Gene–environment interaction modulated by allelic heterogeneity in inflammatory diseases (original) (raw)
Abstract
CARD15 is a major susceptibility gene for a frequent multifactorial chronic inflammatory bowel disorder, Crohn disease (CD). By using NF-κB activation assays, the cytosolic CARD15 was shown to efficiently detect bacterial peptidoglycan (PGN), reminiscent of the PGN recognition protein surveillance mechanism in Drosophila. The 3 CD-associated variants and 13 additional variants carried by CD patients demonstrated impaired PGN-dependent response revealing null, hypomorphic, or dominant-negative properties. Quantitative parametrization of this response, computed from the patients' CARD15 genotypes, was predictive of several variable CD manifestations. In contrast,CARD15 alleles associated with Blau's syndrome promoted PGN-independent NF-κB activation, an observation that accounts for the minimal microbial input in the etiology of this dominant, monogenic inflammatory disorder affecting solely aseptic sites.
Although the precise etiology of Crohn disease (CD; Online Mendelian Inheritance in Man 266600) is unknown, biochemical and clinical observations (1) suggest a breakdown of tolerance to luminal bacteria (2–5) in the digestive tract of susceptible individuals. CARD15, formerly called_NOD2,_ has recently been identified as a CD-susceptibility gene (6, 7). Compared with wild-type (WT) homozygotes, heterozygotes for one of three uncommon variants have a moderate genotype relative risk of 3 to develop CD. However, individuals homozygous or compound heterozygous for these variants present a risk of 40, suggesting that the functional defect causing CD susceptibility is inherited recessively (6). Compared with unaffected individuals, an excess of rare mutations was shown among CD patients, indicating that additional disease-susceptibility alleles may exist (6). Three other_CARD15_ mutations have been implicated in Blau's syndrome (BS), a monogenic, dominantly inherited early-onset granulomatous arthritis (8). CARD15 is a member of a family of human cytosolic, non-TIR NACHT-LRR proteins (TIR = Toll/IL-1 receptor; NACHT = neuronal apoptosis inhibitor protein, MHC class 2 transactivator, HET-E, TP1; LRR = leucine-rich repeats) (9, 10), akin to plant R proteins. Like other members of this family, CARD15 not only might enhance apoptosis but also inflammation through the RIP-like interacting CLARP kinase/Iκ-B kinase/nuclear factor-κB (RICK/IKK/NF-κB) signaling pathway (11). By using NF-κB_in vitro_ assays, the LRR domain of CARD15 WT was shown to confer responsiveness to various bacterial components, including commercial lipopolysaccharides (LPS; ref. 12). One of the CD-associated variants, 1007fs-CARD15 [single nucleotide polymorphism 13 (SNP13)], was reported to be deficient in triggering responsiveness to commercial LPS (7). Here, we investigate the functional ability of CARD15 variants to activate the NF-κB pathway.
Materials and Methods
Eukaryotic Expression Vectors.
A peripheral blood leukocyte cDNA library (λ Zap Express/Eco_RI vector, no. 938202, Stratagene), kindly provided by S. Gisselbrecht (Institut National de la Santé et de la Recherche Médicale U363, Paris), was screened for pBKCMV-CARD15-expressing clones (EMBL accession no. CAC42117). A full-length CARD15-expressing vector was isolated and verified by complete automatic sequencing. Thirty-five nucleotide variations of the_CARD15 gene (6, 8, 14) were subsequently introduced by using QuikChange XL site-directed mutagenesis kit (Stratagene). The entire coding region was verified by sequencing, and the size of the encoded product was verified by immunoblotting for all constructs. Variants are referred to the nomenclature proposed by the HUGO committee.
NF-κB Luciferase Assays.
Human embryonic kidney (HEK) 293 cell lines were cultured in DMEM (Life Technologies, Rockville, MD) supplemented with 10% (vol/vol) FBS. Transfection was performed by using Fugene 6 (Roche Diagnostics), according to the manufacturer's directions. In each experiment, the HEK 293 cells were cotransfected with a constant amount of DNA that included 100 ng of Igκ luciferase reporter plasmid, 10 ng of normalizing constructs pnull-Renilla (Promega), the indicated amount of pBKCMV-CARD15-construct complemented with the corresponding empty pBKCMV. HEK 293 cells were plated in 24-well culture dishes and left unexposed or exposed to relevant sonicated pathogen-associated molecular pattern (PAMP) at the time of transfection. Cell lysates were obtained 24 h after transfection/PAMP exposure. NF-κB luciferase assays were performed on one-tenth of the total lysates by using Dual-Luciferase Reporter Assay System (Promega); in a series of experiments, the remaining lysates were used to verify (by measuring_Renilla_ luminescence) that CARD15 molecules were expressed at a comparable level of expression after normalization. Means and SEM are representative of at least three independent experiments performed in duplicate or triplicate.
