Potent inhibition of scrapie prion replication in cultured cells by bis-acridines (original) (raw)
Abstract
Prion diseases are characterized by an accumulation of PrPSc, a misfolded isoform of the normal cellular prion protein, PrPC. We previously reported the bioactivity of acridine-based compounds against PrPSc replication in scrapie-infected neuroblastoma cells and now report the improved potency of bis-acridine compounds. Bis-acridines are characterized by a dimeric motif, comprising two acridine heterocycles tethered by a linker. A library of bis-(6-chloro-2-methoxy-acridin-9-yl) and bis-(7-chloro-2-methoxy-benzo[_b_][1,5]naphthyridin-10-yl) analogs was synthesized to explore the effect of structurally diverse linkers on PrPSc replication in scrapie-infected neuroblastoma cells. Structure–activity analysis revealed that linker length and structure are important determinants for inhibition of prion replication in cultured scrapied cells. Three bis-acridine analogs, (6-chloro-2-methoxy-acridin-9-yl)-(3-{4-[3-(6-chloro-2-methoxy-acridin-9-ylamino)-propyl]-piperazin-1-yl}-propyl)-amine, N,_N_′-bis-(6-chloro-2-methoxy-acridin-9-yl)-1,8-diamino-3,6-dioxaoctane, and (1-{[4-(6-chloro-2-methoxy-acridin-9-ylamino)-butyl]-[3-(6-chloro-2-methoxy-acridin-9-ylamino)-propyl]-carbamoyl}-ethyl)-carbamic acid _tert_-butyl ester, showed half-maximal inhibition of PrPSc formation at 40, 25, and 30 nM, respectively, and were not cytotoxic to uninfected neuroblastoma cells at concentrations of 500 nM. Our data suggest that bis-acridine analogs may provide a potent alternative to the acridine-based compound quinacrine, which is currently under clinical evaluation for the treatment of prion disease.
The prion protein (PrP) is an essential element in the pathogenesis of a devastating group of neurodegenerative disorders called prion diseases or transmissible spongiform encephalopathies (1). At a molecular level, the prion diseases are characterized by an accumulation of PrPSc, an abnormal, β-rich isoform of the host-encoded α-helix-rich PrP (PrPC; refs. 2 and 3). PrPSc is the only known component of the infectious prion particle. Accumulation of PrPSc in the CNS precedes spongiform degeneration and astrocytic gliosis (4). Patients with prion disease develop progressive neurologic dysfunction that results in death typically within 1 yr after the first clinical symptoms appear.
Prion diseases are unique in that they present as spontaneous, inherited, or infectious maladies. In humans, sporadic Creutzfeldt–Jakob disease (CJD) is the most prevalent form, presenting with ataxia and dementia (5). Familial forms of prion disease account for 10–15% of cases and include Gertsmann–Sträussler–Scheinker syndrome, familial CJD, and fatal familial insomnia (6). The emergence of variant CJD (vCJD) in Europe heightened public awareness of prion disorders. Compelling evidence links vCJD to exposure to beef infected with bovine spongiform encephalopathy prions (7). Recent studies have suggested that blood-borne transmission of CJD is possible (8).
Although animal models have been essential in developing an understanding of prion diseases, the long incubation times have forced drug-discovery efforts to focus on scrapie-infected neuroblastoma (ScN2a) cells. Many groups have successfully used ScN2a cells to identify antiprion compounds (9), including Congo red (10), polyamine dendrimers (11), tetrapyrroles (12), and pyridine-based compounds (13). Several compounds have been shown to prolong the incubation period in prion-inoculated rodents (14). However, the protective effect of these compounds is observed only when administered around the time of prion inoculation, or after a period of exposure to the inoculum before administration of the compound. To be clinically useful, a compound must be effective when administered around the time that neurologic symptoms develop. No compound has met this test. Furthermore, many of the compounds identified to date have limited clinical applications due to unacceptable toxicity as well as poor penetration into the brain parenchyma.
The ability of compounds to cross the blood–brain barrier (BBB) in part determines their efficacy in treating neurologic disorders. In searching for compounds active against PrPSc formation, we chose those known to transit the BBB to drive our discovery efforts. We screened structurally distinct psychoactive pharmaceuticals for their ability to reduce PrPSc concentrations in ScN2a cells (15) and found that tricyclic acridine- and phenothiazine-based compounds inhibit prion replication (EC50 = 300–5,000 nM). Quinacrine, an acridine-based compound, was identified as a potent inhibitor of PrPSc formation (EC50 = 300 nM), confirming an earlier independent report on the antiprion activity of this compound (16). Since this observation, clinical studies have been initiated to test the efficacy of quinacrine in the treatment of human prion disease (17). Recently, we focused our medicinal chemistry efforts on further characterizing acridine-based compounds as a novel class of antiprion compounds. Emphasis was placed on characterizing analogs of quinacrine to derive detailed structure–activity data and to arrive at superior compounds with which to treat prion disease. Due to the propensity of PrPSc to assemble into multimers, we postulated that covalent dimers of quinacrine could be more potent inhibitors of prion replication because of an increased local concentration of the active moiety. We have identified bis-acridine compounds that are 10 times more effective (EC50 = 25–40 nM) than quinacrine in ScN2a cells. We report here the bioactivity of bis-(6-chloro-2-methoxy-acridin-9-yl) and bis-(7-chloro-2-methoxy-benzo[_b_][1,5]naphthyridin-10-yl) compounds, tethered via alkyl, polyamine, heterocyclic, and alkyl-ether linkers. A comparison of the bioactivity of these second-generation compounds revealed the importance of the linker length and structure for activity against PrPSc formation. Our data define bis-acridine compounds as a novel class of compounds capable of reducing PrPSc concentrations in ScN2a cells and with a potentially acceptable therapeutic index.
