role in the ethylene signaling pathway (Solano et al., 1998). Transcription of AtERF1 and AtERF2 is induced by ethylene and wounding, and these factors are able to activate gene expression through the GCC box (Fujimoto et al., 2000). In addition to the genes that belong to the AP2/EREBP gene family (five genes in total in this group, including two additional putative AP2/EREBP genes), we found three genes encoding WRKY-type transcription factors, 10 genes encoding various other types of zinc finger proteins, and two genes encoding Myb proteins.
Group V contains five genes that also are activated by all types of pathogens but differ from those in group IV in their response to viral attack. Genes in group IV appear to be repressed by virus infection at an early time point, whereas genes in group V are activated under viral attack at these same time points (Figure 1; see supplemental data). AtWRKY6 and RAP2.6 belong to this group. This extends the previous finding that AtWRKY6 is induced by virulent and avirulent bacterial pathogens (Robatzek and Somssich, 2002). The expression of the RAP2.6 gene was not well characterized, although it has been shown to contain the highly conserved AP2/EREBP domain (Okamuro et al., 1997).
Although genes that are induced specifically by one or several particular type(s) of stress treatment(s) could be identified, a number of genes that are repressed by different conditions also were found; these are shown in green in Figure 1. Thirty-five genes that were not expressed at a detectable level under any conditions used in this study also were identified; these genes are not included in Figure 1 (data not shown).
In addition to classifying different genes according to their specific expression patterns under various stress conditions, we also distinguished the putative functions among genes in the same gene family. For example, the Arabidopsis TGA subfamily of bZIP transcription factors was found previously to contain six closely related members (Kawata et al., 1992; Schindler et al., 1992; Zhang et al., 1993; Miao et al., 1994; Xiang et al., 1997). Despite the high similarity (>72%) at the amino acid level, the expression characteristics of these four genes are quite different (Figure 1). TGA1 is induced preferentially by infection with Cauliflower mosaic virus and Botrytis cinerea at later time points (60 and 84 hr). TGA2 is repressed after a 27-hr cold treatment. TGA4 is repressed by a number of stresses, including 27 hr of cold treatment, 9 hr after infection with the bacterium P. syringae pv tomato DC3000, and 1 day after infection with Cucumber mosaic virus. TGA5 is induced mainly by infections with several different bacteria, such as P. syringae pv tomato DC3000/ avrRpt2 and P. syringae pv maculicola ES4326/avrRpt2, and by infection with Cauliflower mosaic virus. Distinct expression patterns of closely related genes imply that these genes would play different roles in the regulation of plant responses to biotic and/or abiotic stress.
Transcription Factor Genes Apparently Are Involved in Salicylic Acid, Jasmonic Acid, and Ethylene Signaling Pathways
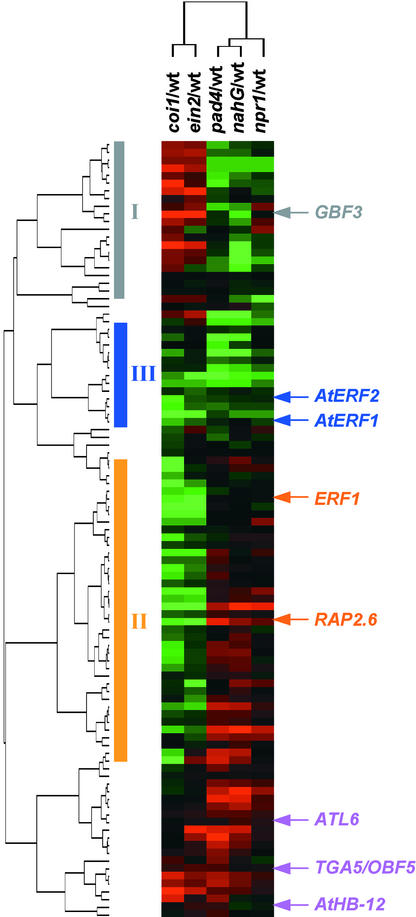
To gain information on how or through which signaling pathways transcription factor genes function in plant defense responses, we studied their expression in response to pathogen infection in different mutants and transgenic plants, namely nahG transgenic plants and the pad4-1, npr1-1, coi1-1, and ein2-1 mutants, in which different signaling pathways controlling plant defense responses are blocked (Delaney et al., 1994; Feys et al., 1994; Cao et al., 1997; Zhou et al., 1998; Alonso et al., 1999). Because of the defects in dif-ferent signaling pathways, all of the mutants or transgenic plants included in this study are more susceptible to some pathogen infections than are wild-type plants (Penninckx et al., 1998; Zhou et al., 1998; Thomma et al., 1999). Among the 95 genes that are activated by P. syringae pv maculicola ES4326 infection in wild-type plants, 80% of them are either less activated or not activated in at least one of the mutants (Figure 2; see supplemental data). On the basis of the altered expression patterns in the mutant and transgenic plants, we categorized the transcription factor genes into three groups (groups I to III).
Figure 2.
Expression of Transcription Factor Genes during Interactions with a Bacterial Pathogen in Mutant and Transgenic Plants That Are Deficient in Salicylic Acid, Jasmonic Acid, and Ethylene Signaling.
Transcription factor genes that were induced more than twofold 30 hr after inoculation with P. syringae pv maculicola ES4326 in wild-type plants were selected. Hierarchical clustering was performed as described in Figure 1, except that the fold change was calculated as average difference in a P. syringae pv maculicola ES4326–infected mutant relative to the infected wild type. Vertical bars at left indicate the clusters of genes that are discussed in the text. Expression of the genes in group I was reduced in the mutant or transgenic plants that are deficient in salicylic acid signaling. Expression of the genes in group II was reduced in the mutants that are deficient in jasmonic acid and/or ethylene signaling. Expression of the genes in group III was reduced in all of the mutants and transgenic plants. Examples of genes in each group are indicated. wt, wild type.
Group I contains genes whose expression relative to wild-type plants is reduced in nahG, pad4, and/or npr1 mutant plants (Figure 2; see supplemental data). GBF3 is in this group. As shown in supplemental data, the expression of GBF3 is induced by both abiotic stress and bacterial pathogens. The abolished induction of GBF3 in response to pathogen infection in the mutant backgrounds suggests that GBF3 functions downstream of salicylic acid and NPR1 in salicylic acid–dependent signaling. Group II contains the genes whose expression is reduced in coi1 and/or ein2 mutant backgrounds relative to wild-type plants (Figure 2; see supplemental data). Genes in this group include ERF1, which has been shown to play a role in ethylene responses. ERF1 is induced by a number of pathogen treatments (see supplemental data). In addition, RAP2.6, a gene that is induced by almost all pathogen treatments (see supplemental data), also belongs to this group. Because the induced expression of these genes is either reduced or abolished in coi1 and ein2 mutants, genes such as ERF1 and RAP2.6 may function in ethylene/jasmonic acid–dependent signaling. Group III contains the genes whose expression is reduced in all mutant and transgenic plant backgrounds relative to wild-type plants (Figure 2; see supplemental data). AtERF1 and AtERF2 are in this group. The presence of ERF1 and AtERF1/2 in different regulons is another example of differences in the regulation of highly homologous transcription factor gene family members.
