Interaction of FK506-binding protein 13 with brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1): Effects of FK506 (original) (raw)
Abstract
BIG1 and BIG2 are brefeldin A-inhibited guanine nucleotide-exchange proteins that activate ADP-ribosylation factors (ARFs), critical components of vesicular trafficking pathways. These proteins can exist in macromolecular complexes and move between Golgi membranes and cytosol. In the BIG1 molecule, a centrally located Sec7 domain is responsible for ARF activation, but functions of other regions are largely unknown. Yeast two-hybrid screens of a human placenta cDNA library with BIG1 cDNA constructs revealed specific interaction of the N-terminal region (amino acids 1–331) with FK506-binding protein 13 (FKBP13). The association was confirmed by immunoprecipitation of both endogenous BIG1 and FKBP13 from Jurkat T cells with antibodies against either one. Binding of BIG1, BIG2, and ARF to cell membranes in vitro was increased by guanosine 5′-[γ-thio]triphosphate, and further increases were induced by FK506. Incubation of Jurkat T cells with FK506 increased binding of BIG1, BIG2, and ARF to Golgi and other membranes in a time- and concentration-dependent manner, without effects on clathrin or γ-adaptin binding. Binding of BIG1, BIG2, and ARF to membranes was also increased by L-732,531, an agonist structurally related to FK506, but was not increased by a related antagonist, L-685,818, nor by cyclosporin A or rapamycin. These findings are consistent with a role for FKBP13 and FK506 in vesicular trafficking, influencing ARF activity through their guanine nucleotide-exchange proteins.
Intracellular vesicular transport depends on numerous proteins that are required to form and move vesicles between intracellular compartments, as well as to select, package, and deliver cargo. Among these are the ADP-ribosylation factors (ARFs), 20-kDa GTPases of the ras superfamily that regulate vesicular transport in eukaryotic cells (1–3). The inactive GDP-bound form of ARF is cytosolic, whereas the active GTP-bound form of ARF associates with membranes. ARFs are divided into three classes: class I (ARF1–3), which functions in endoplasmic reticulum–Golgi trafficking; the much less studied class II (ARF4 and 5); and class III (ARF6), which has critical roles in endocytosis and cytoskeletal dynamics near the cell surface (1–3). Regulators of ARF activity include guanine nucleotide-exchange proteins (GEPs), which activate ARF by catalyzing the replacement of bound GDP with GTP (4), and the GTPase-activating proteins, which accelerate hydrolysis of bound GTP and, thus, ARF inactivation (5).
All known ARF GEP molecules contain Sec7 domains, which are regions of ≈200 aa that are responsible for the acceleration of GTP binding to ARF (6). Based on their structural and functional properties, GEPs can be classified into four families (4): two with smaller molecular sizes are the ≈47-kDa cytohesins, of which four are known (7), and the EFA6 family, which preferentially acts on ARF6 (8). These families are resistant to inhibition by the fungal fatty acid metabolite brefeldin A and have, C-terminal to the Sec7 domain, a pleckstrin homology domain that binds specific inositol phospholipids. The Sec7/BIG and Gea/GBF/GNOM families (4) are of larger molecular size, and, with the exception of GBF1 (9), are all inhibited by brefeldin A. BIG1 and BIG2 were initially purified together from bovine brain cytosol on the basis of their brefeldin A-sensitive activation of class I ARFs (10). The proteins appeared to be, at least partially, colocalized in Golgi membranes and, on gel filtration of cytosol, were eluted together with an apparent molecular size of ≈670 kDa (11–13). They were also immunoprecipitated together by antibodies specific for either BIG1 or BIG2, which led to the conclusion that they exist to a large extent as parts of the same macromolecular complexes, at least in HepG2 cytosol (14).
There is little functional information regarding regions of protein interaction in the BIG molecules outside of the Sec7 domain with its guanine nucleotide-exchange activity (15, 16). We undertook, therefore, to identify novel proteins that interacted with BIG1 by using a yeast two-hybrid system. Here, we report an interaction between an N-terminal fragment of BIG1 (residues 1–331) and FK506-binding protein 13 (FKBP13) that was confirmed by immunoprecipitation of both endogenous proteins from Jurkat T cell membranes by antibodies against either one. FKBP13 (17) is a member of the FK506-binding protein family of immunophilins, but unlike the better-known FKBP12, does not bind or inhibit calcineurin (18). Our studies suggest an effect of FK506 on vesicular trafficking mediated via FKBP13 and ARF activation.
Materials and Methods
Materials.
FK506 and rapamycin were purchased from Calbiochem and cyclosporin A from Sigma. FK506 analogues were kindly supplied by Merck.
Yeast Two Hybrid.
