Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease (original) (raw)
Abstract
The congenital polycystic kidney (cpk) mutation is the most extensively characterized mouse model of polycystic kidney disease (PKD). The renal cystic disease is fully expressed in homozygotes and is strikingly similar to human autosomal recessive PKD (ARPKD), whereas genetic background modulates the penetrance of the corresponding defect in the developing biliary tree. We now describe the positional cloning, mutation analysis, and expression of a novel gene that is disrupted in cpk mice. The cpk gene is expressed primarily in the kidney and liver and encodes a hydrophilic, 145–amino acid protein, which we term cystin. When expressed exogenously in polarized renal epithelial cells, cystin is detected in cilia, and its expression overlaps with polaris, another PKD-related protein. We therefore propose that the single epithelial cilium is important in the functional differentiation of polarized epithelia and that ciliary dysfunction underlies the PKD phenotype in cpk mice.
Introduction
Polycystic kidney disease (PKD) represents a set of hereditary nephropathies characterized by progressive cyst formation, massive renal enlargement, and often, progression to end-stage renal disease. Autosomal dominant PKD (ADPKD) occurs in 1 in 1,000 individuals and, in addition to renal cystic disease, is associated with cyst formation in other epithelial organs, most notably the liver, as well as connective tissue defects, such as intracranial aneurysms, aortic dissection, and cardiac valve defects (1). Mutations in either of two genes, PKD1 and PKD2, cause ADPKD phenotypes that are virtually indistinguishable (2). In comparison, autosomal recessive PKD (ARPKD) is much less frequent (1 in 20,000 live births). The clinical phenotype is dominated by renal collecting duct ectasia, biliary dysgenesis, and portal tract fibrosis (3). The principal disease locus, PKHD1, has been mapped to chromosome 6p21.1-p12 (4, 5), but the gene has yet to be identified.
In the mouse, several distinct, recessively acting mutations cause PKD phenotypes that mimic human disease (6). Among these models, the congenital polycystic kidney (cpk) mutation is the most extensively characterized. The cpk locus on chromosome 12 is defined by a single recessive mutation that arose spontaneously in the C57BL/6J strain (7). The renal phenotype is fully expressed in homozygotes and is strikingly similar to human ARPKD (8, 9), whereas genetic background modulates the penetrance of the corresponding defect in the developing biliary tree (10, 11).
Multiple cellular and extracellular matrix abnormalities have been described in cpk/cpk kidneys. These changes include: (a) enhanced expression of the proto-oncogenes, c-myc, c-fos, c-Ki-ras (12–14); (b) increased expression of the transcriptional repressor, Cux-1, a putative inhibitor of terminal differentiation (15); (c) enhanced growth factor expression (16); (d) apical mislocation of a functional EGF receptor (17); (e) increased expression of basement membrane constituents, laminin β1, and γ1, α1/α2 chains of collagen IV, collagen I, and fibronectin (18, 19); (f) overexpression of the basement membrane remodeling enzymes, matrix metalloproteinases (MMPs), and their specific tissue inhibitors, TIMPs (20); (g) abnormal expression of epithelial cell adhesion molecules (21, 22); and (h) alterations in steroid metabolism and lipid composition (23–25). These numerous abnormalities involving a wide range of developmentally regulated cellular processes suggest that cpk/cpk mutant kidneys are unable to complete the terminal phases of tubuloepithelial differentiation (26).
Here we describe the positional cloning, mutation analysis, and expression of a novel gene that is disrupted in cpk mice. When expressed exogenously in polarized renal epithelial cells, the 145 amino acid protein product, cystin, is detected in cilia in a pattern that overlaps with the expression of polaris, another PKD-related protein.
Methods
Genotyping.
Genotyping was performed of informative meioses from our previous crosses (27) and 150 test meioses generated in an intercross of F1 progeny from a cross between C57BL/6J–_+/cpk_ heterozygotes and DBA/2J mice.
The bacterial artificial chromosome (BAC) end–derived markers from the cpk critical interval, 221A10-SP6, 182L8-T7, 06H17-SP6, 49G13-T7, and 319I11-T7, and the _Rrm2_-derived polymorphic marker have been described (27). Single-strand conformation polymorphisms were amplified in the presence of [α-32P] dCTP, electrophoresed on nondenaturing MDE gels (FMC BioProducts, Rockland, Maine, USA), and detected by autoradiography.
One recombinant was identified between cpk and the proximal flanking marker, 221A10-SP6, and four recombinants were detected between cpk and the distal flanking marker, Rrm2.
Large-scale sequencing of BAC 221A10.
DNA from the mouse BAC clone, 221A10, from the RPCI-22 mouse BAC library (Research Genetics, Huntsville, Alabama, USA) was nebulized and size-fractionated on a 0.8% agarose gel. Fragments 2–3 kb and 3–4 kb in size, subcloned into pUC18, were shotgun sequenced to produce threefold coverage of the BAC clone. Library DNA preparation and sequencing on an ABI377 sequencer were performed by the Australian Genome Research Facility (Brisbane, Australia). Assembly and analysis of genomic sequence data was performed using PHRED, PHRAP, and GAP4 software (28–30).
The final sequence was analyzed using the basic local alignment search tool (BLAST; http://www.ncbi.nlm.nih.gov/BLAST); REPEATMASKER (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker); GENSCAN (http://CCR-081.mit.edu/GENSCAN.html); and GrailPRO (http://www.genomix.com/grailpro.htm).
cDNA characterization and sequence analysis.
Total RNA was extracted from kidneys and livers of 2-week-old B6–wild-type (B6-WT) and B6-cpk/cpk mice as well as from fetal brain, lung, liver, and kidney of B6 mice at embryonic day 15 (E15) using the RNAgents Total RNA Isolation System (Promega Corp., Madison, Wisconsin, USA). Poly(A)+ RNA was prepared using the Oligotex mRNA Midi kit (QIAGEN Inc., Valencia, California, USA).