Some genes are induced by pathogen treatment but appear not to be affected strongly in any of the mutants tested in this study, such as TGA5, Athb-12, and ATL6, a gene that encodes a RING-H2 zinc finger protein and has been shown to be induced rapidly by elicitor (Salinas-Mondragon et al., 1999), and several genes encoding putative transcription factors (Figure 2; see supplemental data). Expression of these genes may be controlled by defense-related signaling pathways that have not yet been identified.
Stress Response and Senescence May Share Overlapping Signaling Pathways
Recent studies suggest that the signaling pathways for leaf senescence and plant defense responses may overlap because several genes are activated both during senescence and by pathogen infection (Quirino et al., 2000). It will be interesting to determine to what extent the pathway activated by senescence shows similarity to the pathway involved in plant defense response and which transcription factors are involved in transcriptional regulation during leaf senescence and plant defense responses.
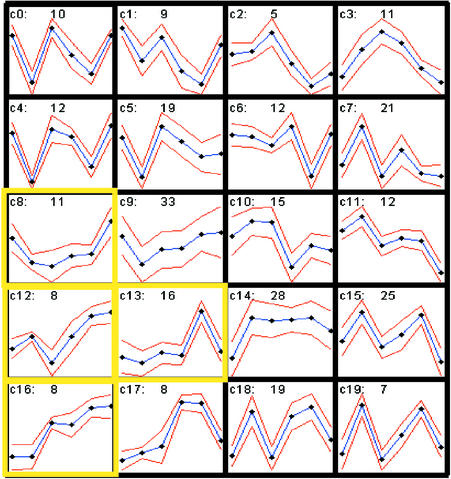
We clustered the transcription factor genes according to their expression profiles at different stages of leaf development. The two-dimensional self-organizing map algorithm (Tamayo et al., 1999) was used to gain an overview of the behavior of each gene relative to the others during the course of leaf development. Genes in clusters c8, c12, c13, and c16 are activated to various degrees in 8-week-old and/or 11-week-old senescent leaves (Figure 3, yellow boxes), suggesting that these genes are associated with senescence. These include the WRKY transcription factor genes AtWRKY4, AtWRKY6, and AtWRKY7, consistent with the previous observation that the expression of these genes is highly induced during senescence (Eulgem et al., 2000). In addition, the expression of genes encoding other types of transcription factors, such as ERF3, AtMyb2, and zinc finger proteins, also is induced during senescence (Table 1). Among the 43 genes in the c8, c12, c13, and c16 clusters, more than two-thirds also are induced after various stress treatments (see supplemental data), including the previously characterized AtWRKY6, ERF3, and AtMyb2. This finding suggests that the signaling pathway activated by senescence may overlap substantially with stress signaling pathways.
Figure 3.
Expression Patterns of the Transcription Factor Genes during Leaf Development.
The average difference was log2 transformed, mean centered for each gene, and subjected to the self-organization map algorithm using a 5 × 4 two-dimensional matrix and 100,000 epochs. The mean expression patterns for 20 distinct gene clusters (blue lines) and the standard deviation for each mean expression level (red lines) are shown. The y axis indicates the relative expression for all of the genes in that cluster, and the x axis indicates the stages during leaf development, following the order of 2-week-old leaf,a 2-week-old leaf,b 5-week-old leaf,a 6.5-week-old leaf,b 8-week-old leaf,b and 11- week-old leafa (a, samples were collected in the afternoon; b, samples were collected in the morning) (see supplemental data for detailed information). The number of genes in each cluster is indicated at the top center of each cluster graph. The clusters for the genes that were expressed at higher levels at 8 and/or 11 weeks are indicated by yellow boxes. c, cluster.
Table 1.
Transcription Factors Induced during the Senescence Process
| Probe Seta | SOMbCluster | Accession Numbers and Descriptionc | Induced by OtherStress Treatments |
|---|---|---|---|
| 12737_f_at | 8 | emb|CAA74603.1 | (Y14207)d R2R3-MYB transcription factor |
| 12908_s_at | 8 | dbj|BAA32422.1 | (AB008107) ethylene-responsive element binding factor 5 |
| 13015_s_at | 8 | emb|CAA67232.1 | (X98674) zinc finger protein |
| 13115_at | 8 | gb|AAB60774.1 | (AC000375) identical to At.WRKY6 |
| 14751_at | 8 | gb|AAC72869.1 | (AF104919) contains similarity to wild oat DNA binding protein ABF2 (GB:Z48431) |
| 15638_s_at | 8 | gb|AAC83582.1 | (AF062860) putative transcription factor (similar to MYB4) |
| 16198_at | 8 | gb|AAD24362.1 | AC007184_2 (AC007184) putative C2H2-type zinc finger protein |
| 16638_s_at | 8 | gb|AAD37511.1 | AF139098_1 (AF139098) putative zinc finger protein |
| 17426_at | 8 | gb|AAC49770.1 | (AF003097) AP2 domain containing protein RAP2.4 |
| 18121_s_at | 8 | gb|AAB63819.1 | (AC002337) MYB transcription factor (AtMyb2) |
| 19696_at | 8 | emb|CAB45059.1 | (AL078637) Arabidopsis WRKY7 |
| 12736_f_at | 12 | emb|CAA90748.1 | (Z50869) MYB-related protein |
| 13722_at | 12 | Contains multiple zinc finger domains: PF00096: zinc finger, C2H2 type | Viruses |
| 14079_at | 12 | emb|CAB39939.1 | (AL049500) similar to RING zinc finger protein, human AF037204 |
| 14852_s_at | 12 | gb|AAC05340.1 | (AC002521) putative MYB family transcription factor |
| 15445_at | 12 | emb|CAB41316.1 | (AL049711) similar to transcription factor PERIANTHIA from Arabidopsis |
| 17514_s_at | 12 | gb|AAD03545.1 | (AF076278) ethylene response factor 1 |
| 18738_f_at | 12 | emb|CAB09173.1 | (Z95741) R2R3-MYB transcription factor |
| 18746_f_at | 12 | emb|CAB09189.1 | (Z95757) R2R3-MYB transcription factor |
| 12522_at | 13 | emb|CAA18764.1 | (AL022605) similar to AP2 domain transcription factor |
| 13293_s_at | 13 | gb|AAB80649.1 | (AC002332) auxin-regulated protein (IAA13) |