Materials and Methods
Equipment and Reagents.
1H and 13C NMR were performed on a Varian Inova-400 NMR spectrometer; shifts are quoted relative to an internal tetramethylsilane standard. Mass spectral analyses were performed at the Mass Spectrometry Facility (University of California, San Francisco) by using a Perkin–Elmer Sciex API300 electrospray MS. Flash-column chromatography was carried out on silica gel 60 (230–400 mesh) from Sigma–Aldrich. Preparative HPLC was performed on a Varian Prostar 210, fitted with a Varian Prostar 345 dual wavelength UV/Vis detector. Injections were made to a Peeke Scientific (Redwood City, CA) Combi-A 5 μm RP-C18 semipreparative column. 6,9-Dichloro-2-methoxyacridine, and 7,10-dichloro-2-methoxy-benzo[_b_][1,5]naphthyridine were purchased from Sigma–Aldrich and diamines from Sigma–Aldrich and ACROS Organics (Geel, Belgium). _N_-Boc-Ala-Gly-OH (Boc, _tert_-butoxycarbonyl) was purchased from Bachem. _N_-Boc-Ala-OH was from Fluka and PyBrop from Novabiochem.
Preparation of Bis-(6-chloro-2-methoxy-acridin-9-yl) and Bis-(7-chloro-2-methoxy-benzo[_b_][1,5]naphthyridin-10-yl) Compounds 1–11, 13, 14, 20, and 21.
With the exception of compounds 13, 15, and 21 (see spectral assignment in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org), all compounds have been reported (18, 19). Compounds were synthesized according to established procedures (20). Compounds 9 and 20 were prepared as hydrochloride salts by adding drops of concentrated hydrochloric acid to a solution of the crude-free base in acetone and were recrystallized from methanol. The structures of the bis-acridine compounds were confirmed by 1H and 13C NMR, electrospray MS, and when possible, from a comparison with previously published spectral data (18, 19).
Preparation of Acylated Bis-(6-chloro-2-methoxy-acridin-9-yl) Compounds 16–19.
All N-acylated compounds have been published and were prepared by established procedures (18). Treatment of compounds 17 and 19 with 50% trifluoroacetic acid/dichloromethane (10 mM) for 4 h at room temperature afforded the _N_-_tert_-butoxycarbonyl deprotected compounds, 16 and 18, respectively (18). Essentially pure samples (>95% by HPLC) were obtained by RP-C18 HPLC (methanol/water/0.1% trifluoroacetic acid). The structures of the acylated analogs were confirmed by 1H and 13C NMR, electrospray MS, and from a comparison of published spectral data (18).
PrPSc Inhibition Screening Assay in ScN2a Cells.
The ScN2a cell-screening assay was adapted from published methods (13, 15). Neuroblastoma (N2a) cells were infected with the Rocky Mountain Laboratory strain of mouse-adapted scrapie prions and subcloned as described (21). Cells were maintained at 37°C in MEM (Cell Culture Facility, University of California, San Francisco) supplemented with 10% FCS (GIBCO/BRL), 1% Glutamax (GIBCO/BRL), and 1% streptomycin–penicillin (GIBCO/BRL). A 100-mm confluent plate of ScN2a cells was split, and a drop of cells was transferred to a 60-mm plate containing MEM (4 ml). Identically seeded six-well plates were used for the screening assay. Stock solutions of compounds 1–21 were prepared fresh in either PBS, DMSO, or methanol at 1 mM and stored at 4°C. Before use, compounds were diluted to 0.1 mM with PBS, then filtered through a 0.2-μm syringe filter to sterilize. Control cells were unaffected when treated with solvent alone. ScN2a cells were incubated with compound at 50, 200, and 400 nM for 3 d, after which cells were lysed with cold lysis buffer (500 μl/10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.5% Nonidet P-40/0.5% deoxychlolate), and the protein was collected. Protein concentration was determined by using the BCA assay (Pierce) and samples normalized to 0.5 mg of protein. Samples were digested with proteinase K (PK; GIBCO/BRL) at 20 μg⋅ml−1 for 1 h at 37°C. Digestion was inhibited with PMSF (2 mM) and the samples ultracentrifuged (4°C; 100,000 × g) for 30 min. Protein pellets were resuspended in lysis buffer (10 μl) and SDS loading buffer (10 μl), then boiled for 5 min. Samples were loaded onto a 12% SDS/PAGE precast gel (Criterion, Bio-Rad). Western blot analysis was performed as described (15).
Cell Viability Assay.