A predicted cpk cDNA sequence was assembled using the sequences in UniGenes Mm.34424 and Mm.52265 as a scaffold. Primary and nested PCRs were performed using the following primers (oriented 5′ to 3′ in the cDNA): C-1F: 5′-CATCTCCGGCTCTCCTTTTCTGT-3′; C-1R: 5′-AGAGTAAGCGGGATGAAGAGAGG-3′; C-2F: 5′- AGATGATTCTTTCGCCCTGACTTC-3′; C-2R: 5′-AGGGG-GATTCTGGAGGAGTGAG-3′; C-3F: 5′-TCCTCCCTCCCTATCTCTCCAT-3′; C-3R: 5′-ATCCAGCAGGCGTAGGG-TCTC-3′; C-4F: 5′-AGACCCTACGCCTGCTGGATCA-3′; C-4R: 5′-TTGTCCAGCTCAGCGGCAGTA-3′; C-5F: 5′-AACAGCCCCAAGAGACCCGAG-3′; and C-5R: 5′-GTTGCTAGCTCTGGGAGGTTTT-3′. To obtain the 5′ end of the cDNA sequence, we amplified cDNA from mouse kidney by 5′ rapid amplification of cDNA ends–PCR (RACE-PCR) using the Marathon cDNA Amplification kit (BD Biosciences Clontech, Palo Alto, California, USA).
Nucleotide comparison of the cpk cDNA sequence with known genes and expressed sequence tags (ESTs) listed in the nonredundant compilation of the GenBank and European Molecular Biology Laboratory databases were performed using BLAST searches (31). We also performed amino acid comparisons with the nonredundant Swiss-Prot databases using the BLASTP program. Protein-domain homologies and motifs were analyzed with the PredictProtein package (http://cubic.bioc.columbia.edu/predictprotein/).
RT-PCR and Northern blot analysis.
Reverse transcription was carried out in a 40-μl reaction volume with 10 μg of total kidney RNA using the Gibco-BRL Superscript RNAse H-Reverse Transcriptase kit (Life Technologies Inc., Gaithersburg, Maryland, USA). Aliquots of 100 ng of cDNA were amplified in PCR reactions using various cDNA primer combinations under standard PCR conditions (30 cycles of 96°C for 30 seconds, 54–64°C for 30 seconds, 72°C for 30 seconds).
Northern blots prepared with either 3 μg of kidney and liver poly(A+) RNA pooled from B6-WT and B6-cpk/cpk mice (n = 5) or 3 μg of brain, lung, liver, and kidney poly(A+) RNA pooled from E15 mice (n = 12) were probed with a 351-bp probe that encompasses the tandem deletion (primers C-3F and C-3R). This probe was also hybridized to a mouse multiple tissue Northern blot (7762-1; CLONTECH Laboratories Inc.) as well as a human fetal Northern blot (7756-1; CLONTECH Laboratories Inc.). To evaluate differential RNA loading, we screened for housekeeping gene expression using _Gapdh_-specific primers: forward 5′-TGGAGCCAAACGGGTCATCATCT-3′ and reverse 5′-GAAGAGTGGGAGTTGCTGTTGAA-3′.
Southern blot analysis.
Using primers C-3F and C-4R, we generated a 500-bp probe from neonatal kidney cDNA that contained the complete open reading form (ORF). This probe was hybridized to _Eco_RI-digested genomic DNAs from multiple species (Zoo-blot, 7753-1; CLONTECH Laboratories Inc.) as well as to a panel of B6-WT and B6-cpk/cpk genomic DNAs digested with _Eco_RI. The blots were washed serially with 2× SSC 0.1% SDS at RT, 0.5× SSC 0.1% SDS at 65°C, and 0.2× SSC 0.1% SDS at 65°C.
Transgene construction and cell culture transfection.
A PCR fragment (_Bam_HI-_Eco_RI) containing the Kozak consensus sequence, and the complete putative ORF was amplified from wild-type kidney cDNA and subcloned into the pEF6/Myc-His A vector (Invitrogen Corp., San Diego, California, USA). The construct was verified by sequence analysis.
Transfection assays for expression and localization of cystin were performed in a cortical collecting duct cell line (mCCD) isolated from the kidney of a mouse transgenic for the early region of SV40 large T antigen (32). Transfection of mCCD cells with the wild-type cystin cDNA expression construct was performed as previously described using Lipofectamine Plus (Life Technologies Inc.) (33). Stable cell lines were established by drug selection with 15 μg ml–1 Blasticidin.
Immunolocalization studies.
The mCCD cells transfected with the wild-type cystin construct were grown in DMEM-F12 containing Earle’s balanced salt solution (Cellgro; Mediatech Inc., Herndon, Virginia, USA) supplemented with 10% FBS (HyClone Laboratories, Logan, Utah, USA), 100 U/ml penicillin, 100 mg/ml streptomycin, in 5% CO2/95% air at 37°C. For expression studies, cells were plated at near confluence on 6-mm Falcon tissue culture inserts (3104; Becton Dickinson and Co., Franklin Lakes, New Jersey, USA).