| 14043_at | 13 | gb|AAD39282.1 | AC007576_5 (AC007576) Arabidopsis WRKY4 |
| 14243_s_at | 13 | emb|CAA49525.1 | (X69900) ocs element binding factor 5 |
| 14802_at | 13 | gb|AAD20087.1 | (AC006532) putative C2H2-type zinc finger protein |
| 15214_s_at | 13 | gb|AAB06611.1 | (U51850) G-box factor 3 |
| 16909_at | 13 | emb|CAA49524.1 | (X69899) ocs element binding factor 4 |
| 17424_at | 13 | gb|AAC49768.1 | (AF003095) AP2 domain containing protein RAP2.2 |
| 17490_s_at | 13 | gb|AAF01532.1 | AC009325_2 (AC009325) homeobox-leucine zipper protein HAT5 |
| 17833_at | 13 | gb|AAD22653.1 | AC007138_17 (AC007138) putative CHP-rich zinc finger protein |
| 18386_at | 13 | gb|AAB86455.1 | (AC002409) putative TGACG sequence–specific bZIP DNA binding protein |
| 18745_f_at | 13 | emb|CAB09188.1 | (Z95756) R2R3-MYB transcription factor |
| 18751_f_at | 13 | gb|AAA33067.1 | (L04497) putative MYB A from cotton |
| 18939_at | 13 | emb|CAA71854.1 | (Y10922) similar to Athb-14 HD-Zip protein |
| 20456_at | 13 | gb|AAB87098.1 | (AC002391) putative AP2 domain transcription factor |
| 20586_i_at | 13 | gb|AAC73042.1 | (AC005824) putative zinc finger protein |
| 12709_f_at | 16 | emb|CAB09204.1 | (Z95772) R2R3-MYB transcription factor |
| 13432_at | 16 | gb|AAD23013.1 | AC006585_8 (AC006585) putative WRKY-type DNA binding protein |
| 16073_f_at | 16 | gb|AAC83630.1 | (AF062908) putative Myb DNA binding transcription factor |
| 16483_at | 16 | emb|CAA48189.1 | (X68053) Arabidopsis TGA1 |
| 17791_s_at | 16 | emb|CAA18200.1 | (AL022198) similar to parsley WRKY1 |
| 19611_s_at | 16 | gb|AAC83596.1 | (AF062874) Myb-related protein Y49 |
| 19646_s_at | 16 | gb|AAC69925.1 | (AC005819) homeodomain transcription factor (Athb-7) |
| 20471_at | 16 | gb|AAC49767.1 | (AF003094) AP2 domain containing protein RAP2.1 |
Roots Express Higher Levels of Some Stress-Related Transcription Factor Genes
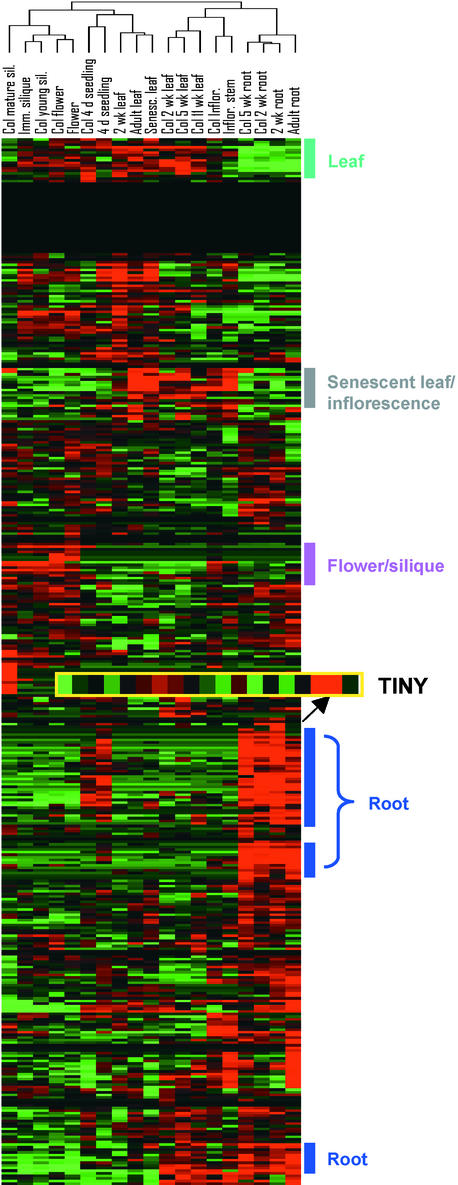
The developmental and organ-specific regulation of the 402 transcription factor genes was examined. As shown in Figure 4, the majority of genes exhibit temporal and spatial variations in expression level. Using clustering analysis, organ-specific genes can be identified within each family of transcription factor genes. For most of the known transcription factor genes, the results from our GeneChip analyses for developmental and organ-specific regulation are consistent with the gene expression patterns identified previously using techniques such as RNA gel blot analysis. For example, among the 63 genes in the AP2/EREBP superfamily, the expression patterns of 22 have been reported previously. These genes include AP2, ANT, TINY, ERF1, ABI4, DREB1A, DREB1B, AtERF-1 to AtERF-5, and _AP2_-related (RAP2) genes (Jofuku et al., 1994; Elliott et al., 1996; Klucher et al., 1996; Wilson et al., 1996; Okamuro et al., 1997; Stockinger et al., 1997; Finkelstein et al., 1998; Liu et al., 1998; Solano et al., 1998; Medina et al., 1999; Fujimoto et al., 2000).
Figure 4.
Expression Profiles for the Arabidopsis Transcription Factor Genes in Different Organs and at Different Developmental Stages.
Clustering was performed as described in Figure 1, except that the expression values, rather than fold changes, were used for cluster analysis. The color for each gene indicates its expression level relative to its mean across all of the experiments. Expression greater than mean level is represented by red, expression less than mean level is represented by green, and expression close to mean level is represented by black. Genes that are expressed preferentially in root, leaf, flower/silique, and senescent leaf/inflorescence are indicated. Genes that do not show detectable expression in any of the experiments are shown as a stretch of black across all experiments. The expression pattern for TINY is enlarged as indicated. Col, Columbia; d, day-old; Imm, immature; Inflo, inflorescence; Sil, silique; wk, week-old.
Expression profiles for well-characterized genes such as AP2, ANT, ABI4, and RAP2 are in good agreement with previous reports in terms of tissue-specific expression patterns. However, TINY gene expression is an exception. It is expressed at relatively high levels in 2-week-old roots but not in other vegetative and floral tissues (Figure 4). TINY is required for both vegetative and floral organogenesis (Wilson et al., 1996), so it ought to be expressed in these tissues. We cannot exclude the possibility that a low expression level of TINY (below the GeneChip detection limit) might be sufficient for its cellular function. The remaining 41 genes identified here have not yet been characterized. Thirty-eight of these genes showed diverse expression patterns in this set of experiments. There are three genes for which no transcripts could be detected in any of the experiments. These three genes showed sequence similarity to the previously identified AtERF4 and RAP2.8 from Arabidopsis and EREBP3 from tobacco. Similar to the AP2/EREBP gene family, sets of organ-specific genes from the AtMyb protein family and the zinc finger transcription factor family also were identified (data not shown). Clustering analysis with genes from each individual (super)family of transcription factors gave a similar experimental classification, supporting the classification generated with all 402 transcription factor genes shown in Figure 4.