The DupLEX-A yeast two-hybrid system (OriGene Technologies, Rockville, MD) was used to screen a human placenta LexA cDNA library (CLONTECH) with human BIG1 cDNA encoding amino acids 1–1834 fused to LexA in plasmid pEG202 as bait. Fragments of this clone, similarly fused in pEG202, are shown in Fig. 1. Expression of bait proteins was confirmed by Western blot by using LexA antibody. According to the protocol supplied by OriGene, bait plasmid phBIG1-202 was tested for its autoactivation potential and ability to enter the nucleus and bind LexA operators. Of ≈1 × 106 colonies screened, 159 were positive. Plasmids recovered from positive clones were transformed into KC8 bacteria to select colonies containing only target plasmids. A search of the GenBank database (accession no. JC1365) with sequences of DNA from positive colonies identified one as FKBP13. To confirm the association and identify the site(s) of interaction of hBIG1 and FKBP13, a mating assay was performed. phBIG1-202 and each of the fragments in pEG202 were transformed with pSH18–34 reporter plasmid into RFY206 cells. pFKBP13 in pJG4.5 (target plasmid) was transformed into EGY48 cells.
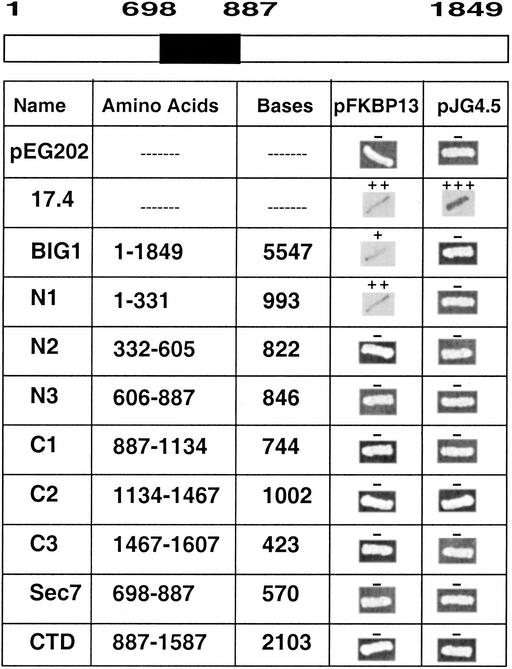
Figure 1.
FKBP13-binding region of BIG1. In the BIG1 molecule diagram, the Sec7 domain (amino acids 698–887) is shaded. The FKBP13-binding region of BIG1 was identified by β-galactosidase filter assays in yeast-mating experiments as described in Materials and Methods. pJG4.5 and pEG202 are vectors for FKBP13 and BIG1, respectively. BIG1 DNA constructs encoded the amino acids and included the numbers of bases indicated. 17.4 is the positive control in the yeast two-hybrid kit (OriGene).
Transformed RFY206 and EGY48 cells were incubated together overnight before plating on 5-bromo-4-chloro-3-indolyl β-D-galactosidase–Leu plates.
Cloning and Expression of FKBP13.
DNA encoding the mature form of FKBP13 was synthesized by PCR by using FKBP13-4.5 (17) as a template and primers 5′-ACGGGGACCGAGGGCAAA-3′ (forward) and 5′-GCTCGAGTTACAGCTCAGTTCGTCGC-3′ (reverse). An Xho site (underlined) was introduced to facilitate cloning. The PCR product (471 bp) was ligated to pCR2.TOPO (Invitrogen) to produce FKBP13-PCR plasmid. Automated sequencing verified the sequence of FKBP13, which was excised with _Eco_RI and _Xho_I and ligated to the 3′ end of GST DNA in pGEX4T-1 (Amersham Pharmacia) for production of GST-FKBP13.
Plasmid GST-FKBP13 was transformed into DE3 Escherichia coli. A single colony was picked and grown overnight (37°C) in LB broth with ampicillin (100 μg/ml), diluted 100-fold with the same medium, and incubated until OD595 ≅ 0.6, when isopropyl β D-thiogalactoside was added (final concentration, 0.2 mM), and incubation was continued for 4 h. Bacteria were pelleted by centrifugation (2,000 × g, 10 min) and dispersed in ice-cold PBS (1/5 culture volume) with 0.5 mM 4-(2-aminoethylbenzenesulfonyl fluoride, leupeptin (10 μg/ml), aprotinin (10 μg/ml), and pepstatin A (1 μg/ml). After incubation for 30 min on ice with lysozyme (20 μg/ml), the bacterial lysate was sonified and centrifuged (8,000 × g, 20 min). Pelleted debris was discarded and Triton X-100 was added to the supernatant (final concentration, 1%), followed by gentle agitation for 1 h at room temperature with reduced glutathione–Sepharose 4B [50% slurry (Amersham Pharmacia), 2 ml/100 ml of supernatant] to adsorb GST-tagged proteins. Beads were pelleted by centrifugation (500 × g, 10 min), dispersed in PBS (1/100 culture volume), transferred to a column, washed with 10 vol PBS, and incubated (gentle agitation, overnight, at room temperature) with thrombin (Amersham Pharmacia, 30 units/ml), 5 ml per 4 liters of culture. The supernatant containing FKBP13, plus four more supernatant fractions (similar but collected after 2-h incubation), were pooled and incubated (at room temperature, overnight) with a mixture of reduced glutathione–Sepharose 4B and benzamidine–Sepharose 6B (10:1 vol/vol, each 50% slurry) to bind, respectively, GST and thrombin. The supernatant, which contained purified FKBP13, was concentrated (Centriprep, Amicon; 2,300 × g, 30 min) and stored in 0.5-ml portions (1 mg/ml) at 4°C in the presence of 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, leupeptin (10 μg/ml), aprotinin (10 μg/ml), and pepstatin A (1 μg/ml; Sigma). This FKBP13 was used to immunize rabbits, and antibodies were affinity purified (14).