Immunolocalization studies were performed on confluent cultures grown for a minimum of 3 days to establish full epithelial polarization. Cells were fixed in 4% paraformaldehyde, 0.2% Triton in PBS for 10 minutes, incubated in blocking buffer (1% BSA in PBS), followed by 60-minute incubation with the primary Ab’s diluted in blocking buffer. The primary Ab’s used in this study were the α-myc mouse mAb (R950-25 diluted 1:250; Invitrogen Corp.), α-his-15 rabbit polyclonal Ab (SC-803, diluted 1:250; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA), α-beta-tubulin mouse mAb (Mu178-UC diluted 1:200; BioGenex Laboratories, San Ramon, California, USA), α-ZO-1 rat mAb (diluted 1:2; gift of D.F. Balkovetz, University of Alabama at Birmingham), and α-polaris rabbit polyclonal Ab (34) (GN593 diluted 1:200). After washing with PBS, cells were incubated for 1 hour with fluorochrome-conjugated secondary Ab’s diluted 1:200 in blocking buffer. The secondary Ab’s were goat anti-mouse IgG (O-6380, Oregon green; Molecular Probes Inc., Eugene, Oregon, USA), donkey anti-rabbit IgG (tetramethyl rhodamine isothiocyanate [TRITC], 711-025-152; Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA), and donkey anti-rat IgG (TRITC, 712-295-153; Jackson ImmunoResearch Laboratories Inc.). Nuclei were stained for 5 minutes using HOECHST 33528 (Sigma Chemical Co., St. Louis, Missouri, USA) diluted in 1:1,000 in PBS.
The complete sequence of BAC 221A10 and the 1.86-kb cpk cDNA sequence have been submitted to GenBank (accession numbers AF390547 for the BAC 221A10 and AF390548 for the 1856 bp mRNA).
Results
Refinement of the cpk candidate interval and generation of a transcript map.
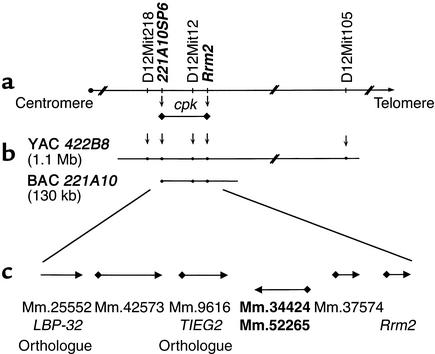
We have constructed an integrated genetic and physical map of the 650-kb region containing the cpk locus (27). Recombinational mapping (data not shown) has allowed us to refine the cpk locus to an approximate 100-kb interval, centered on D12Mit12 and bounded by the markers 221A10-SP6 and Rrm2 (Figure 1a). This interval is contained within a single BAC clone, 221A10 (Figure 1b).
Figure 1.
Integrated genetic and physical map of the cpk candidate interval. Recombination analyses refined the cpk locus to an approximately 100-kb interval, centered on D12Mit12 and delimited by 221A10SP6 and Rrm2 (indicated in bold italics; a). These data positioned cpk on a single BAC, 221A10, within our yeast and bacterial artificial cromosome (YAC/BAC)-based physical map (b). Computational analyses identified six putative transcriptional units within the cpk interval, and each predicted gene corresponded, at least in part, to a mouse UniGene cluster (c).
Computational analyses of the BAC sequence, obtained using a random shotgun method, identified six putative transcriptional units that corresponded to seven mouse UniGene clusters (Figure 1c). Previously, we excluded mouse Rrm2 as a cpk candidate gene (27). Comparative sequence analysis revealed no base pair variations in the mouse orthologues of LBP-32 and TIEG2 amplified from wild-type and cpk/cpk kidney cDNA (data not shown). RT-PCR indicated that the mRNAs corresponding to UniGene Mm.37574 were not expressed in the kidney or liver. While similar analyses with Mm.42573 demonstrated expression in kidney and liver, no transcripts were identified by Northern blotting. Therefore, no further analyses were performed for either UniGene cluster.
Identification of the cpk transcript and characterization of the genomic sequence.
Using RT-PCR, 5′ RACE-PCR of a mouse kidney adaptor-ligated cDNA library, and direct sequence analysis, we determined that the UniGene clusters, Mm.34424 and Mm.52265, represent the 5′ and 3′ ends, respectively, of a single transcript. We compiled a 1,856-bp full-length cDNA as well as a 1,786-bp splice variant, in which 70 nucleotides corresponding to residues 1422–1492 were removed. An ATG codon situated 905 nt downstream from the first nucleotide lies within a Kozak consensus sequence (35), followed by an predicted ORF of 435 bp and either a 513-bp or 443-bp 3′ UTR that contains an atypical polyadenylation signal (ATTAAA) 22 nt upstream of the poly(A+) tail. The 1,856-bp cpk cDNA shares 99% nucleotide homology through the ORF and 3′ UTR with a recently reported cDNA, AK013490, from the RIKEN full-length enriched library.
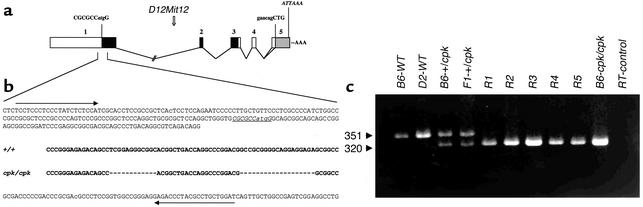
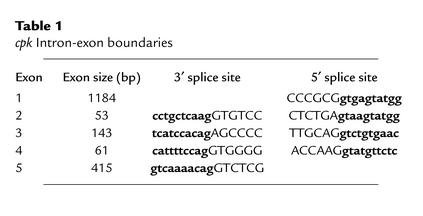
Alignment of the candidate cpk cDNA and BAC genomic DNA sequences demonstrated that the gene is encoded in five exons spanning 14.4 kb of genomic DNA (Figure 2a). The exon-intron boundaries for these five exons (Table 1) conform to the consensus sequences for 5′ and 3′ splice sites (36). Analysis of the genomic sequence 3,000 bp upstream of the ATG codon, using the TSSG and TSSW programs, did not identify a robust transcription start site. Exon 1 contains the ATG start site and has 100% homology with all 18 ESTs contained in UniGene Mm.34424 as well as 15 of the 38 ESTs that belong to Mm.52265. The ORF extends into exon 3, whereas exons 4 and 5 are apparently untranslated. A putative cryptic splice site within exon 5 would account for the 1,856-bp and 1,786-bp splice variants.
Figure 2.