The most striking finding from the clustering analysis is that ∼15% of the 402 transcription factor genes are expressed at relatively high levels in roots, compared with 6% that are leaf specific and 3% that are flower/silique specific (Figure 4; see supplemental data). These root-specific/preferential genes belong to different families of transcription factors, including members of the AP2/EREBP family, the Myb family, the HD-ZIP and bZIP families, the IAA/AXR gene family, and the zinc finger family. More than half of these genes also can be induced after attack by different pathogens (data not shown). Because the root samples analyzed were grown under several different conditions, including sterile liquid culture medium and soil mix, it is unlikely that the upregulation of such a large number of genes is attributable to either biotic or abiotic stress.
Identification of Putative Downstream Target Genes for Transcription Factors
In an attempt to identify putative downstream targets for transcription factors, we examined the promoters of genes that are induced at late time points upon cold stress and pathogen attack for potential and known binding sites for these transcription factors.
In the case of cold response, we first identified those transcription factors whose expression is activated by cold stress at an early time point (3-hr cold treatment) (Figure 5A). The early transcription factor gene cluster comprises 18 genes that encode bZIPs (GBF3), AP2/EREBP (DREB1A, DREB1B), Myb proteins (AtMyb2), zinc finger proteins, and IAA12 (see supplemental data). The previously defined conserved binding sites for some of these transcription factors are listed in Table 2. We then identified genes that are activated after a 27-hr cold treatment (late-response genes) from all of the genes on the Arabidopsis GeneChip using the hierarchical clustering methods of Eisen et al. (1998), as described in Methods. One cluster was found that includes Cor15b, Cor47, Cor78, RD29a/RD29b (the GeneChip probes cannot distinguish these genes), Iti29, and Kin1, some of which are known to be induced by cold (Figure 5A; supplemental data) (Shinozaki and Yamaguchi-Shinozaki, 2000). The available sequences 1.2 kb upstream from known or predicted translation start sites (ftp://ftp.tigr.org) for the 57 genes in the late-response cluster were searched for the occurrence of the cis elements listed in Table 2.
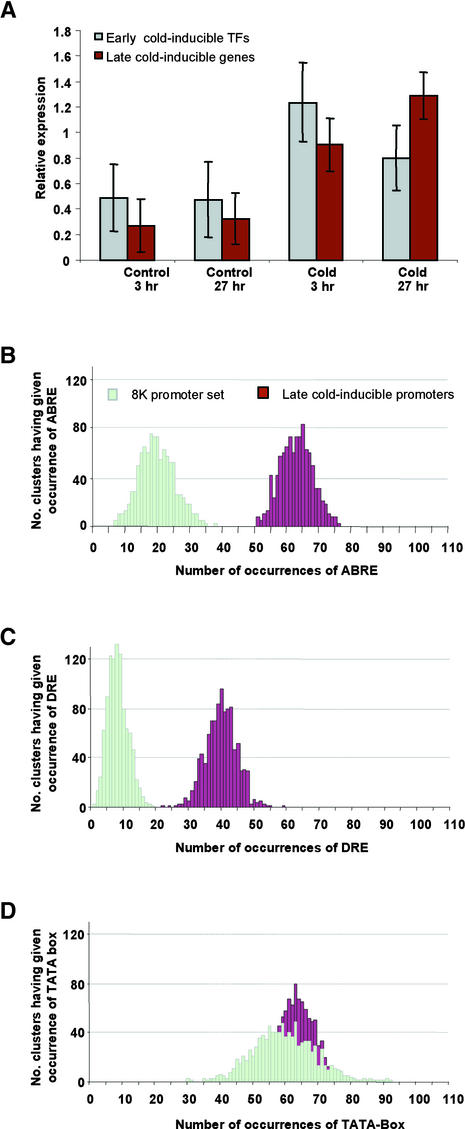
Figure 5.
ABRE-like and DRE-like Elements Are Enriched among the Promoters for Late Cold Response Genes.
(A) Expression of transcription factor genes and other genes that are induced by cold treatment. Transcription factor genes and other genes that are induced during 4°C cold treatment were selected based on self-organizing map analysis and hierarchical cluster analysis as described in Methods. The cluster shown contains transcription factor genes and the genes on the Arabidopsis GeneChip that are activated by cold treatment at either 3 hr (early) or 27 hr (late). The y axis indicates the relative expression for the genes in each cluster, with sd values indicated at the top of each bar.
(B) to (D) Occurrences of the ABRE-like element (B), the DRE-like element (C), and the TATA box (D) among the bootstrapped sets of late cold response promoters (brown bars) were compared with those among the bootstrapped control promoter sets (light green bars).
TFs, transcription factors.
Table 2.
Conserved Binding Motifs and Their Percentage of Occurrence for Different Types of Transcription Factors in the Promoter Region of Cold-Inducible, Pathogen-Inducible, and Randomly Selected Gene Clusters
| Type ofTranscriptionFactor | Name of the_cis_ Element | Sequence of the cis Element | Percentage of Occurrence in Cold-Inducible Cluster of Interest | Percentage of Occurrence in Pathogen-Inducible Cluster of Interest | Percentage of Occurrence in8K GeneChip | Reference |
|---|---|---|---|---|---|---|
| AP2/EREBP | GCC-box | GCCGCC | 5.26 | 4.87 | 7.53 | Shinozaki and Yamaguchi- Shinozaki, 2000 |
| AP2/EREBP | DRE-like | (A/G/T)(A/G) CCGACN(A/T) | 45.60 | 26.83 | 12.76 | Shinozaki and Yamaguchi- Shinozaki, 2000 |
| Myb | AtMyb1 | (A/C)TCC(A/T)ACC | 5.26 | 9.76 | 6.31 | Martin and Paz-Ares, 1997 |
| Myb | AtMyb2 | TAAC(G/C)GTT | 3.51 | 2.44 | 4.87 | Martin and Paz-Ares, 1997 |
| Myb | AtMyb3 | TAACTAAC | 14.04 | 12.20 | 5.28 | Martin and Paz-Ares, 1997 |
| Myb | AtMyb4 | A(A/C)C(A/T)A(A/C)C | 71.93 | 85.36 | 74.78 | Rushton and Somssich, 1998 |
| bZIP, TGA type | as-1/ocs element-like | TGACG | 56.14 | 63.41 | 54.50 | Schindler et al., 1992 |
| bZIP, GBF type | G-box | CACGTG | 43.86 | 12.20 | 17.11 | Schindler et al., 1992 |
| bZIP | ABRE-like | (C/G/T)ACGTG(G/T) (A/C) | 68.40 | 17.07 | 23.68 | Shinozaki and Yamaguchi- Shinozaki, 2000 |
| WRKY | W-box | TTGAC(C/T) | 73.68 | 80.49 | 67.24 | Eulgem et al., 2000 |
Two elements, the ABRE-like element and the DRE-like element (Shinozaki and Yamaguchi-Shinozaki, 2000, and references therein), occur at significantly higher frequencies in the promoters from genes in this cluster than their average frequency in all of the promoters of the genes on the Arabidopsis GeneChip (Table 2). To confirm that the differences shown in Table 2 are statistically significant, a bootstrapping analysis (Efron and Tibshirani, 1994) was performed with 1000 control promoter sets, each of which contains 57 promoters from genes that were selected randomly from the Arabidopsis GeneChip. In parallel, the late cold response cluster also was bootstrapped to generate 1000 late cold response promoter sets. A histogram then was generated to visualize the frequency distribution for the ABRE-like element and the DRE-like element in each promoter set.