Uninfected N2a cells were identically seeded onto six-well plates containing MEM (3 ml, supplemented as above). Compounds 1–21 were added at 50, 200, and 500 nM concentrations. Medium was exchanged, together with the compound, every 3 d. After 7 d, thiazolyl blue (300 μl of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, Sigma–Aldrich) in PBS (5 mg⋅ml−1), previously sterile filtered, was added. Cells were incubated at 37°C for 5 h. The medium was removed, and DMSO (3 ml) was added and repeatedly aspirated to solubilize the converted dye. Absorbance was recorded in duplicate at 562 nm, from two independent experiments.
Curing and Reinfection of Scrapie-Infected Mouse Hypothalamus (ScGT1) Cells.
ScGT1 cells were grown in DMEM (4 ml, supplemented as above) on 60-mm plates in the presence of compound 11 at concentrations of 0.5 and 1.0 μM. Control cells were not treated with compound. Media were exchanged every 3 d (together with compound, where appropriate), and cells were split after 1 wk by a 1:20 dilution onto a 60-mm plate. Samples were retained for Western blot analysis. Cells were grown for an additional 2 wk in the absence of compound 11, with media exchanged every 3 d and cells split every 7 d. Samples were retained after this period for later analysis.
“Cured” ScGT1 cells were split 1:10 onto 24-well plates in DMEM (1 ml). A scrapied cell homogenate was prepared from a mixture of ScN2a and ScGT1 (1:1) as described (21). Scrapied cell inoculum (30 μl) was added to the cured cells and incubated for 4 d. Cells were maintained in DMEM, split every 3–4 d onto 24-well plates, for a total of 29 d. After this time, cells were split to a 60-mm plate and maintained for 7 d. Cell lysates were collected, and all samples were analyzed by Western blot, as described.
Molecular Modeling.
Molecular modeling was carried out on an O2 Silicon Graphics (Mountain View, CA) workstation by using ACCELRYS CERIUS2 4.7. Diamines were drawn in extended staggered conformation and energy minimized by using the Steepest Descent function. The distance between distal primary nitrogens was measured in angstroms.
Results
PrPSc and PrPC differ in many ways, including secondary structure content, protease resistance, and oligomeric state. Many therapeutic efforts have focused on breaking the β-sheet structure of PrPSc (22) or stabilizing PrPC (23). Although the mechanism by which some acridine compounds block PrPSc formation is unclear, we reasoned that the potency of acridine compounds could be improved by forming covalent acridine dimers. In effect, dimeric analogs could increase the local concentration of the active moiety and would exploit the propensity of PrPSc to form multimers. To explore this hypothesis, we synthesized a library of bis-acridines and bis-aza-acridines, with which to determine the effect of linker length and conformational constraint on bioactivity (20). We incorporated different acridine linkers, including alkyl (e.g., 1–4, Table 1), polyamine (e.g., 5–10), alkyl ether (e.g., 13) and heterocyclic moieties (e.g., 11 and 12). To understand whether additional activity could arise from linker substituents, we also synthesized a series of N-acylated and N-alkylated analogs (14–19, Table 1).
Table 1.
Efficacy and cellular toxicity screen of bis-acridine compounds for PrPSc inhibition in ScN2a cells
| Compound* | % PrPSc (±SEM), nM | % Cell viability (±SEM), nM | ||||
|---|---|---|---|---|---|---|
| 50 | 200 | 400 | 50 | 200 | 500 | |
| Alkyl | ||||||
| 1 | 72 (±1) | 65 (±2) | 54 (±1) | 100 (±9) | 100 (±9) | 100 (±6) |
| 2 | 84 (±6) | 50 (±7) | 41 (±3) | 90 (±3) | 96 (±3) | 82 (±3) |
| 3 | 93 (±7) | 71 (±3) | 78 (±2) | 100 (±4) | 100 (±2) | 97 (±6) |
| 4 | 100 (±2) | 58 (±4) | 20 (±1) | 99 (±21) | 95 (±21) | 90 (±17) |
| Polyamine | ||||||
| 5 | 90 (±7) | 58 (±1) | 44 (±10) | 100 (±20) | 100 (±9) | 100 (±18) |
| 6 | 98 (±5) | 59 (±6) | 46 (±8) | 98 (±5) | 33 (±1) | 1 (±1) |
| 7 | 61 (±3) | 26 (±2) | 13 (±3) | 26 (±1) | 8 (±1) | 1 (±1) |
| 8 | 72 (±6) | 19 (±3) | 9 (±4) | 100 (±13) | 23 (±10) | 1 (±1) |
| 9 | 85 (±1) | 32 (±3) | 13 (±5) | 92 (±7) | 16 (±7) | 10 (±2) |
| 10 | 72 (±3) | 71 (±5) | 31 (±6) | 100 (±4) | 43 (±5) | 1 (±1) |
| Heterocyclic | ||||||
| 11 | 61 (±3) | 19 (±5) | 4 (±2) | 94 (±11) | 89 (±9) | 79 (±1) |
| 12† | 80 (±2) | 75 (±6) | 67 (±1) | 100 (±10) | 100 (±6) | 100 (±11) |
| Alkyl ether | ||||||
| 13 | 41 (±8) | 29 (±8) | 15 (±7) | 97 (±8) | 90 (±9) | 98 (±11) |
| _N_-Alkylated polyamine | ||||||
| 14 | 78 (±9) | 49 (±8) | 38 (±9) | 73 (±2) | 59 (±8) | 1 (±1) |
| 15 | 88 (±2) | 41 (±6) | 11 (±1) | 100 (±5) | 77 (±4) | 0 (±1) |
| _N_-Acylated polyamine | ||||||
| 16 | 100 (±6) | 80 (±12) | ‡ | 100 (±22) | 59 (±13) | 0 (±1) |
| 17 | 84 (±10) | 36 (±1) | 7 (±1) | 98 (±12) | 85 (±6) | 84 (±3) |
| 18 | 90 (±5) | 83 (±5) | 79 (±3) | 96 (±4) | 93 (±1) | 0 (±1) |
| 19 | 97 (±2) | 84 (±6) | 72 (±10) | 88 (±18) | 78 (±13) | 70 (±12) |
| Bis-aza-acridines | ||||||
| 20 | 88 (±3) | 82 (±8) | 65 (±1) | 100 (±15) | 100 (±9) | 88 (±8) |
| 21 | 100 (±6) | 88 (±8) | 77 (±3) | 95 (±3) | 100 (±6) | 96 (±5) |
A qualitative ScN2a cell screening assay was used to derive trends in bioactivity across the compound library and to identify potential lead bis-acridine compounds. Antiprion activity was determined from Western blot densitometry of the PK-resistant PrPSc and is expressed as an average percent reduction of PrPSc compared with untreated control cells (Table 1). Control cells treated with solvent alone showed no detectable reduction in PrPSc concentrations.