Organization of the cpk genomic sequence and identification of the cpk mutation. (a) Alignment of the cpk cDNA, the UniGene consensus sequences, and the BAC genomic sequence demonstrated that the cpk gene is encoded in five exons spanning 14.4 kb of genomic DNA. The first nucleotide of the cDNA corresponds to the first nucleotide of exon 1, which spans 1,184 bp and is the largest of the five exons. This exon contains an ATG start site that lies within a Kozak consensus sequence (CGCGCC_atg_G). The 435-bp ORF extends into exon 3. Exons 4 and 5 are apparently untranslated, and a putative cryptic splice site (gaacagCTG) within exon 5 appears to account for the 1,856-bp and 1,786-bp (gray box) splice variants. An atypical polyadenylation signal (ATTAAA) lies 22-nt upstream of the poly(A+) tail. Of note, the microsatellite marker, D12Mit12, lies within intron 1 of the cpk gene. (b) PCR amplification and direct sequence analysis identified tandem 12-bp and 19-bp deletions in exon 1 of the cpk gene. The comparative sequence is indicated in bold text. The resulting frameshift truncates the predicted protein. The position of the PCR primers is identified by arrows. The putative Kozak sequence is underlined and italicized. (c) Primers flanking the tandem deletion in the cpk mutant allele amplify a 351-bp product from wild-type (B6 and D2) DNA, a 320-bp product from B6-cpk/cpk DNA, and both bands from B6_-+/cpk_ and F1 +/cpk heterozygotes. In the key F2 cpk/cpk recombinants (R1–R5), only the 320-bp mutant allele was amplified.
Table 1.
cpk Intron-exon boundaries
Mutation analysis.
Comparative sequence analysis of the candidate cpk kidney cDNA amplified from each of eight, 2-week-old B6-WT and B6-cpk/cpk mice revealed a tandem deletion of 12 bp (992delTCGGAGGGCGGC) and 19 bp (1017delGCCGCGGGGCAGGAGGAGA) in the mutant cDNA resulting in a frameshift within exon 1 that truncates the protein (Figure 2b). The occurrence of this tandem deletion on the B6 background provides strong supportive evidence that this candidate cDNA is indeed associated with the cpk phenotype.
The exon 1 primer set amplified a 351-bp product from B6-WT genomic DNA, whereas a 320-bp product was amplified from B6-cpk/cpk genomic DNA (Figure 2c). Both products were amplified from DNAs of B6_-+/cpk_ and D2 × B6_-+/cpk_ F1 heterozygotes, but only the mutant 320-bp allele was amplified from all five key F2 cpk/cpk recombinants.
We have yet to define the molecular mechanisms leading to this complex mutation. While the sequence flanking the tandem deletion is very GC-rich, extensive genomic analysis failed to reveal homologous sequences at the breakpoints. We therefore speculate that, as proposed for the complex genomic rearrangements described in a mouse fld mutant allele (37), the mutation event in this gene involved a nonhomologous recombination mechanism.
Expression of the cpk transcript and characterization of the protein product.
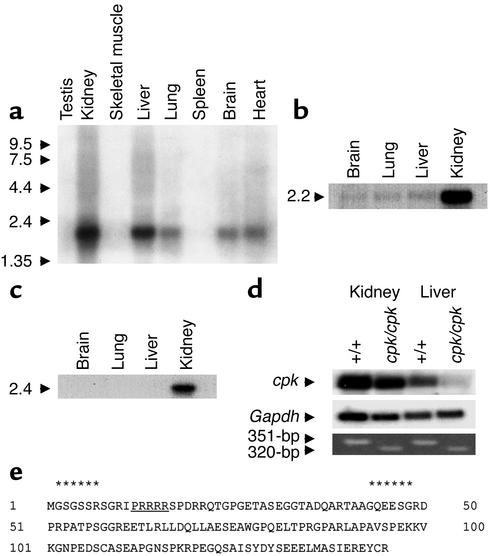
The cpk phenotype primarily involves renal cysts and biliary tract dysgenesis. In adult mice, we detected a 2.2-kb transcript that was strongly expressed in the kidney (Figure 3a). A similar 2.2-kb transcript was also detected in the liver, with lower expression evident in lung, brain, and heart. We also analyzed the expression of cpk in mouse and human fetal tissues. Northern blot analysis revealed that the 2.2-kb transcript was strongly expressed in mouse fetal kidney, but barely perceptible in fetal brain, lung, and liver (Figure 3b). Similarly, a somewhat larger transcript of 2.4-kb was expressed predominantly in human fetal kidney (Figure 3c).
Figure 3.
Expression pattern of the cpk transcript and characterization of the predicted protein product. (a) The tissue distribution of the 2.2-kb cpk transcript is shown in adult mouse tissues. The size of the polyadenylated transcript is consistent with the 1,856-bp full-length cDNA. The size markers are indicated on the left and expressed as kilobases. (b) Northern blot analysis of mouse fetal poly(A+) RNA revealed a 2.2-kb transcript in the fetal kidney. (c) Northern blot analysis of human fetal poly(A+) RNA revealed a 2.4-kb transcript in the fetal kidney. (d) The relative expression of the 2.2-kb cpk transcript is shown in kidney and liver poly(A+) RNA from 2-week-old B6-WT (n = 5 mice) and B6-cpk/cpk (n = 5 mice) (top panel). Rehybridization with Gapdh cDNA served as a loading control (middle panel). RT-PCR was performed using the deletion-flanking primer pairs on the same poly(A+) RNAs (bottom panel). (e) Amino acid sequence of the predicted 145-AA protein product. Two potential myristoylation sites (residues 2–7, 43–48, indicated by asterisks) are predicted, the first of which is coupled to a polybasic domain (underlined).