Cell Fractionation and Western Blotting.
Jurkat T cells purchased from the American Type Culture Collection were grown (37°C, 5% CO2/95% air) in RPMI medium 1640 with 10% FCS and amphotericin B (10 μg/ml). Cells (≈2 × 106 cells in 50 ml of medium) were sedimented by centrifugation (4,000 × g, 5 min) and homogenized [10 strokes in a Dounce tissue grinder (Wheaton Scientific)] in 4 ml of TENDS buffer (20 mM Tris⋅HCl, pH 8.0/1 mM EDTA/1 mM NaN3/1 mM DTT/250 mM sucrose) containing 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, leupeptin (10 μg/ml), aprotinin (10 μg/ml), and pepstatin A (1 μg/ml). The homogenate was centrifuged (400 × g, 10 min), and the postnuclear supernatant was centrifuged (100,000 × g, 1.5 h) to separate cytosol and membrane fractions. Membranes were homogenized in 0.5 ml of TENDS buffer containing protease inhibitors as in homogenizing buffer. Samples of proteins in cytosol (100 μg) and membrane (10 μg) fractions were separated by SDS/PAGE in 4–12% gel and transferred to nitrocellulose membranes, which were divided for reactions with antibodies against BIG1 (0.5 μg/ml) and/or BIG2 (1 μg/ml), γ-adaptin (Sigma; 1:5,000), clathrin (Transduction Laboratories, Lexington, KY; 1:5,000), or ARF (Affinity BioReagents, Golden, CO; 1:1,000). Secondary antibodies, goat anti-rabbit and goat anti-mouse IgG conjugated to horseradish peroxidase (Promega), were detected by using SuperSignal Chemiluminescent substrate (Pierce). Immunoreactive bands were quantified by ChemiImager 5500 (Alpha Innotech, San Leandro, CA). Antibodies against BIG1 and BIG2 were prepared and purified as described (14).
Immunoprecipitation.
Cytosol and microsomes were prepared from Jurkat T cells as described above, except that membranes were homogenized in TENDS buffer with protease inhibitors (as in homogenizing buffer) plus 2 mM MgCl2 and 0.5% (wt/vol) (octylphenoxy)polyethoxyethanol (IGEPAL CA-630), which is chemically identical to Nonidet P-40). Before immunoprecipitation (all procedures at 4°C), samples of membrane proteins (400 μg/ml in 500 μl) were precleared (incubated overnight) with 50 μl of 50% (vol/vol) slurry of protein A Sepharose CL-4B in PBS (Amersham Pharmacia), incubated without or with FK506 as indicated and then incubated for 4 h with appropriate antibodies, before adding 60 μl of protein A Sepharose [50% (vol/vol) slurry]. After incubation overnight, beads were washed (500 μl) three times with each of three buffers: 50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate; 50 mM Tris⋅HCl, pH 7.5/500 mM NaCl/0.1% Nonidet P-40/0.05% sodium deoxycholate; and 10 mM Tris⋅HCl, pH 7.5/0.1% Nonidet P-40/0.5% sodium deoxycholate. All buffers contained the same protease inhibitors as the homogenizing buffer. Bound proteins were eluted by boiling beads in 100 μl of loading buffer, separated by SDS/PAGE in 8–16% gel, transferred to nitrocellulose, and detected by using horseradish peroxidase-conjugated antibodies with chemiluminescent substrate.
Results
FKBP13 and BIG1 Interaction in Yeast.
The β-galactosidase filter assay was used to assess interaction of eight fragments of BIG1 with FKBP13 in a yeast-mating assay (Fig. 1). Positive interactions were observed only with full-length BIG1 and with the most N-terminal sequence (amino acids 1–331). No interactions of the vectors used for FKBP13 (pJG4.5) or BIG1 (pEG202) were detected.
Coimmunoprecipitation of BIG1 and FKBP13 from Jurkat Cell Membranes.