Activity data derived from this initial screen revealed that structural features of the linker contribute to activity against PrPSc formation. Compounds tethered by polyamine (e.g., 5–10) or alkyl-ether linkers (e.g., 13) showed improved activity relative to alkyl-linked bis-acridines (e.g., 1–4). The increase in activity may result from additional hydrogen-bonding interactions between the heteroatoms of the linker and the target receptor. Additionally, the increased hydrophilicity of a heteroalkyl linker may improve the localization of these compounds to the subcellular compartment, where their effect on PrPSc concentration is exerted. N-Alkylated and N-acylated analogs (e.g., 14–19, Table 1) had variable activities depending on the size and polarity of the _N_-substituent. The data suggest that linker substituents can contribute favorably to bioactivity; however, this activity can be compromised by modest increases in the molecular volume of the substituent, presumably due to steric constraints.
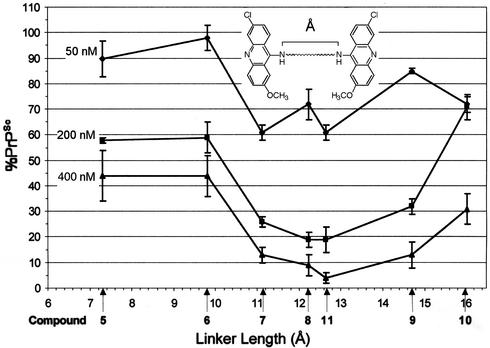
The activity of polyamine-linked compounds 5–11 allowed us to correlate the length of the polyamine linker with activity against PrPSc formation. The energy minimized staggered conformation of polyamine linkers used in compounds 5–11 was modeled in compuo and the distance between distal nitrogens measured. The lengths of the polyamines ranged from 7.4 to 16.0 Å (Fig. 1), and the bioactivity of compounds 5–11 correlated with the interacridine distance. Optimal activity was observed when the acridine heterocycles were separated by more than ≈10.0 Å (Fig. 1).
Figure 1.
The observed bioactivity of polyamine-linked bis-acridines in ScN2a cells correlates to the length of the polyamine linker. The energy-minimized staggered conformations of polyamine linkers from compounds 5–11 were modeled in compuo. The linker length (Å) was measured as the distance between distal nitrogens. Average percent reduction in PrPSc concentrations in ScN2a cells (Table 1) is plotted versus the length of the polyamine linker (bars represent standard errors from at least three independent immunoblots). Optimal bioactivity was observed when pendant acridine heterocycles were separated by more than ≈10 Å.
Bis-acridines are known to be cytotoxic due to DNA bis-intercalation of the acridine heterocycles according to the nearest-neighbor exclusion principle (24). Accordingly, we used cell cytotoxicity as additional selection criteria in our study of bis-acridine compounds. Uninfected N2a cells were incubated with individual compounds, and cell viability was determined by using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide assay (Table 1). The results obtained reflect previously reported cytotoxicity data for this class of compounds (18, 25). Notably, we observed that polyamine-linked bis-acridines were generally cytotoxic to N2a cells. For example, the spermidine-linked analog, compound 7, a known bis-intercalator, proved to be cytotoxic to N2a cells, with only 26% of cells remaining viable at 50 nM. However, bis-intercalation requires both a permissible linker length and conformational flexibility; thus cells remained viable (>85%) when incubated with bis-acridines comprising alkyl (e.g., 1–4), alkyl ether (13), and certain sterically hindered linkers (e.g., 11 or 17).