The 2.2-kb cpk transcript was detected in kidney and liver from both 2-week-old B6-WT and B6-cpk/cpk mice, but the band intensities were relatively diminished in tissues from mutant mice (Figure 3d). These data indicate that both the wild-type and the mutant cpk transcripts are more abundantly expressed in the kidney than in the liver and the truncating mutation reduces transcript expression in both tissues. Loading controls and RT-PCR analysis confirmed the specificity of these Northern blot results and demonstrated the presence of small amounts of cpk transcript in mutant liver (Figure 3d).
Characterization of the protein product.
The cpk gene encodes a novel, hydrophilic protein of 145 amino acids that we have termed cystin (Figure 3e). Secondary structural predictions failed to identify significant alpha helical or beta sheet structures within the protein. Motif searches have identified two potential myristoylation sites (amino acids [AA] 2–7 and AA 43–48), the first of which is coupled to a polybasic domain (AA 12-16). A potential PEST — polypeptide enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) — sequence is predicted at position 101–118 (PEST score = 6.98). There is no evidence for specific DNA-binding motifs, but the protein is proline rich (10%) and is predicted to have at least two potential nuclear localization signals (PRRRRSP at AA 12–18 and PEKKVKG at AA 96–104).
Southern analyses indicate that cpk homologues exist as single-copy genes in mammals including humans, monkeys, rats, dogs, and cows, as well as in chickens (data not shown). However, no significant homology to previously characterized proteins or protein domains was revealed in searches of human, Caenorhabditis elegans, Drosophila, or yeast databases. Of note, the genomic interval flanked by TIEG2 and RRM2 that includes the human cpk orthologue is not contained in the draft sequence currently available from the International Human Genome Sequence Consortium (http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html).
Northern analysis in human fetal tissues identified a 2.4-kb transcript in the kidney but not liver, brain, or lung (Figure 3c). A human fetal library–derived EST, AA446394, has 85% identity with cpk over 88 nucleotides and 71% amino acid identity with the carboxy terminus of the cystin protein (data not shown). However, the full-length human cpk orthologue has yet to be identified.
Transfection studies and subcellular localization.
Subcellular localization of the cystin protein was analyzed in mCCD cells transfected with an expression construct containing the complete cystin coding region cloned in frame with carboxyl terminal c-myc (myc) and histidine (his) tags, under the control of the EF-1α promoter. Stable cell cultures were established by selection in Blasticidin. The cells were then grown on cell culture inserts for a minimum of 3 days after confluence to allow cilial development. These polarized cells are not a clonal cell line, but rather represent a cultured cell population that has been subjected to transfection with the cystin transgene. Because the transfection efficiency in such experiments is typically less that 100%, some cells in this population did not express the transgene.
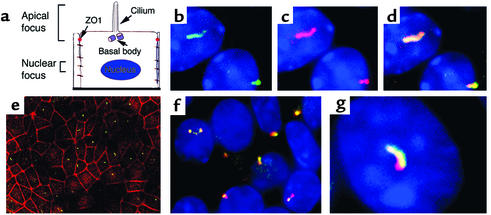
Immunofluorescence analysis revealed that the epitope-tagged cystin protein localized to a small domain on the apical cell surface (Figure 4, b–d). In a broad-field view, cystin was positioned in the center of this apical domain when compared with tight junction staining by α-ZO-1 (38) (Figure 4e). The distribution of cystin overlapped with β-tubulin IV, a known component of the single apical cilium (39) (data not shown), as well as polaris (Figure 4, f and g), a protein that is encoded by the Tg737 gene and associated with motile and immotile cilia (34). However, unlike polaris, which is expressed primarily in the ciliary basal bodies as well as in the axoneme, cystin expression appeared to be enriched in the ciliary axonemes, with minor staining in the basal bodies (Figure 4g). The immunofluorescence images are representative of n = 5 experiments.
Figure 4.
Localization of exogenously expressed cystin in stably transfected mCCD cells. To analyze cystin localization, wild-type mCCD cells were transfected with a myc and his epitope–tagged cystin construct. Stable cell cultures were established by selection in Blasticidin. Immunofluorescence analysis was conducted in cells grown on cell culture inserts for a minimum of 3 days after confluence to allow cilial development. (a) Schematic diagram of a principal cell with its primary apical cilium. The two focal planes used in immunofluorescence imaging are indicated. The apical focal plane was used to capture the cilium and the position of the tight junction, indicated by α-ZO-1 staining, and the nuclear focal plane identified the HOECHST-stained nuclei (blue). (b) Immunofluorescence localization of cystin (green) as determined using the anti-his rabbit polyclonal Ab. (c) Cystin (red) localization in the same mCCD cells as shown in b when probed with the anti-myc mAb. (d). Merged image of b and c demonstrated colocalization (yellow) of the myc and his epitope–tagged cystin. (e) In a broad-field view, staining with the anti-his polyclonal Ab (green) indicated that cystin localized to the center of mCCD cells relative to the tight junctions stained for ZO-1 (red). (f and g) Broad-field and representative high-magnification views demonstrated the colocalization of the exogenously expressed cystin (red, anti-myc mAb) and endogenous polaris (green, rabbit polyclonal Ab) in cilia of mCCD cells.
Discussion
We have identified by positional cloning a novel gene that is mutated in the cpk mouse model. Southern analyses indicate that the cpk gene exists as a single copy in the mouse and other mammals. The mouse gene encodes a novel, hydrophilic protein of 145 amino acids that has no significant similarity to previously characterized proteins or protein domains. However, several putative functional motifs are present in the protein, including two potential myristoylation sites, the more N terminal of which is coupled to a polybasic domain.
Recent in vitro studies have demonstrated that N-terminal myristoylation acts as an intracellular membrane-associating signal. However, stable anchoring of _N_-myristoylated proteins to membranes requires, among other factors, linkage to a second signal, such as a series of positively charged residues adjacent or distal to the protein lipidation site (40). This combined _N_-myristoylation site/polybasic residue motif is used by proteins such as c-SRC, c-Ki-ras, and myristolated alanine-rich C-kinase substrate (MARCKS) proteins for membrane anchoring (41–43), leading us to speculate that cystin would be associated with the plasma membrane and/or membranes of intracellular organelles. We therefore examined the subcellular localization of cystin exogenously expressed in mCCD cells.