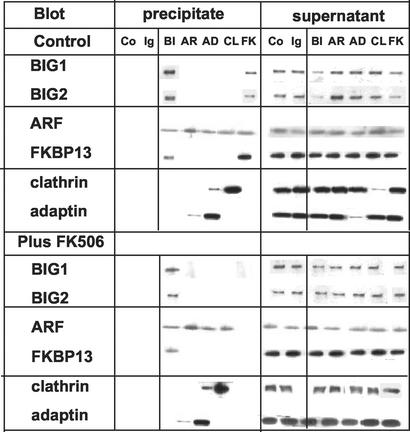
Antibodies against BIG1 also precipitated BIG2, ARF, and FKBP13 from Jurkat cell membranes (Fig. 2). Antibodies against FKBP13 precipitated (in addition to FKBP13) BIG1, BIG2, and ARF. After incubation of membranes with 100 nM FK506, however, none of those proteins was precipitated by anti-FKBP13 antibodies, although antibodies against BIG1 still precipitated FKBP13 along with BIG1, BIG2, and ARF (Fig. 2). Binding of FK506 by FKBP13 evidently interfered with its reaction with the antibody, but not with its binding to BIG1, or perhaps when bound to BIG1, FKBP13–FK506 was not immunoreactive. FK506 did not alter precipitation by antibodies against ARF (which precipitated a small amount of γ-adaptin, against clathrin, which precipitated some ARF) or against γ-adaptin, which precipitated also small amounts of clathrin and ARF (Fig. 2).
Figure 2.
Effect of FK506 on immunoprecipitation of BIG1, FKBP13, and other proteins from Jurkat cell membranes. Samples of membranes (400 μg of protein) in 500 μl of TENDS buffer containing protease inhibitors and 0.5% Nonidet P-40 were incubated (30 min at 37°C) without or with 100 nM FK506 before addition of antibodies against BIG1 (BI), ARF (AR), FKBP13 (FK), clathrin (CL), or γ-adaptin (AD), nonspecific rabbit IgG (Ig), or no antibodies (Co). Immunoprecipitated proteins were collected on protein A Sepharose CL-4B, separated by SDS/PAGE in 8–16% gel, and reacted on Western blots with antibodies against BIG1, BIG2, ARF, FKBP13, clathrin, or γ-adaptin. Data were similar in two other experiments.
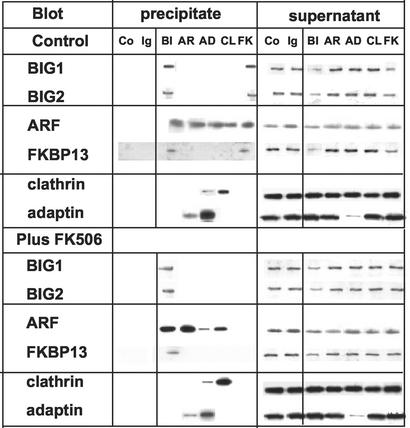
Incubation of intact Jurkat cells with FK506 had effects on immunoprecipitation of proteins from the isolated membrane fraction rather similar to those observed after exposure of membranes to FK506 in vitro. Antibodies against FKBP13 failed to precipitate any proteins from membranes isolated from cells that had been incubated with FK506, but FKBP13 was precipitated (along with BIG1, BIG2, and ARF) from the same membrane preparations by antibodies against BIG1 (Fig. 3). No consistent effects of FK506 treatment on immunoprecipitation with other antibodies were found.
Figure 3.
Effect of incubation of Jurkat cells with FK506 on immunoprecipitation of BIG1, FKBP13, and other proteins from isolated membranes. Cells were incubated (30 min at 37°C) without or with 100 nM FK506 before washing, homogenization, and preparation of membranes. Samples of membranes (400 μg) in TENDS buffer with protease inhibitors and 0.5% Nonidet P-40 (total volume, 500 μl) were incubated with antibodies, and experiments were completed as described in the legend for Fig. 2. Data were similar in two other experiments.
Effects of FK506 on Membrane Association of BIG1 and Other Proteins in Jurkat Cells.