We previously showed the importance of the tricyclic acridine scaffold for activity against PrPSc formation in ScN2a cells (15). Phenothiazine- or quinoline-based analogs were shown to give reduced bioactivity in ScN2a cells relative to acridine-based compounds. Bis-acridine analogs 20 and 21 (Table 1) incorporate a substituted benzo[_b_][1,5]naphthyridine (“aza-acridine”) heterocycle. Benzo[_b_][1,5]naphthyridines differ from acridines by having an additional ring nitrogen. The activity of bis-aza-acridine analog 20 was reduced relative to the equivalently linked bis-acridine, 7. Additionally, 20 was nontoxic to N2a cells at 500 nM, whereas we observed 99% cell death with an equivalent concentration of the bis-acridine 7. We have also observed that a monomeric aza-quinacrine analog (“azacrine”) (26), in which the acridine heterocycle of quinacrine was replaced by a benzo[_b_][1,5]naphthyridine heterocycle, displayed reduced activity relative to quinacrine. Azacrine was inactive in ScN2a cells at concentrations between 100 nm and 400 nM, whereas quinacrine has an EC50 of 300 nM (B.C.H.M., S.B.H., L.W.D., S.B.P., and F.E.C., unpublished data). Thus, the bioactivity of acridine-based compounds can be compromised by small structural changes to the heterocyclic scaffold, even a carbon-to-nitrogen substitution.
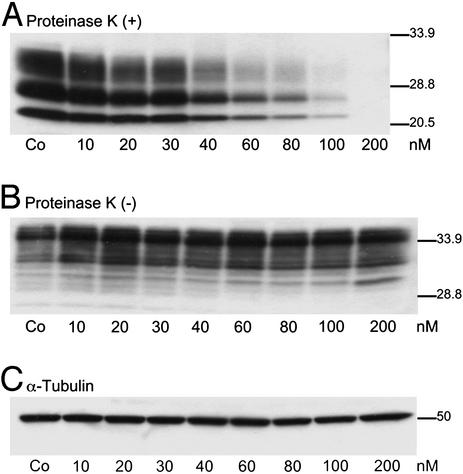
From these bioactivity and cytotoxicity screens, we identified three bis-acridine compounds (11, 13, and 17) that warranted further characterization. Compounds 11, 13, and 17 showed similar bioactivity to that of flexible polyamine-based analogs of a similar linker length (e.g., 7 and 8) in a 3-d incubation with ScN2a cells. N2a cells remained viable (>85%) when incubated with up to 500 nM concentrations of compounds 11, 13, or 17, whereas other bis-acridine compounds were cytotoxic at this concentration. The linker lengths of compounds 11, 13, and 17 were calculated at 12.63, 10.68, and 11.11 Å, respectively, and thus lie outside the observed minimal distance constraint of ≈10.0 Å for optimal bioactivity against PrPSc formation, as observed for polyamine-linked analogs (Fig. 1). Compound 11 was shown to reduce PrPSc levels in ScN2a cells in a dose-dependent manner (Fig. 2A), without affecting PrPC (Fig. 2B). In contrast to other polyamine-based antiprion compounds (e.g., polyamine dendrimers; ref. 11), the bioactivity of compound 11 was not compromised when incubated with the lysosomotropic agent, chloroquine, in cultures of ScN2a cells (results not shown).
Figure 2.
Piperazine-based bis-acridine, 11, reduces PrPSc concentration from ScN2a cells in a dose-dependent manner. ScN2a cells were incubated with compound 11 at the concentrations indicated (10–200 nM) for 7 d. Control cells (Co) were untreated. Cell lysates were harvested and either PK-digested (A) or undigested (B) before immunoblotting with anti-PrP Fab D13. (A) Dose-dependent reduction of PrPSc concentration in ScN2a after incubation with compound 11. (B) PrPC levels remained unchanged by treatment. (C) Immunoblot of undigested cell lysate probed with antitubulin.
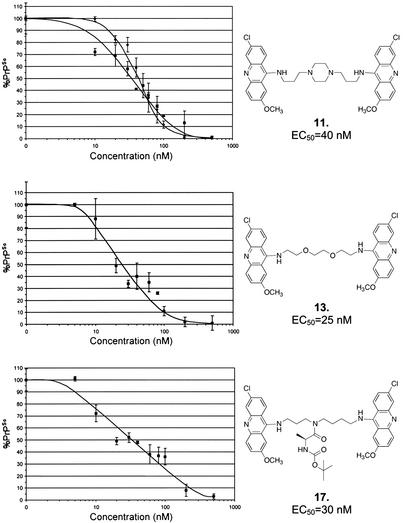
Dose–response curves were derived after incubating ScN2a cells with compounds 11, 13, and 17 for 7 d at concentrations between 5 and 500 nM (Fig. 3). Cell lysates were analyzed by ELISA (D. Peretz, personal communication). The EC50 of compounds 11, 13, and 17 were 40, 25, and 30 nM, respectively. A ScN2a cell dose–response curve for compound 11 was also derived by Western blot densitometry. Results from both Western blot and ELISA methods were consistent. The bioactivity of these compounds is ≈10-fold greater relative to the monomeric acridine-based compound, quinacrine (EC50 = 300 nM).
Figure 3.
Dose–response curves for bis-acridine compounds 11, 13, and 17, nanomolar inhibitors of PrPSc replication in ScN2a cells. ScN2a cells were incubated for 7 d with compound 11, 13, or 17 at concentrations between 5 and 500 nM. PK-digested cell lysates were analyzed by ELISA (squares, lower curve), by using the procedure of D. Peretz (personal communication). The EC50 of 11, 13, and 17 were 40, 25, and 30 nM, respectively (bars represent standard error from three independent experiments). A dose–response curve for compound 11 was also derived by Western blot densitometry of PK-digested cell lysates (diamonds, upper curve). Both the Western blot and ELISA methods were in good agreement.