In polarized mCCD cells stably transfected with wild-type cystin, the epitope-tagged protein localized in the single apical cilia with β-tubulin IV and polaris. But, unlike polaris, which is expressed both in the ciliary basal bodies and the axoneme, cystin expression appeared to be enriched in the ciliary axoneme. This subtle difference in protein distributions may indicate that cystin is not involved in a ciliogenic pathway like polaris, but rather plays a novel role in ciliary function.
We hypothesize that cystin is bound to the axonemal membrane and functions as part of a molecular scaffold that stabilizes microtubule assembly within the ciliary axoneme. In support of this speculation, we note that Woo et al. have demonstrated a significant attenuation in renal cystic disease progression in cpk/cpk mice treated with paclitaxel (Taxol) and related taxanes, which promote microtubule assembly (44). In comparison, paclitaxel treatment was ineffective in Tg737orpk homozygotes (45), consistent with the model that polaris and cystin play distinct, perhaps sequential, roles in ciliary formation and function. Further studies of the interactions of polaris, cystin, tubulins, and other ciliary proteins will be required to test this model.
There is a growing body of evidence that links ciliary dysfunction, embryonic left-right (LR) patterning defects, and cystic disease of visceral organs, e.g., kidney, liver, and pancreas. Polaris is among several proteins that appear to function in ciliogenesis and LR body axis determination (46). Cilia formation is initiated in basal bodies and involves a process called intraflagellar transport in which ciliogenic proteins assemble into complexes that migrate up the ciliary axoneme (47). Targeted deletions of these genes in mice, including Tg737, typically cause loss of embryonic nodal cilia, disruption of LR axis specification, and embryonic lethality (34, 48). In contrast, homozygosity for the Tg737orpk hypomorphic allele causes an ARPKD-like phenotype, with renal, biliary, and pancreatic cysts (49). There are no associated defects in LR specification, although Tg737orpk homozygotes have attenuation of the renal epithelial cilia (50) and loss of cilia in the ventricular ependymal cell layer (34). The inversion of embryo turning (inv) gene encodes a novel, ankyrin repeat–containing protein that is ubiquitously expressed in early mouse embryos (48). Homozygous mutants have both reversal of LR visceral asymmetries, as well as severely dilated renal collecting ducts, pancreatic abnormalities, including dilatation of the acinar ducts, and anomalies of the extrahepatic biliary system (48, 51). The nodal cilia in inv mutants are present and motile, but can produce only very weak leftward nodal flow (52). The monocilia in renal, biliary, and pancreatic ductal cells have not been well examined in these mutants, and their functionality remains uncharacterized.
LR patterning defects have not been identified in cpk mutants, and recent ultrastructural studies of renal and biliary cysts have not demonstrated defects in ciliary structure (11). The collecting duct cysts appear to be lined by a single layer of principal-like cells that have a single apical cilium and short microvilli. Similarly, the intrahepatic biliary duct cysts are lined by epithelial cells with numerous microvilli and a single apical cilium. These data are permissive for our hypothesis that cystin may be important in cilia function, but the protein is probably not involved in ciliogenesis, a process critical in embryo patterning. Cystin homologues appear to be present only in chordates, and not in primitive ciliated eukaryotes such as C. elegans. These data suggest that cystin has a later phylogenetic origin than proteins associated with intraflagellar transport, such as polaris, and that this novel protein may have a very specialized function in ciliated epithelia of higher vertebrates.
Single nonmotile cilia are expressed on the epithelia lining of much of the nephron, the biliary tract, and the pancreatic ducts (53). These cilia are less well characterized than the motile cilium present on embryonic node cells. However, recent data suggest that the apical cilium on renal epithelial cells has a mechanosensory function (54). While cilia are expressed on a broad spectrum of mammalian cell types, in the nematode worm, C. elegans, cilia are present only in the specialized neurons where they function as sensory organelles. Interestingly, the nematode homologues of PKD1 and PKD2 (lov-1 and pkd-2) (55) and Tg737 (osm-5) (56, 57) are all expressed in the same subset of ciliated sensory neurons, suggesting that PKD-related proteins may function in common chemosensory or mechanosensory pathways in these cilia.
Extrapolating from these data, Barr and colleagues (57) have proposed that cilia are required for the function of vertebrate polycystins and that any gene mutation that causes disruption of cilia assembly or function would result in PKD. Given this model, the PKD phenotype in Tg737orpk homozygous mice may be attributed to failure to properly target polycystins due to aberrant cilia structure, whereas a similar phenotype in cpk/cpk mice may result from ciliary dysfunction. Further studies to evaluate the potential colocalization of the polycystins, polaris, and cystin in epithelial cilia are required to evaluate this model.
Finally, studies by our group and others have strongly implicated a role for cystin in tubuloepithelial differentiation within the embryonic kidney, biliary tract, and pancreas (10, 11, 58). In preliminary studies, we also have observed structural changes in basement membranes surrounding early dilating renal tubules in fetal cpk/cpk mice (L. Guay-Woodford, unpublished data). Therefore, further analysis of cystin should permit a more thorough investigation of the molecular interactions between cilia function, differentiation and maintenance of the polarized epithelial phenotype, and macromolecular organization of the basement membrane, all processes that are critical for normal organogenesis as well as cystogenesis.