As shown in Figures 5B and 5C, the frequency for the DRE-like element in each of 1000 control promoter sets ranged from 0 to 19 times, whereas the frequency for the ABRE-like element ranged from six to 40 times. We found that the frequencies for the DRE-like element and the ABRE-like element in the promoters of 57 genes in the late cold response cluster was 40 and 63 times, with a bootstrapped value for each of 1000 promoter sets from 22 to 59 and from 48 to 82, respectively. Given a 99.9% confidence interval, the DRE-like element and the ABRE-like element both are significantly overrepresented among the promoters for the late cold response cluster. As a comparison, we applied this same statistical analysis to the TATA box. As shown in Figure 5D, the frequency for the TATA box in 1000 promoter sets ranged from 30 to 92 times in each control promoter set and from 41 to 80 times in each of the 1000 bootstrapped late cold response promoters.
It is clear that there is no significant difference in the frequency of the TATA box between the control and late cold response promoter sets. So it is likely that the ABRE-like element and the DRE-like element are two major elements that are important for the transcriptional regulation of genes in the late cold response cluster. Proteins that bind to these two elements belong to the bZIP and AP2/EREBP transcription factor families, and we have found these types of known or putative transcription factor genes within the cluster of early cold-inducible genes (see supplemental data). Thus, it is possible that genes in the early transcription factor gene cluster may participate in the regulation of genes that belong to the late response gene cluster through binding to specific cis elements, which is consistent with the results reported by Stockinger et al. (1997) and Liu et al. (1998).
A similar approach was used to identify target genes in plant defense responses, including those to bacteria, fungi, oomycetes, and viruses (see supplemental data). Cluster analysis of all of the genes on the Arabidopsis GeneChip identified a cluster of 41 genes that are pathogen inducible and that encode proteins involved in metabolism, transportation, and transcription (see Methods and supplemental data). We scanned 1.2-kb upstream sequences from the known or predicted translation start sites for all of the genes within the pathogen-inducible cluster for the known cis elements listed in Table 2 and found that none of these cis elements occurs at a frequency that is statistically different from that occurring in randomly selected promoters. However, with MotifSampler (http://sphinx.rug.ac.be:8080/PlantCARE/ cgi/index.html), we found that the (T/C/G) (T/C/G) (A/T)G-AC(C/T)T sequence occurs at a much higher frequency in our pathogen-inducible cluster.
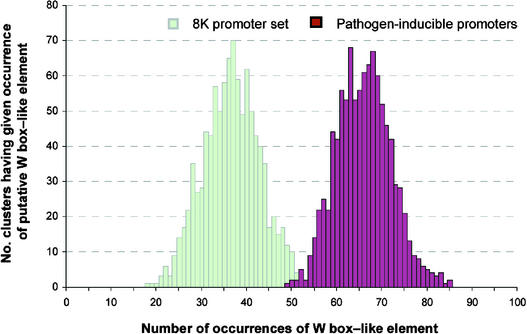
As shown in the histogram in Figure 6, the frequency for this element among 1000 control promoter sets ranged from 18 to 60, whereas the frequency for this element in the promoters of the pathogen-inducible cluster was 66, with a bootstrapped value from 49 to 85. The nonparametric U test delivers a P value < 0.001, meaning that the frequency for this element in the pathogen-inducible cluster was statistically different from that of the randomly selected cluster. When we examined this element in more detail, we found that a part of it is quite similar to the consensus binding site of WRKY transcription factors (W box). However, in W box elements, the third position is always a T, whereas in the case of the element we have identified through MotifSampler, the nucleotide at the corresponding position may be either A or T. Although the core TGAC sequence has been shown to be conserved absolutely in WRKY binding sites (Eulgem et al., 2000), it is not clear if the change from T to A would affect W box activity. It is possible that this element could be a degenerate binding site for WRKY proteins. However, it is possible as well that another type of transcription factor might recognize this site.
Figure 6.
A W Box–like Element Is Overrepresented among the Promoters of Genes in the Pathogen-Inducible Gene Cluster.
The pathogen-inducible gene cluster was selected based on self-organizing map analysis and hierarchical clustering as described in Methods. Histograms were generated as described in Figure 5 for the control promoters of all of the genes on the Arabidopsis GeneChip (light green bars) and the promoters of all of the genes within the pathogen-inducible gene cluster (brown bars).
DISCUSSION
Microarrays have been shown to be powerful tools for generating large amounts of data for parallel gene expression analysis. However, controlling the data quality remains a challenge. In this study, we analyzed the GeneChip data obtained from experiments performed under multiple conditions. To improve data quality and comparability, the following strategies were used. (1) To control the biological variation that can interfere with data interpretation, all of the samples included in the study were pooled from at least eight individual plants receiving the same treatment or from plants receiving the same treatment in replicate experiments. Therefore, the detected gene expression will be the common (average) response of the biological replicates. (2) To test the biological reproducibility of the samples used here, 11 experiments with biological replicates were performed (see supplemental data). The results demonstrated high correlations between members of each pair, with correlation coefficients > 0.90 for each pair (see supplemental data).
(3) Relatively stringent criteria were used to select differentially expressed genes. In most cases, differentially expressed genes were selected based on multiple time points of the same treatment or multiple similar treatments (such as infection by different pathogens), as illustrated in Figure 1. The twofold cutoff was chosen because when the same sample was hybridized to two chips, the false-positive rate at twofold was 0.2% (Zhu and Wang, 2000). (4) Although it is nearly impossible to confirm the measurement for each gene and each condition included in this study by other means, RNA gel analyses have confirmed GeneChip data for a limited number of genes under several different conditions studied (Zhu et al., 2001b). The strategies mentioned above are valid, because our GeneChip results are consistent with the expression patterns of several stress-inducible genes studied previously. However, confirmation of the data by other means is recommended to overcome the technical limitations of the microarrays (such as cross-hybridization between closely related genes) as well as biological variance.