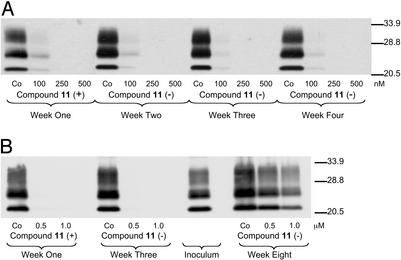
Incubation of ScN2a cells with compound 11 at concentrations of 250 and 500 nM for 1 wk resulted in the complete clearance of protease-resistant PrPSc, as determined by Western blot analysis (Fig. 4A). After this treatment, cells were serially passaged for an additional 3 wk in the absence of compound 11. Cell lysates were collected at 1-wk intervals and analyzed for PK-resistant PrPSc by Western blot. PrPSc could not be detected in cell lysates after treatment with compound 11, suggesting that treatment permanently cured ScN2a cells of PrPSc.
Figure 4.
(A) Treatment of ScN2a cells with compound 11 for 7 d clears existing PrPSc and cells remain clear of PrPSc 3 wk after discontinuation of treatment. ScN2a cells were incubated with compound 11 at 100, 250, and 500 nM for 1 wk. After treatment, the cells were serially passaged for an additional 3 wk in the absence of compound 11. Cell lysates were collected at the end of each week and PK-digested before Western blot analysis (anti-PrP Fab D13). Three weeks after discontinuation of treatment, protease-resistant PrPSc could not be detected in cell lysates, suggesting that bis-acridine, 11, permanently cured ScN2a cells after treatment for 1 wk. (B) ScGT1 cells were incubated for 1 wk with compound 11 at concentrations of 0.5 and 1.0 μM. Cell lysates were collected and analyzed by Western blot after 1 wk of treatment to reveal the absence of PK-resistant PrP. The cells were maintained in supplemented culture medium for an additional 2 wk in the absence of 11, then reinfected with a prion inoculum. Cells were grown in the presence of the inoculum for 4 d, then maintained for an additional 5 wk. After this period, cell lysates were collected and analyzed for PK-resistant PrP. PrPSc was detected in cells that had previously been cured by compound 11.
The mouse hypothalamic cell line, GT1, can be infected with mouse scrapie prions and can produce stable levels of PrPSc (ScGT1) (27). To validate the observed reduction in PrPSc concentrations in ScN2a cells on treatment with bis-acridines, we also treated ScGT1 cells with compound 11. Bis-acridine compound 11 reduced PrPSc in a dose-dependent manner at concentrations between 100 and 500 nM, as determined by Western blot analysis of PK-digested cell lysates (data not shown). Additionally, ScGT1 cells could be cured of PrPSc after a 1-wk incubation with compound 11 at either 0.5 or 1.0 μM (Fig. 4B). Similar to the observed curing of ScN2a cells by compound 11 (Fig. 4A), PrPSc was not detected in ScGT1 cell lysates 2 wk after discontinuation of treatment. These cured ScGT1 cells could subsequently be reinfected by incubation with a prion inoculum (Fig. 4B). However, the prion titer obtained in these reinfected cells depended on the concentration of compound 11 originally used to cure the cells. Thus, treatment with compound 11 appears to decrease the capacity of ScGT1 cells to be reinfected with prions, relative to controls.
Discussion
The protein deposition diseases, including the prion diseases, Alzheimer's and Parkinson's diseases, involve multimerization and aggregation of specific misfolded proteins (28, 29). As expected, in vitro and in vivo models have demonstrated that increases in protein concentration can initiate or accelerate misfolding and aggregation. Therapeutic strategies targeting these diseases have focused on either increasing the lability of the aggregated state or inhibiting initial multimerization of the aberrant, misfolded protein (30). In the latter case, removal of the supply of the oligomeric precursor proteins is often sufficient to reverse aggregation as the concentration equilibrium shifts to favor plasma-soluble species. Oligomerization of PrP seems to be central to the mechanism of PrPSc formation (31), suggesting that high local protein concentrations are achieved in all steps of multimerization leading to the aggregated endpoint. Thus, we reasoned that a covalently linked dimer of a compound that reduced PrPSc concentration in ScN2a cells would be more potent than its monomeric counterpart. This multivalency strategy may be generally applicable to other diseases of protein conformation, in which species along the pathway to the aggregated endpoint are characterized by high protein concentrations.
Bis-acridines are well characterized in medicinal chemistry due to their cellular toxicity (24). This has been exploited in their evaluation as compounds to treat cancer (20, 25). The challenge in targeting bis-acridine-based therapies is to separate the desirable bioactivity of these compounds from their DNA bis-intercalative cytotoxicity. Therapeutic indices have been used in pharmacology to express the ratio between the efficacious and injurious concentrations of a compound. For compounds designed to treat moderate chronic diseases, these ratios overwhelmingly favor efficacy. For most cancer chemotherapeutics, the therapeutic index is unfortunately close to unity.