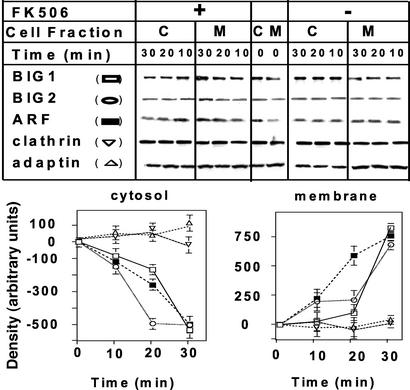
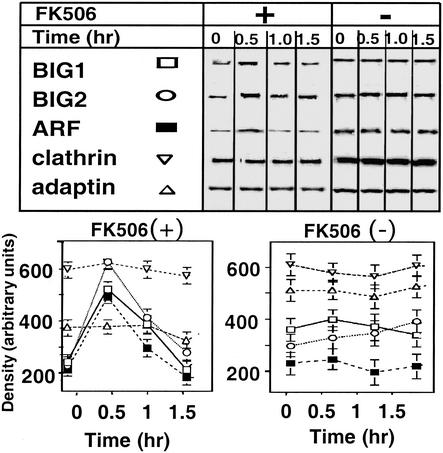
During incubation of Jurkat cells with 100 nM FK506, amounts of membrane-associated BIG1, BIG2, and ARF increased and reached a maximum at 30 min (Fig. 4). There was a concomitant, reciprocal decline in the amounts of those proteins in the cytosol. The distribution of clathrin and γ-adaptin did not change during this time (Fig. 4). In the absence of FK506, no net movement of any of the proteins was observed (data not shown). Although amounts of membrane-bound BIG1, BIG2, and ARF were markedly increased after 30 min of incubation of cells with FK506, 30–60 min later they had returned, essentially, to zero time levels (Fig. 5). Amounts of membrane-associated clathrin and γ-adaptin were unchanged throughout the 90-min incubation.
Figure 4.
Effects of incubation of Jurkat cells with FK506 on association of BIG1 and other proteins with membranes. Cells were incubated at 37°C for 0, 10, 20, or 30 min without or with 100 nM FK506 before separation of membrane (M) and cytosol (C) fractions and protein analysis as in Fig. 3. Representative Western blots of the indicated proteins are shown. Below are means ± half the range of densitometric values (after subtraction of zero time values) from duplicate samples in one experiment representative of three.
Figure 5.
Effects of FK506 on association of BIG1 and other proteins with membranes. Experiment was performed as described in the Fig. 4 legend with longer incubation times. Immunoblots of membrane proteins are shown above graphic presentation of means ± half the range of densitometric values from duplicate samples in one experiment representative of three.
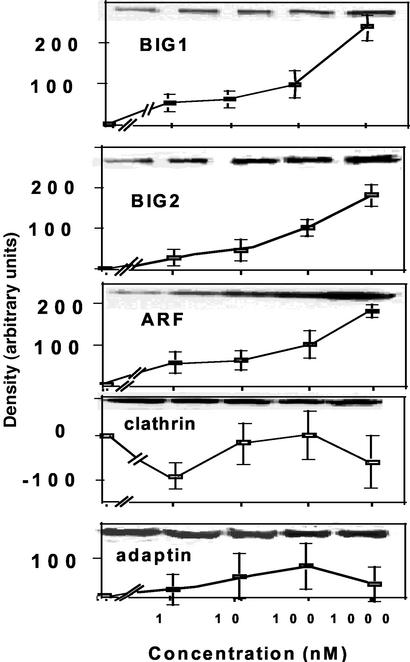
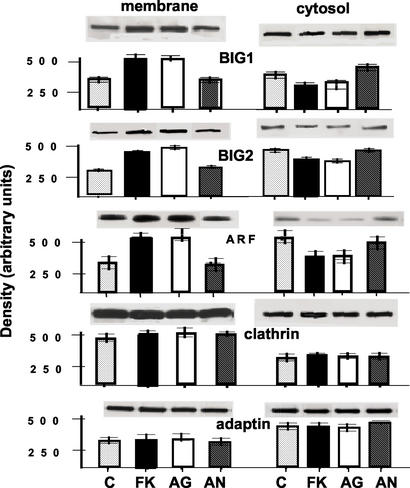
Effects of FK506 on intact cells were concentration dependent. Amounts of membrane-bound BIG1, BIG2, and ARF were increased considerably more by 1 μM than by 100 nM FK506 (used in most experiments), and effects of 10 or 1 nM FK506 were distinctly smaller. Fig. 6 presents means of densitometry values of duplicate samples with representative blots in one experiment. At concentrations of 100 nM, an analogue with agonist activity (L-732,531) had effects similar to those of FK506, whereas an analogue with antagonist activity (L-685,818) was without effect on membrane-bound BIG1, BIG2, and ARF (Fig. 7). Reciprocal changes in the amounts of those proteins in cytosol fractions were observed, and, like FK506, neither of the analogues altered the distribution of clathrin or γ-adaptin.
Figure 6.
Effect of FK506 concentration on membrane association of BIG1 and other proteins in Jurkat cells. Cells were incubated (30 min at 37°C) with the indicated concentration of FK506 before separation of fractions and protein analysis. Data are means ± half the range of densitometric values (after subtraction of zero time values) from duplicate samples in one experiment representative of three.
Figure 7.