It has been estimated that the Arabidopsis genome codes for at least 1533 transcription factors, ∼5.9% of its total estimated genes (Riechmann et al., 2000). Among these, ∼800 genes encode AP2/EREBP, bZIP, Myb proteins, zinc finger proteins, HD-ZIP, and AUX/IAA types of transcription factors. Because the GeneChip used covers only approximately one-third of the Arabidopsis genome (Zhu and Wang, 2000), the overall number of transcription factor genes included in this study is smaller than predicted. In spite of incomplete coverage, the expression profile matrix provides detailed information on the expression pattern of a large number of transcriptional factor genes in response to various signals. Compared with conventional methods for RNA-level analysis, such as RNA gel blot analysis, global RNA profiling methods such as GeneChip analysis represent a powerful approach to assigning possible functions to different members in each gene family. As exemplified by the case of the TGA subfamily, the matrix distinguishes genes by their expression patterns from other genes closely related at the sequence level, which normally presents a serious challenge when using other approaches, because the Arabidopsis genome contains many large gene families.
We did not apply any expression threshold to the selection of transcription factors for the data analysis because the expression levels of a number of transcription factors are not high enough to be detected with a high degree of confidence under certain conditions, although they may be expressed at a high level in other conditions. As a result of the inclusion of such low expression levels, some of our conclusions may require further validation. The expression of 18 genes that show similarity to genes encoding Myb proteins, AP2/EREBPs, or zinc finger proteins could not be detected (called “absent”) in all of the samples used in this study (Table 3). Although the samples used in this study cover broad developmental stages and various stress conditions, it is possible that these genes are expressed at a high level only in very specific situations not covered in this analysis. It also is possible that the mRNA levels of these transcription factors are very low or limited to a small number of cells in the tissue samples. In addition, we cannot exclude the possibility of gene prediction errors.
Table 3.
Genes Whose Expression Could Not Be Detected in This Study
Overlap among different signaling pathways is implied by the upregulation of common transcription factor genes. A number of transcription factor genes characterized previously as being activated by abiotic stress also were found to be activated after pathogen infection (Figures 1 and 2). These genes include GBF3, Athb-12, AtERF1, and AtWRKY6. These observations support the hypothesis that different stress signaling pathways may overlap or converge at specific points (Ingram and Bartels, 1996). It is possible that some transcription factors could represent those points of convergence. For example, the overexpression of tobacco Tsi, which encodes an AP2/EREBP-type transcription factor, enhances resistance to a bacterial pathogen as well as salt tolerance by activating genes such as PR-1 and RD29a (Park et al., 2001).
The importance of transcription factors in plant–pathogen interactions was further implied by the altered expression of transcription factor genes in mutants that have defects in salicylic acid, jasmonic acid, and ethylene signal transduction pathways. Mutant analysis of the transcription factors with reduced or abolished expression strongly suggests their participation in the salicylic acid and/or jasmonic acid/ethylene signaling network. Although ethylene has been shown to act synergistically with jasmonic acid in activating defense gene expression in many cases (Penninckx et al., 1998), the interaction of the ethylene/jasmonic acid signaling pathway with the salicylic acid signaling pathway remains controversial. Both agonistic and antagonistic interactions have been reported previously (for review, see Glazebrook, 2001). Nevertheless, communication among different signaling pathways is thought to render plants capable of defending themselves against a variety of pathogen infections (Glazebrook, 2001). Indeed, we have observed both negative and positive interactions between ethylene/jasmonic acid and salicylic acid signaling pathways in this study. For example, the expression of genes in group I in Figure 2, such as GBF3, is induced after pathogen infection and is reduced by mutations in salicylic acid signaling but is enhanced by mutations in both ethylene and jasmonic acid signaling. Conversely, the expression of genes in group II, such as RAP2.6, is reduced by mutations in both ethylene and jasmonic acid signaling but is enhanced by mutations in salicylic acid signaling.
Although this observation illustrates a negative interaction between ethylene/jasmonic acid and salicylic acid signaling, we also have identified a number of genes whose expression is reduced by all of the tested mutations that block the salicylic acid, jasmonic acid, and ethylene signaling pathways (group III; Figure 2). Similar results from microarray experiments were obtained in the study of Arabidopsis response to the fungal pathogen Alternaria brassicicola (Schenk et al., 2000). These results suggest that the plant defense signaling network is quite complex, and both positive and negative interactions among different signaling pathways may contribute to resistance to different types of pathogens. The existence of genes whose expression is not affected strongly by any of the mutations (Figure 2) may suggest additional pathways to salicylic acid and ethylene/jasmonic acid through which the corresponding encoded proteins function in response to pathogen infection. Alternatively, the pathways may play redundant roles in the regulation of these genes.
Recently, some of the Arabidopsis WRKY proteins, such as AtWRKY4, -6, -7, and -11, have been suggested to play a role in leaf senescence, because the expression of these genes is increased in senescent leaves (Eulgem et al., 2000). However, reports of transcription factors that have been suggested to be involved in leaf senescence are limited. Previous studies have shown that signals such as pathogen infection and ethylene can induce senescence, and similar sets of genes have been identified as being induced during both senescence and defense responses (for review, see Quirino et al., 2000, and references therein). We found that leaf senescence is correlated with the expression of a number of different types of transcription factor genes, including genes encoding AP2/EREBP, Myb proteins, and bZIPs as well as some of the WRKY genes (Table 1, Figure 3). Interestingly, an analysis of the promoter regions for the 23 genes that are induced during leaf senescence (Zhu et al., 2001a) showed that the consensus WRKY protein binding site is enriched significantly within the promoters of these genes (data not shown), suggesting that some of the transcription factors in this group may play a role during senescence by regulating the expression of senescence-related genes.
The majority (88%) of the 402 transcription factor genes studied here are expressed at different levels in different organs and/or at different developmental stages, suggesting that their expression is regulated at the transcriptional level. Among them, ∼15% are expressed highly in roots, and some of them are root specific (Figure 4). Although the exact biological meaning of this expression localization is unclear, the high relative expression levels of these genes in roots suggests an evolutionary adaptation of roots in response to continuous exposure to various stresses. Thus, a steady transcription level of these transcription factor genes is required to fulfill a number of requirements such as detoxification and defense response in roots. In support of this hypothesis, 50% of known or putative transcription factor genes that are highly expressed in all of the root samples tested also are induced after different types of stress treatments. In addition to the transcription factor genes, many root-specific genes identified in previous GeneChip analyses also are defense related (Zhu et al., 2001a). The fact that all types of transcription factors examined are present in this cluster expressed preferentially in roots suggests that they are required to function coordinately to control the spatial expression of root-specific genes.