Acknowledgments
The authors thank Gail A. P. Bruns and James Schafer for helpful discussions and Vicente Torres for providing the principal cell schema. Alison Gardner, William J. Green, and Alicia Lakings provided technical assistance. This work was supported by the NIH grants DK-55534 (L. Guay-Woodford) and DK-55007 (B.K. Yoder). The University of Alabama at Birmingham and the Brigham and Women’s Hospital are fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
References
- 1.Gabow P. Autosomal polycystic kidney disease. N Engl J Med. 1993;329:332–342. doi: 10.1056/NEJM199307293290508. [DOI] [PubMed] [Google Scholar]
- 2.Watnick T, Germino G. Molecular basis of autosomal dominant polycystic kidney disease. Semin Nephrol. 1999;19:327–343. [PubMed] [Google Scholar]
- 3.Guay-Woodford, L., Jafri, Z., and Bernstein, J. 1999. Other cystic diseases. In Comprehensive clinical nephrology. R. Johnson and J. Feehally, editors. Mosby International. London, United Kingdom. 50.1–50.2.
- 4.Zerres K, et al. Mapping of the gene for autosomal recessive polycystic kidney disease (ARPKD) to chromosome 6p21-cen. Nat Genet. 1994;7:429–432. doi: 10.1038/ng0794-429. [DOI] [PubMed] [Google Scholar]
- 5.Guay-Woodford L, et al. The severe perinatal form of autosomal recessive polycystic kidney disease (ARPKD) maps to chromosome 6p21.1-p12: implications for genetic counseling. Am J Hum Genet. 1995;56:1101–1107. [PMC free article] [PubMed] [Google Scholar]
- 6.Schieren G, Pey R, Bach J, Hafner M, Gretz N. Murine models of polycystic kidney disease. Nephrol Dial Transplant. 1996;11(Suppl.):38–45. doi: 10.1093/ndt/11.supp6.38. [DOI] [PubMed] [Google Scholar]
- 7.Russell E, McFarland E. Cystic kidneys, ck. Mouse News Letter. 1977;56:40. [Google Scholar]
- 8.Preminger G, et al. Murine congenital polycystic kidney disease: a model for studying development of cystic disease. J Urol. 1982;127:556–560. doi: 10.1016/s0022-5347(17)53911-7. [DOI] [PubMed] [Google Scholar]
- 9.Fry J, et al. A genetically determined murine model of infantile polycystic kidney disease. J Urol. 1985;134:828–833. doi: 10.1016/s0022-5347(17)47448-9. [DOI] [PubMed] [Google Scholar]
- 10.Guay-Woodford L, Green W, Lindsey J, Beier D. Germline and somatic loss-of-function of the mouse cpkgene causes biliary ductal pathology that is genetically modulated. Hum Mol Genet. 2000;9:769–778. doi: 10.1093/hmg/9.5.769. [DOI] [PubMed] [Google Scholar]
- 11.Ricker J, Gattone V, Calvet J, Rankin C. Development of autosomal recessive polycystic kidney disease in BALB/c-cpk/cpkmice. J Am Soc Nephrol. 2000;11:1837–1847. doi: 10.1681/ASN.V11101837. [DOI] [PubMed] [Google Scholar]
- 12.Cowley BJ, Smardo F, Grantham J, Calvet J. Elevated c-myc protooncogene expression in autosomal recessive polycystic kidney disease. Proc Natl Acad Sci USA. 1987;84:8394–8398. doi: 10.1073/pnas.84.23.8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowley BJ, Chadwick L, Grantham J, Calvet J. Elevated proto-oncogene expression in polycystic kidneys of the C57BL/6J (cpk) mouse. J Am Soc Nephrol. 1991;1:1048–1053. doi: 10.1681/ASN.V181048. [DOI] [PubMed] [Google Scholar]
- 14.Harding M, Gattone VN, II, Grantham J, Calvet J. Localization of overexpressed c-myc mRNA in polycystic kidneys of the cpk mouse. Kidney Int. 1992;41:317–325. doi: 10.1038/ki.1992.44. [DOI] [PubMed] [Google Scholar]
- 15.vanden Heuvel G, Bodmer R, McConnell K, Nagami G, Igarashi P. Expression of a cut-related homeobox gene in developing and polycystic mouse kidney. Kidney Int. 1996;50:453–461. doi: 10.1038/ki.1996.336. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura T, et al. Growth factor expression in the kidney of murine polycystic kidney disease. J Am Soc Nephrol. 1993;3:1378–1386. doi: 10.1681/ASN.V371378. [DOI] [PubMed] [Google Scholar]
- 17.Orellana S, Avner E. Cystic maldevelopment of the kidney. Semin Nephrol. 1995;15:341–352. [PubMed] [Google Scholar]
- 18.Ebihara I, et al. Altered mRNA expression of basement membrane components in a murine model of polycystic kidney disease. Lab Invest. 1988;58:262–269. [PubMed] [Google Scholar]
- 19.Taub M, Laurie G, Martin G, Kleinman H. Altered basement membrane protein biosynthesis by primary cultures of cpk/cpk mouse kidney. Kidney Int. 1990;37:1090–1097. doi: 10.1038/ki.1990.90. [DOI] [PubMed] [Google Scholar]
- 20.Rankin C, et al. Matrix metalloproteinases and TIMPS in cultured C57BL/6J-cpk kidney tubules. Kidney Int. 1996;50:835–844. doi: 10.1038/ki.1996.383. [DOI] [PubMed] [Google Scholar]
- 21.Harding M, Chadwick L, Gattone V, Calvet J. The SGP-2 gene is developmentally regulated in the mouse kidney and abnormally expressed in collecting duct cysts in polycystic kidney disease. Dev Biol. 1991;146:483–490. doi: 10.1016/0012-1606(91)90249-3. [DOI] [PubMed] [Google Scholar]
- 22.Rocco M, Neilson E, Hoyer J, Ziyadeh F. Attenuated expression of epithelial cell adhesion molecules in murine polycystic kidney disease. Am J Physiol. 1992;262:F679–F686. doi: 10.1152/ajprenal.1992.262.4.F679. [DOI] [PubMed] [Google Scholar]
- 23.Aziz N, Maxwell M, Jacques BS, Brenner B. Downregulation of Ke 6, a novel gene encoded within the major histocompatibility complex, in murine polycystic kidney disease. Mol Cell Biol. 1993;13:1847–1853. doi: 10.1128/mcb.13.3.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aziz N, Brown D, Lee W, Naray-Fejes-Toth A. Aberrant 11 beta-hydroxysteriod dehydrogenase-1 activity in the cpk mouse: implications for regulation by the Ke 6 gene. Endocrinology. 1996;137:5581–5588. doi: 10.1210/endo.137.12.8940387. [DOI] [PubMed] [Google Scholar]