We initially targeted a small library of bis-acridines to explore anti-PrPSc activity and cellular cytotoxicity, with the aim of identifying bis-acridines with an acceptable therapeutic index. Our previous structure–activity data on monoacridine compounds (e.g., quinacrine) revealed a heavy dependence on the length and composition of the dibasic alkyl substituent peri to the ring nitrogen (15). Thus, we reasoned that bis-acridine compounds may have a similar side-chain dependence to their monoacridine counterparts. In this instance, the side-chain serves as a linker to tether two pendant acridine heterocycles. The focused library of bis-acridine compounds explored tolerances for linker length and composition and defined a strong correlation between structural features of the acridine linker and bioactivity against PrPSc formation. We have also explored the dependence on the heterocyclic scaffold for activity against PrPSc formation. The reduced bioactivity of aza-acridine compounds 20 and 21 illustrated the specific contribution to activity made by the acridine scaffold. Such a dramatic reduction in activity by transforming a scaffold carbon to a nitrogen atom is surprising. These observations add to the accumulating structure–activity relationship data for acridine compounds, the subtlety of which suggests that these compounds do not work via a generalized mechanism (e.g., lysosomotropic). Given the parallel structure dependence, both for the side-chain and acridine scaffold, it is interesting to speculate that mono- and bis-acridine compounds access an equivalent mechanism to reduce PrPSc load in ScN2a and ScGT1 cells.
Bis-intercalative cytotoxicity has been well characterized for bis-acridine compounds. To achieve a balance between desirable bioactivity and cytotoxicity, we targeted sterically constrained bis-acridine analogs. Intercalation of bis-acridines can be mitigated by using rigidified or sterically constrained linkers to tether the acridine heterocycles (32). Following this rationale, we targeted the piperazine-linked analog compound 11, and N-acylated analog compound 17. In a previous study, compound 11 was shown to be less cytotoxic to MRC-5 fibroblast cells than nonconstrained polyamine-linked bis-acridines (e.g., 7; ref. 18). This suggests that inclusion of the piperazine heterocycle disfavors DNA complexation, either through steric hindrance or by restricting the conformational flexibility of the acridine heterocycles, such that bis-intercalation is mitigated. Additionally, we considered that the conformational restriction imparted by the piperazine heterocycle or bulky _N_-substituents may be beneficial to activity against PrPSc replication. Conformational restriction has been used extensively in drug design to preorganize ligands into a bioactive conformation. A rigid scaffold can correctly position and orient key structural features without having to undergo an entropically costly conformational rearrangement before binding. Thus, constrained ligands can have improved bioactivity relative to their unrestrained counterparts, if the constraints are compatible with the ultimate receptor-relevant conformation. A balance between cellular efficacy and toxicity was observed with compounds 11 (EC50 = 40 nM) and 17 (EC50 = 30 nM, Fig. 3). We are encouraged by the improved therapeutic index of compounds 11 and 17 relative to flexible polyamine-linked analogs, and plan to synthesize a third-generation compound library to explore constrained bis-acridine analogs.
It is interesting to speculate about the mechanism of action of bis-acridine compounds. The linker dependence observed for the polyamine series (5–11, Fig. 1) hints at a possible binding mechanism whereby both acridine heterocycles can occupy independent binding sites to affect PrPSc load in scrapied cells. Analogs linked by less than ≈10 Å had reduced bioactivity, suggesting that in these instances, the pendant acridine heterocycles could not bind cooperatively due to the distance constraint. Similarly, as the linker length increased above the observed optimum (≈12.5 Å), bioactivity decreased, possibly due to entropic considerations of the longer and hence more flexible analogs. Determining the mechanism of action of acridine compounds will greatly aid in developing this class of compound for the effective treatment of prion disease.
Complex cellular pathways have been dissected by using small molecule agonists or antagonists of biological function. Selective inhibitors have been used to probe the proteome, to elucidate function, and to identify directly novel receptor molecules. Because the conversion of PrPC to PrPSc is not clearly understood at either a molecular or cellular level, compounds that exert a dramatic effect on PrPSc concentrations, such as bis-acridines, can serve as tools to probe prion biology. The subtleties in structure–activity dependence of both mono- and bis-acridine-based compounds suggest that this class of compounds has a specific binding interaction with a novel receptor molecule that participates in either PrPSc formation or clearance. We have synthesized labeled acridine analogs with photolyzable crosslinking moieties that are bioactive against PrPSc load in ScN2a cells.
Given that high protein concentrations are inherent to the mechanism of prion aggregation, we rationalized that covalent acridine dimers could be more potent than monomeric equivalents. We have used a traditional medicinal chemistry approach to explore this notion. The data presented here define bis-acridines as a potent class of compounds that demonstrate an acceptable therapeutic index in a cell-based model of prion disease. We have identified three lead compounds, 11, 13, and 17, that reduce PrPSc levels at nanomolar concentrations. Although we currently do not understand the mechanism by which acridine compounds affect PrPSc formation, these compounds offer unique tools to study the mechanism of prion replication.