In an attempt to identify downstream genes that could be possible targets for known or putative transcription factors in the plant stress response, we examined genes that are activated by different stresses at later time points compared with genes encoding various transcription factors that are upregulated at earlier time points. Several known or putative transcription factors that belong to the AP2/EREBP family, the bZIP family, the Myb family, and the zinc finger protein family are induced rapidly after cold treatment (Figure 5A; see supplemental data). Two cis elements, the ABRE-like element and the DRE-like element, which are the likely binding sites for bZIP-type and AP2/EREBP-type transcription factors, occur at significantly higher frequencies within the promoters of a cluster of genes that are activated by cold treatment at a later time point (Figures 5B and 5C). Genes in this late cold response cluster could be potential targets for the transcription factor genes in the early cold response cluster. ABRE and DRE are two elements that have been well characterized previously and that have been demonstrated to be critical for the expression of genes such as Rd or Cor in response to cold treatment (Shinozaki and Yamaguchi-Shinozaki, 2000, and references therein).
Recently, a cDNA microarray experiment with 1300 full-length Arabidopsis cDNA clones showed that the DRE-like element, including the CCGAC sequence, occurs within the promoters of 11 of 12 DREB1A target genes, and the ABRE element was found in the promoters of six of 12 DREB1A target genes (Seki et al., 2001). The statistical significance for the frequency of either of these two elements among the 1300 cDNAs was not tested, and whether or not these two elements are overrepresented in the DREB1A target gene cluster, which contains only 12 genes, is not known. Through a bootstrapping approach, we demonstrated that both the DRE-like element and the ABRE-like element are overrepresented in our late cold response gene cluster. However, 26 genes (of 57) included in this cluster contain the DRE-like element, 39 genes contain the ABRE-like element, and 46 genes contain either one, suggesting that there could be other variations of these elements. The hexanucleotide CCGAAA, which is very similar to the CCGAC core sequence of the DRE-like element, has been shown to be important in mediating low-temperature response of the barley blt4.9 gene (Dunn et al., 1998). We also identified a cluster of genes whose expression is induced by cold treatment at an early time point (3 hr) (early cold response gene cluster) and analyzed the promoters of genes in this cluster. Although the ABRE-like element was found to be enriched slightly (at a 95% confidence interval, the enrichment was not statistically significant), the DRE-like element is not enriched in this early cold response cluster.
A number of cis elements have been shown previously to be important in the plant defense response to pathogens, including the TGA binding site (as-1/ocs element) and the WRKY binding site (W box) (Rushton and Somssich, 1998). We found that although TGA and WRKY binding sites occur at relatively higher frequencies in the promoters for the pathogen-inducible gene cluster, the frequencies are not statistically greater than those occurring in randomly selected clusters. One explanation is that none of the elements listed in Table 2 is the common element that confers the response to various pathogen infections, so some other as yet unidentified element(s) could serve this function. Indeed, using the MotifSampler program, we were able to identify a sequence that occurs at a frequency significantly greater than that in control promoter sets (Figure 6). It is possible that this element could serve as a common element for plants in response to all types of pathogen infections and that the specificity for each pathogen is achieved by the combinatorial interactions among this element and other cis elements and their corresponding transcription factors (for review, see Singh, 1998). Alternatively, distinct permutations of this consensus sequence, which may be enriched in more strictly defined subclusters, may be recognized by specific members of a transcription factor family. Interestingly, this element is quite similar to the WRKY binding site found to be important in the plant stress response and to be a common element in systemic acquired resistance (Maleck et al., 2000). It needs to be determined if the element we have identified acts as a WRKY binding site and is important for plant defense responses.
In conclusion, we have identified a number of genes encoding known or putative transcription factors that are expressed specifically in particular organs, expressed at particular developmental stages, and/or induced under particular stress conditions. These genes were classified according to their expression in response to various stress treatments. The potential involvement of a number of transcription factors in different stress pathways has been illustrated by the induced expression of genes during stress treatments. Furthermore, their importance was further implied by their reduced or abolished induction in mutants that are defective in plant defense signaling. However, the roles they play in development and in plant stress responses need to be verified using other approaches, such as reverse genetics. With the blossoming of Arabidopsis functional genomics, the role for each transcription factor suggested through this expression profiling analysis can be tested rapidly.
METHODS
Identification of 402 Known and Putative Transcription Factors
Potential stress-related genes that encode known or putative transcription factors on the Arabidopsis GeneChip (Affymetrix, Santa Clara, CA) were identified based on the annotation associated with probe sets on the chip. Additional genes were identified by searching for conserved domains. The nucleotide and amino acid sequences from conserved domains for AP2/EREBPs, Myb proteins, bZIPs, and WRKY zinc finger proteins were used to blast against the TIGR Arabidopsis thaliana database (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/PSEUDOMOLECULES/), using the BLASTN, BLASTX, and BLASTP programs (Altschul et al., 1997), to generate the entire list of known or putative transcription factor genes of these families. Homologs (E value < 1E − 20) of the list members represented on the GeneChip were included in the analysis.
Data Set Collection, Data Processing, and Data Analysis
All of the RNA samples used in this study are described in the supplemental data. For Arabidopsis GeneChip experiments, RNA samples were extracted and subsequent cDNA synthesis, array hybridization, and overall intensity normalization for all of the arrays for the entire probe sets were performed as described by Zhu et al. (2001a). The average difference (expression level) for the selected 402 genes then was extracted from the data. Any average difference that was <5 was floored to 5. Then, to generate Figure 1, the fold change was calculated for each gene by dividing the average difference from various stress-treated samples by the average difference from the corresponding mock-treated control samples. Genes with average differences equal to 5 (which were called “absent”) across all of the experiments were eliminated from further analysis. In these stress response experiments, the logarithms (base 2) of the fold change values for each gene were subjected to normalization across all of the samples. In the case of studying developmental control and organ-specific gene expression (Figures 3 and 4), the floored average difference, rather than the fold change, was subjected directly to log2 transformation followed by mean centering across each gene. All of the processed data then were subjected to the self-organizing map algorithm followed by complete linkage hierarchical clustering of both genes and experiments, using Cluster/Treeview (Eisen et al., 1998) (Figures 1, 2, and 4), or to the self-organizing maps algorithm for genes, using GeneCluster 1.0 (Tamayo et al., 1999) (Figure 3).
Mutant Analysis
Four-week-old Arabidopsis Columbia wild-type plants and various mutant and transgenic plants were infected with Pseudomonas syringae pv maculicola ES4326 (106 colony-forming units) for 30 hr. The infected leaf samples then were collected and subjected to GeneChip analysis. Transcription factor genes that are induced by at least twofold in wild-type plants after infection with P. syringae pv maculicola ES4326 (with an Affymetrix present call) were identified. To generate Figure 2, the fold change was calculated by dividing the average difference from mutant or transgenic plant samples by the average difference from the wild-type plant sample. Then, the logarithms (base 2) of the fold change values were subjected to cluster analysis as described above.