Effects of FK506 analogues on binding of BIG1 and other proteins to membranes in Jurkat cells. Cells were incubated (30 min at 37°C) without (C) or with 100 nM FK506 (FK), agonist analogue L-732,531 (AG), or antagonist analogue L-685,818 (AN) before washing and preparation of membrane and cytosol fractions. Data are means ± half the range of densitometric values (after subtraction of zero time values) from duplicate samples with Western blots above, in one experiment representative of three.
No FKBP13 was detected in cytosol, and amounts in the membrane fraction were not altered by incubation of cells without or with FK506 (1–1,000 nM) for up to 3 h (data not shown).
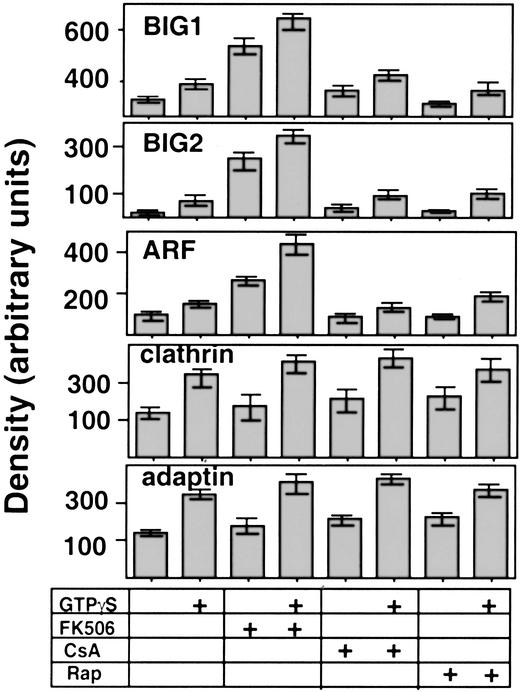
Cyclosporin A and rapamycin, like FK506, are used clinically as immunosuppressants. Cyclosporin A binds to cyclophilin A with resulting inhibition of calcineurin signaling, whereas rapamycin interacts with mTOR (mammalian target of rapamycin) as well as FKBPs. Unlike FK506, 100 nM cyclosporin A or rapamycin had no effects on membrane association of BIG1, BIG2, and ARF during incubation of Jurkat cell fractions in vitro, whether or not guanosine 5′-[γ-thio]triphosphate was present (Fig. 8). FK506, on the other hand, markedly increased membrane-associated BIG1, BIG2, or ARF, even in the absence of guanosine 5′-[γ-thio]triphosphate, with or without other additions.
Figure 8.
Effects of cyclosporin A (CsA) and rapamycin (Rap) on binding of BIG1 and other proteins to membranes from Jurkat cells. Samples of proteins from cytosol (400 μg in 100 μl) and membranes (200 μg in 50 μl) were incubated (30 min at 37°C) with 100 nM FK506, CsA, or Rap, as indicated, plus 100 μM guanosine 5′-[γ-thio]triphosphate added at zero time (total volume, 750 μl). After centrifugation (12,000 × g, 20 min, 4°C), samples of membrane (50 μg) proteins were separated by SDS/PAGE in 8–16% gel and transferred to nitrocellulose for Western blotting. Blots are representative of those from duplicate samples in three experiments.
Discussion
While trying to better understand the functions of BIG1 and their regulation, we also consider BIG2 because the two proteins, apparently so similar in structure and GEP activity, were initially purified together in a macromolecular complex of ≈670 kDa from bovine brain cytosol and were associated in HepG2 cells (14). More recently, however, Shinotsuka et al. (19) presented evidence, based on overexpression of epitope-tagged BIG2, that BIG2 functions in the recruitment of adaptor protein-1 for formation of clathrin-coated vesicles from the trans-Golgi network. These authors implicated BIG2 in transport of lysosomal enzymes between trans-Golgi network and late endosomes, as well as in the association of GGA1 with trans-Golgi network membranes (20). BIG1, on the other hand, seems clearly to be involved in the recruitment of coatomer to cis-Golgi membranes (21). Li et al. (22), in our laboratory, recently found that BIG2 contains three AKAP (A kinase-anchoring protein) domain sequences with different specificities for binding R subunits of PKA. This finding presents the possibility of a role(s) for BIG2 at the intersection of cAMP signaling and vesicular trafficking pathways, which are now being investigated. Perhaps a complex containing BIG1 and BIG2 can function as a scaffold to transmit and integrate messages among immunoactive agents and agonists that act via cAMP. At the same time, BIG1 and BIG2 may each have effects that are independent of the other.
When we confirmed the interaction of BIG1 and FKBP13 by coimmunoprecipitation of endogenous proteins from Jurkat cells, we observed that BIG2 and ARF were among the proteins precipitated by antibodies against BIG1 or FKBP13. Clathrin and γ-adaptin (a component of the adaptor protein-1 complex), which are also involved in vesicular trafficking together with ARF (23), were not detected, however, and served as negative controls. In yeast-mating experiments, the N-terminal 331-aa segment of BIG1 was responsible for FKBP13 association. This observation and the report of Mansour et al. (12) that Golgi-localization signals reside in the first 560 aa of BIG1, that overexpressed hemagglutinin-BIG1 (560–890) was not excluded from the nucleus, and that the sequence between 562 and 591 “may be required for protein folding and thermostability” are the only clues to locations of functional domains in BIG1, other than sequences in the Sec7 domain that are responsible for GEP activity and its brefeldin A inhibition (15, 16).