Supplementary Material
Supporting Information
Acknowledgments
We thank Joanne Lee (Institute for Neurodegenerative Diseases, University of California, San Francisco) for technical assistance. B.C.H.M. is supported by an award from The John Douglas French Alzheimer's Foundation. This work is supported by National Institutes of Health Grants AG02132 and AG10770 as well as by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations
PrP
prion protein
PrPC
host-encoded PrP
PrPSc
abnormal isoform of PrP
N2a
mouse neuroblastoma
ScN2a
scrapie-infected N2a
ScGT1
scrapie-infected mouse hypothalamus
PK
proteinase K
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–133683. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 3.Cohen F E, Pan K M, Huang Z, Baldwin M, Fletterick R J, Prusiner S B. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 4.DeArmond S J, Prusiner S B. Curr Top Microbiol Immunol. 1996;207:125–146. doi: 10.1007/978-3-642-60983-1_9. [DOI] [PubMed] [Google Scholar]
- 5.Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen S G, Farlow M, Dickson D W, Sima A A, Trojanowski J Q, et al. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner S B, Hsiao K K. Ann Neurol. 1994;35:385–395. doi: 10.1002/ana.410350404. [DOI] [PubMed] [Google Scholar]
- 7.Scott M R, Will R, Ironside J, Nguyen H-O B, Tremblay P, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1999;96:15137–15142. doi: 10.1073/pnas.96.26.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter N, Foster J, Chong A, McCutcheon S, Parnham D, Eaton S, MacKenzie C, Houston F. J Gen Virol. 2002;83:2897–2905. doi: 10.1099/0022-1317-83-11-2897. [DOI] [PubMed] [Google Scholar]
- 9.Supattapone S, Nishina K, Rees J R. Biochem Pharmacol. 2002;63:1383–1388. doi: 10.1016/s0006-2952(02)00874-2. [DOI] [PubMed] [Google Scholar]
- 10.Rudyk H, Vasiljevic S, Hennion R M, Birkett C R, Hope J, Gilbert I H. J Gen Virol. 2000;81:1155–1164. doi: 10.1099/0022-1317-81-4-1155. [DOI] [PubMed] [Google Scholar]
- 11.Supattapone S, Wille H, Uyechi L, Safar J, Tremblay P, Szoka F C, Cohen F E, Prusiner S B, Scott M R. J Virol. 2001;75:3453–3461. doi: 10.1128/JVI.75.7.3453-3461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caughey W S, Raymond L D, Horiuchi M, Caughey B. Proc Natl Acad Sci USA. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrier V, Wallace A C, Kaneko K, Safar J, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 2000;97:6073–6078. doi: 10.1073/pnas.97.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown P. Neurology. 2002;58:1720–1725. doi: 10.1212/wnl.58.12.1720. [DOI] [PubMed] [Google Scholar]
- 15.Korth C, May B C, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 2001;98:9836–9841. doi: 10.1073/pnas.161274798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doh-Ura K, Iwaki T, Caughey B. J Virol. 2000;74:4894–4897. doi: 10.1128/jvi.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love R. Lancet. 2001;358:563. doi: 10.1016/S0140-6736(01)05748-8. [DOI] [PubMed] [Google Scholar]
- 18.Girault S, Grellier P, Berecibar A, Maes L, Mouray E, Lemiere P, Debreu M A, Davioud-Charvet E, Sergheraert C. J Med Chem. 2000;43:2646–2654. doi: 10.1021/jm990946n. [DOI] [PubMed] [Google Scholar]
- 19.Barbet J, Roques B P, Le Pecq J B. C R Hebd Seances Acad Sci D. 1975;281:851–853. [PubMed] [Google Scholar]
- 20.Chen T K, Fico R, Canellakis E S. J Med Chem. 1978;21:868–874. doi: 10.1021/jm00207a006. [DOI] [PubMed] [Google Scholar]
- 21.Bosque P J, Prusiner S B. J Virol. 2000;74:4377–4386. doi: 10.1128/jvi.74.9.4377-4386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto C, Kascsak R J, Saborio G P, Aucouturier P, Wisniewski T, Prelli F, Kascsak R, Mendez E, Harris D A, Ironside J, et al. Lancet. 2000;355:192–197. doi: 10.1016/s0140-6736(99)11419-3. [DOI] [PubMed] [Google Scholar]
- 23.Peretz D, Williamson R A, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn I R, Legname G, Wormald M R, Rudd P M, et al. Nature. 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 24.Le Pecq J B, Le Bret M, Barbet J, Roques B. Proc Natl Acad Sci USA. 1975;72:2915–2919. doi: 10.1073/pnas.72.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen J B, Langvad E, Frandsen F, Buchardt O. J Med Chem. 1983;26:1510–1514. doi: 10.1021/jm00364a028. [DOI] [PubMed] [Google Scholar]
- Besly, D. M. & Goldberg, A. A. (1954) J. Chem. Soc. 2448–55.
- 27.Schatzl H M, Laszlo L, Holtzman D M, Tatzelt J, DeArmond S J, Weiner R I, Mobley W C, Prusiner S B. J Virol. 1997;71:8821–8831. doi: 10.1128/jvi.71.11.8821-8831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson C M, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 29.Kirkitadze M D, Bitan G, Teplow D B. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 30.Sacchettini J C, Kelly J W. Nat Rev Drug Discov. 2002;1:267–275. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- 31.Baskakov I V, Legname G, Baldwin M A, Prusiner S B, Cohen F E. J Biol Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 32.Denny W A, Atwell G J, Baguley B C, Wakelin L P G. J Med Chem. 1985;28:1568–1574. doi: 10.1021/jm00149a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information