Identification of the Cold Response Cluster and the Pathogen-Inducible Cluster
The cluster of cold-inducible genes was selected based on only one time point, either early or late cold treatment. Three-week-old Columbia wild-type plants grown on sterilized Murashige and Skoog (1962) (MS) agar medium at 22°C under a 12-hr/12-hr light/dark cycle were transferred to fresh MS liquid medium for several days of equilibration before treatment. Salt, osmotic, and cold stresses then were applied by replacing the medium with new MS medium containing 100 mM NaCl or 200 mM mannitol or incubating at 4°C. Tissues from the aerial or root portions of control and treated plants were collected, RNA purified, and subjected to GeneChip analysis. Genes from the Arabidopsis GeneChip that were induced by at least twofold after any stress treatment described above (with an Affymetrix present call) were selected. Then, the average differences of these genes were log2 transformed, mean centered, and subjected to the Cluster/Treeview program as described above. Clusters of genes whose expression was induced preferentially by 3-hr (early transcription factor gene cluster) and 27-hr (late response gene cluster) cold treatments were identified; they are described in the supplemental data.
Genes within the pathogen-inducible cluster were identified as follows. First, genes from the Arabidopsis GeneChip that were induced by at least twofold after any pathogen infection and at any time point (with an Affymetrix present call) were selected. Then, the fold change was calculated for each gene by dividing the average difference from various pathogen-treated samples by the average difference from the corresponding mock-treated samples, followed by log2 transformation. The processed data were subjected directly to the Cluster/Treeview program as described above. Clusters of genes whose expression was induced by all pathogens at all time points were identified; they are described in the supplemental data.
Statistical Analysis of Frequency for Elements That Occur within Promoters
Arabidopsis genomic sequence was obtained from TIGR (ftp://ftp.tigr.org). Sequences 1.2 kb upstream from known or predicted coding sequences that are present on the chip were extracted and used to search for the cis elements listed in Table 2 using a custom Perl script. Then, bootstrapping was performed by generating 1000 control promoter sets from genes on the chip and 1000 bootstrapped promoter sets from genes either in the late cold response gene cluster or in the pathogen-inducible gene cluster. Bootstrapped sets were generated using another custom Perl script.
Supplementary Material
[Supplemental Data]
Acknowledgments
We thank Devon Brown and Bin Han for technical assistance with preparing samples used in the microarray experiments, Bin Han for help in conducting the microarray experiments, and Dr. Rongling Wang for providing the Perl script for searching for elements within the promoters of genes from the Arabidopsis GeneChip and for the suggestion of using the bootstrap method for promoter analysis. T.E. is a recipient of a Deutsche Forschungsgemeinschaft postdoctoral fellowship.
W⃞ Online version contains Web-only data.
References
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284**,** 2148–2152. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaeffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**,** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis athb-8 hd-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126**,** 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden, K.L. (2000). RING domains: Master builders of molecular scaffolds? J. Mol. Biol. 295**,** 1103–1112. [DOI] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88**,** 57–63. [DOI] [PubMed] [Google Scholar]
- Capili, A.D., Schultz, D.C., Rauscher, I.F., and Borden, K.L. (2001). Solution structure of the PHD domain from the KAP-1 corepressor: Structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20**,** 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rekka, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266**,** 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dunn, M.A., White, A.J., Vural, S., and Hughes, M.A. (1998). Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol. Biol. 38**,** 551–564. [DOI] [PubMed] [Google Scholar]
- Efron, B., and Tibshirani, R.J. (1994). Random samples and probability. In An Introduction to the Bootstrap: Monographs on Statistics and Applied Probability No. 57, pp. 17–28.
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95**,** 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8**,** 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (1999). Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18**,** 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5**,** 199–206. [DOI] [PubMed] [Google Scholar]
- Feys, B., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6**,** 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10**,** 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12**,** 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4**,** 301–308. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47**,** 377–403. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280**,** 104–106. [DOI] [PubMed] [Google Scholar]
- Jensen, R.B., Jensen, K.L., Jespersen, H.M., and Skriver, K. (1998). Widespread occurrence of a highly conserved RING-H2 zinc finger motif in the model plant Arabidopsis thaliana. FEBS Lett. 436**,** 283–287. [DOI] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19**,** 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G., Van Montagu, M., and Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6**,** 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17**,** 287–291. [DOI] [PubMed] [Google Scholar]
- Kawata, T., Imada, T., Shiraishi, H., Okada, K., Shimura, Y., and Iwabuchi, M. (1992). A cDNA clone encoding HBP-1b homologue in Arabidopsis thaliana. Nucleic Acids Res. 20**,** 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8**,** 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.H., and Chun, J.Y. (1998). A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol. Biol. 37**,** 377–384. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10**,** 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G., Paul, A.L., McCarty, D.R., and Ferl, R.J. (1996). Transcription factor veracity: Is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 8**,** 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26**,** 403–410. [DOI] [PubMed] [Google Scholar]
- Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13**,** 67–73. [DOI] [PubMed] [Google Scholar]
- Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119**,** 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Z.H., Liu, X., and Lam, E. (1994). TGA3 is a distinct member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 25**,** 1–11. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15**,** 473–497. [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94**,** 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.M., Park, C.J., Lee, S.B., Ham, B.K., Shin, R., and Paek, K.H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13**,** 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10**,** 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino, B.F., Noh, Y.S., Himelblau, E., and Amasino, R.M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5**,** 278–282. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290**,** 2105–2110. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence and defense related processes. Plant J., in press. [DOI] [PubMed]
- Rushton, P.J., and Somssich, I.E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1**,** 311–315. [DOI] [PubMed] [Google Scholar]
- Salinas-Mondragon, R.E., Garciduenas-Pina, C., and Guzman, P. (1999). Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol. Biol. 40**,** 579–590. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97**,** 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Beckmann, H., and Cashmore, A.R. (1992). TGA1 and G-box binding factors: Two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4**,** 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi- Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13**,** 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3**,** 217–223. [PubMed] [Google Scholar]
- Singh, K.B. (1998). Transcriptional regulation in plants: The importance of combinatorial control. Plant Physiol. 118**,** 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman, E., Mattsson, J., and Engstrom, P. (1996). The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 10**,** 375–381. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12**,** 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a _cis_-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94**,** 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E.S., and Golub, T.R. (1999). Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96**,** 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121**,** 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93**,** 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8**,** 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Miao, Z., and Lam, E. (1997). DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34**,** 403–415. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., and Schmidt, R.J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17**,** 209–214. [DOI] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13**,** 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Foley, R.C., and Singh, K.B. (1993). Isolation and characterization of two related Arabidopsis ocs-element bZIP binding proteins. Plant J. 4**,** 711–716. [DOI] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10**,** 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T., and Wang, X. (2000). Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol. 124**,** 1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T., Budworth, P., Han, B., Brown, D., Chang, H.S., Zou, G., and Wang, X. (2001. a). Toward elucidating the global gene expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by high-density oligonucleotide probe array. Plant Physiol. Biochem. 39**,** 221–242. [Google Scholar]
- Zhu, T., Chang, H.-S., Schmeits, J., Gil, P., Shi, L., Budworth, P., Zou, G., Chen, X., and Wang, X. (2001. b). Gene expression microarrays: Improvements and applications towards agricultural gene discovery. J. Assoc. Lab. Automat. 6**,** 95–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]