FKBP13 is one of a family of proteins that bind FK506 (tacrolimus), an immunosuppressive drug isolated from the culture medium of Streptomyces tsukubaenis (18). FK506-binding proteins have intrinsic peptidyl-prolyl _cis_-_trans_-isomerase (rotamase) activity that is postulated to facilitate protein folding (18, 24). Rotamase activity, which is inhibited by FK506, is not responsible for immunosuppression (24, 25). FKBP13 is believed to be involved in protein folding in the endoplasmic reticulum lumen (26, 27) and interacted with an erythrocyte membrane cytoskeletal protein (28). It did not interact with calcineurin and failed to mediate effects of FK506 in Jurkat cells (29). FKBP12, which is 43% identical in amino acid sequence to FKBP13 (17), is the prototype of this protein family and interacts in cells with receptors for ryanodine, inositol tris-phosphate and transforming growth factor β (18). Marx et al. (30) isolated from cardiac muscle a macromolecular complex that contained FKBP12.6, a ryanodine receptor, an A kinase-anchoring protein, an A kinase, and two phosphatases. This finding may have a parallel in the association of BIG2 and its A kinase-anchoring protein functions (22) with BIG1, although we have not yet ascertained whether FKBP13 is present in that complex.
The immunosuppressive action of FK506 results from its interaction with FKBP12 to produce a complex that binds to and inhibits the protein phosphatase activity of calcineurin. This inhibition prevents dephosphorylation and translocation of NFATc (nuclear factor of activated T cells cytoplasmic unit) into the nucleus, thereby blocking induction of IL-2 synthesis, which depends on association of NFAT with the IL-2 promoter enhancer (18). It was initially believed that FKBP12 did not interact with calcineurin in the absence of FK506. Cardenas et al. (31) were the first to report an FK506-independent interaction, which they detected in a yeast two-hybrid system (as we did with BIG1 and FKBP13) and showed was enhanced by FK506. FK506 also increased binding of calcineurin to immobilized FKBP12 (31). We postulate that the BIG1–FKBP13 interaction and effects of FK506 on this interaction may be analogous to those of calcineurin and FKBP12. If so, it should be possible to demonstrate differences in the BIG1–FKBP13 interaction dependent on FK506 binding.
In searching for less toxic therapeutic alternatives to FK506, pharmaceutical companies have prepared drugs that are similar in structure. Of the analogues that we tested, a pharmacological antagonist to FK506, L685,818, was without effect on BIG1 binding to membranes in Jurkat cells, whereas L-732,531, an agonist (18), increased binding. To explore possible relationships between ARF activation and immunophilins other than FKBP13, effects of other immunosuppressive drugs were also tested. Cyclosporin A binds to cyclophilin A and the drug–immunophilin complex inhibits calcineurin (32), whereas rapamycin interacts with FKBPs, in addition to producing effects via its association with target-of-rapamycin (18). Neither of these drugs enhanced membrane association of BIG1 in Jurkat cells.
The GNOM protein of Arabadopsis thaliana, which resembles BIG1 and BIG2 in structure and has ARF GEP activity, interacted with cyclophilin 5 in a yeast two-hybrid system (33). Cyclophilin 5 has rotamase and protein-refolding activities that are inhibited by cyclosporin A. Amino acids 18–360 in GNOM are required for its homodimerization and cyclophilin 5 binding. BIG1 and BIG2 molecules lack sequence corresponding to this N-terminal segment, and their homo- or heterodimerization has not been systematically investigated. Cyclophilin 5 is expressed in Arabadopsis during embryogenesis (31), a time when gross distortions of embryonic architecture result from GNOM mutations (34), and a role in regulation of GNOM function was suggested (33).
When investigating the effects of FK506 on coimmunoprecipitation of BIG1 and FKBP13, we found that the drug completely prevented the precipitation of FKBP13 (and other proteins) by anti-FKBP13 antibodies. It appears that bound FK506, either directly or indirectly, interfered with the antigen–antibody reaction, but FKBP13 was still precipitated (along with BIG2 and ARF) by antibodies against BIG1. Because we have not yet quantified FK506, the presence of FKBP13–FK506 in the BIG1 immunoprecipitate was not established. It seems likely, however, that the increased binding of BIG1 (and BIG2 and ARF) to Jurkat cell membranes induced by FK506 reflects differences in the BIG1–FKBP13 interaction dependent on FK506 binding, as shown by Cardenas et al. (31) for the calcineurin–FKBP12 association. We expect that there will be concomitant changes in the structure and function of other components of the multiprotein complex, e.g., utilization of ARF as substrate by the BIG1 Sec7 domain. Effects on BIG2 are also, of course, of major interest. With our recent recognition of a potential AKAP function of BIG2 (22), we must now consider, in addition, possible intersections of those functions and nuclear processes in which BIG1 may have a role.