- 25.Deshmukh G, Radin N, Gattone VN, II, Shayman J. Abnormalities of glycosphingolipid, sulfatide, and ceramide in the polycystic (cpk/cpk) mouse. J Lipid Res. 1994;35:1611–1618. [PubMed] [Google Scholar]
- 26.Calvet JP. Polycystic kidney disease: primary extracellular matrix abnormality or defective cellular differentiation? Kidney Int. 1993;43:101–108. doi: 10.1038/ki.1993.17. [DOI] [PubMed] [Google Scholar]
- 27.Mrug M, et al. An integrated genetic and physical map of the 650-kb interval containing the congenital polycystic kidney (cpk) locus on mouse Chromosome 12. Cytogenet Cell Genet. 2001;94:55–61. doi: 10.1159/000048783. [DOI] [PubMed] [Google Scholar]
- 28.Ewing B, Hillier L, Wendl M, Green P. Base-calling of automated sequencer traces using PHRED. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 29.Ewing B, Green P. Base-calling of automated sequencer traces using PHRED. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 30.Staden R, Beal K, Bonfield J. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S, Gish W, Miller W. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Kizer N, Lewis B, Stanton B. Electrogenic sodium absorption and chloride secretion by an inner medullary collecting duct cell line (mIMCD-K2) Am J Physiol. 1995;268:F347–F355. doi: 10.1152/ajprenal.1995.268.2.F347. [DOI] [PubMed] [Google Scholar]
- 33.Yoder B, et al. Differential rescue of the renal and hepatic disease in an autosomal recessive polycystic kidney disease mouse mutant. A new model to study the liver lesion. Am J Pathol. 1997;150:2231–2241. [PMC free article] [PubMed] [Google Scholar]
- 34.Taulman P, Haycraft C, Balkovetz D, Yoder B. Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol Biol Cell. 2001;12:589–599. doi: 10.1091/mbc.12.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 36.Mount S. A catalogue of slice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterfly M, Phan J, Xu P, Reue K. Lipodystrophy in the fldmouse results from mutation of a new gene encoding a nuclear protein, lipin. Nature Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 38.Anderson J, Stevenson B, Jesaitis L, Goodenough D, Mooseker M. Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J Cell Biol. 1988;106:1141–1149. doi: 10.1083/jcb.106.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renthal R, Schneider B, Miller M, Luduena R. Beta IV is the major beta-tubulin isotype in bovine cilia. Cell Motil Cytoskeleton. 1993;25:19–29. doi: 10.1002/cm.970250104. [DOI] [PubMed] [Google Scholar]
- 40.McCabe J, Berthiaume L. Functional roles for fatty acid acylated amino-terminal domains in subcellular localization. Mol Biol Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancock J, Paterson H, Marshall C. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 42.Resh M. Myristoylation and palmitoylation of SRC family members: the fats of the matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 44.Woo D, Tabancay A, Jr, Wang C. Microtubule active taxanes inhibit polycystic kidney disease progression in cpkmice. Kidney Int. 1997;51:1613–1618. doi: 10.1038/ki.1997.222. [DOI] [PubMed] [Google Scholar]
- 45.Sommardahl C, Woychik R, Sweeney W, Avner E, Wilkinson J. Efficacy of taxol in the orpk mouse model of polycystic kidney disease. Pediatr Nephrol. 1997;11:728–733. doi: 10.1007/s004670050376. [DOI] [PubMed] [Google Scholar]
- 46.Murcia N, et al. The Oak Ridge polycystic kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 47.Cole D, et al. Chylamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis eleganssensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan D, et al. Inversin, a novel gene in the vertebrate left-right axis pathway is partially deleted in the invmouse. Nature Genet. 1998;20:149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]
- 49.Moyer J, et al. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. doi: 10.1126/science.8191288. [DOI] [PubMed] [Google Scholar]
- 50.Pazour G, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazziotti M, et al. Anomalous development of the hepatobiliary system in the invmouse. Hepatology. 1999;30:372–378. doi: 10.1002/hep.510300223. [DOI] [PubMed] [Google Scholar]
- 52.Okada Y, et al. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell. 1999;4:459–468. doi: 10.1016/s1097-2765(00)80197-5. [DOI] [PubMed] [Google Scholar]
- 53.Wheatley D, Wang A, Strugnell G. Expression of primary cilia in mammalian cells. Cell Biol Int. 1996;20:73–81. doi: 10.1006/cbir.1996.0011. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz E, Leonard M, Bizios R, Bowser S. Analysis and modeling of the primary cilium bending response to fluid shear. Am J Physiol. 1997;272:F132–F138. doi: 10.1152/ajprenal.1997.272.1.F132. [DOI] [PubMed] [Google Scholar]
- 55.Barr M, Sternberg P. A polycystic kidney disease gene homologue required for male mating behavior in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 56.Haycraft C, Swoboda P, Taulman P, Thomas J, Yoder B. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- 57.Qin H, Rosenbaum J, Barr M. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegansciliated neurons. Curr Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- 58.Avner E, et al. Congenital murine polycystic kidney disease. I. The ontogeny of tubular cyst formation. Pediatr Nephrol. 1987;2:210–218. doi: 10.1007/BF00862593. [DOI] [PubMed] [Google Scholar]