Acknowledgments
We thank Carol Kosh and Dorothy Honemond for expert secretarial assistance and Dr. Vincent Manganiello for valuable discussions and manuscript review.
Abbreviations
ARF
ADP-ribosylation factor
GEP
guanine nucleotide-exchange protein
FKBP
FK506-binding protein
References
- 1.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 2.Moss J, Vaughan M. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 3.Chavrier P, Goud B. Curr Opin Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 4.Jackson C L, Casanova J E. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson J G. Proc Natl Acad Sci USA. 2000;97:3792–3794. doi: 10.1073/pnas.97.8.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzusoff A, Schekman R. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss J, Vaughan M. Arch Biochem Biophys. 2002;397:156–161. doi: 10.1006/abbi.2001.2661. [DOI] [PubMed] [Google Scholar]
- 8.Franco M, Peters P J, Boretto J, Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claude A, Zhao B-P, Kuziemsky C E, Dahan S, Berger S J, Yan J-P, Arnold A D, Sullivan E M, Melançon P. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- 10.Morinaga N, Tsai S-C, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morinaga N, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1997;94:12926–12931. doi: 10.1073/pnas.94.24.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansour S J, Skaug J, Zhao X-H, Giordano J, Sherer S W, Melançon P. Proc Natl Acad Sci USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 14.Yamaji R, Adamik R, Takeda K, Togawa A, Pacheco-Rodriguez G, Ferrans V J, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1999;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson C L. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 16.Sata M, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1999;96:2752–2757. doi: 10.1073/pnas.96.6.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Y-J, Albers M W, Lane W S, Bierer B E, Schreiber S L, Burakoff S J. Proc Natl Acad Sci USA. 1991;88:6677–6681. doi: 10.1073/pnas.88.15.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumont F J. Curr Med Chem. 2000;7:731–748. doi: 10.2174/0929867003374723. [DOI] [PubMed] [Google Scholar]
- 19.Shinotsuka C, Yoshida Y, Kawamoto K, Takatsu H, Nakayama K. J Biol Chem. 2002;277:9468–9472. doi: 10.1074/jbc.M112427200. [DOI] [PubMed] [Google Scholar]
- 20.Shinotsuka C, Waguri S, Wakasugi M, Uchiyama Y, Nakayama K. Biochem Biophys Res Commun. 2002;294:254–260. doi: 10.1016/S0006-291X(02)00456-4. [DOI] [PubMed] [Google Scholar]
- 21.Presley J F, Ward T H, Pfeifer A C, Siggla E D, Phair R D, Lippincott-Schwartz J. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Adamik R, Pacheco-Rodriguez G, Moss J, Vaughan M. Proc Natl Acad Sci USA. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchhausen T. Cell. 2002;109:413–416. doi: 10.1016/s0092-8674(02)00751-1. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber S L. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- 25.Bierer B E, Mattila P S, Standaert R F, Herzenberg L A, Burakoff S J, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigam S, Jin Y-J, Jin M-J, Bush K, Bierer B E, Burakoff S. Biochem J. 1993;294:511–515. doi: 10.1042/bj2940511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partaledis J, Berlin V. Proc Natl Acad Sci USA. 1993;90:5450–5454. doi: 10.1073/pnas.90.12.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walensky L, Gascard P, Fields M, Blackshaw S, Conboy J, Mohandas N, Snyder S H. Proc Natl Acad Sci USA. 1998;141:143–153. doi: 10.1083/jcb.141.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bram R J, Hung D T, Martin P K, Schreiber S L, Crabtree G R. Mol Cell Biol. 1993;13:4760–4769. doi: 10.1128/mcb.13.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx S O, Reiken S, Hisamatsu Y, Jayamaran T, Burkhoff D, Rosemblit N, Marks A R. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 31.Cardenas M E, Hemenway C, Muir R S, Ye R, Fiorentino D, Heitman J. EMBO J. 1994;13:5944–5957. doi: 10.1002/j.1460-2075.1994.tb06940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho S, Clipstone N, Timmerman L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree G R. Clin Immunol Immunopathol. 1996;80:S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 33.Grebe M, Gadea J, Steinman T, Kientz M, Rahfeld J-U, Salchert K, Koncz C, Jürgens G. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinman T, Geldner N, Grebe M, Mangold S, Jackson C L, Paris S, Gälweiler L, Palme K, Jürgens G. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]