Human Cell Exposure to Bacterial Cell-Wall Components.
Several commercial LPS (Sigma) were used in this work (Escherichia coli O111:B4, Serratia marcescens,E. coli O55:B5, E. coli O26:B6,Salmonella typhimurium, Salmonella enteritidis, and E. coli O127:B8). The E. coli deep rough mutant strain F515 hexaacylated LPS was purified and resuspended in endotoxin-free sterile saline. Lipoteichoic acid (LTA) was isolated from Bacillus subtilis (Sigma). Lipid A (Sigma) was isolated from E. coli. Insoluble (Fluka) and soluble peptidoglycan (PGN) were both derived from Staphylococcus aureus. E. coli K-12 lipoproteins, used in this work, correspond to a recombinant “PGN-associated lipoprotein” (encoded by excC) devoid of the first cysteine residue (with six additional N-terminal histidines), which is soluble and nonacylated (provided by Roland Lloubes, Centre National de la Recherche Scientifique UPR9027, Marseille, France). The synthetic bacterial lipopeptide (PAM3-CSK4), initiating proinflammatory signal transduction through Toll-like receptor 2 (TLR2), was also used. All the above cited PAMPs were used at 10 μg/ml. Finally, the unmethylated immuno-stimulatory bacterial DNA was mimicked by CpG-ODN (5′-TCC-ATG-ACG-TTC-CTG-ATG-CT-3′) with, as negative control, the non-CpG-ODN (5′-TCG-TCG-TTC-CCC-CCC-CCC-CC-3′). They were both used at 5 μM in the assays.
Immunoblotting and Antisera.
For CARD15 Western blotting, a CARD15 polyclonal antiserum raised against the specific CARD15 region (1030-KLGCRDTRLLL-1040) and a commercial CARD15 polyclonal antibody (Cayman Chemical, Ann Arbor, MI) were used at a 1:500 dilution in 5% skim milk/TBST (25 mM Tris, pH 7.5/150 mM NaCl/0.1% Tween 20). Equivalent protein amounts of lysed cell extracts were resolved on SDS/8% polyacrylamide gel, and the proteins were transferred to nitrocellulose membranes. CARD15 immunocomplexes were revealed by using a horseradish peroxidase-conjugated anti-rabbit IgG used at a 1:2,000 dilution and visualized by enhanced chemiluminescence ECL Plus (Amersham Pharmacia).
Results
CARD15 Is a Sensor of Bacterial PGN but Not of LPS.
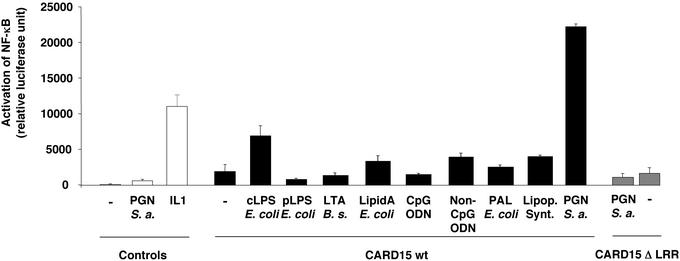
Because CARD15 confers differential response to commercial LPS preparations (12 and data not shown), the putative implication of biologically active trace contaminants was addressed (Fig.1). Although proficient in activating NF-κB in macrophages, a protein-free fraction of the most active LPS from E. coli deep rough mutant strain F515 and lipid A, which is the active component of LPS, were ineffective in activating NF-κB in CARD15-transfected HEK 293 cells. Thus, LPS is not a relevant PAMP for CARD15. On the contrary, commercial and endotoxin-free purified bacterial PGN triggered NF-κB in CARD15-transfected cells. The PGN-induced activation was CARD15-dose dependent (data not shown) and requires the LRR domain. Moreover, compared with PGN, CARD15-mediated responses with other bacterial components were inconspicuous. TLR2, a major PGN-membrane recognition receptor, is not expressed at a detectable level in HEK 293 (13). In conclusion, CARD15 confers responsiveness to PGN, but not to LPS. It may be complementary and nonredundant with CARD4/NOD1 for the detection of bacterial infection (12, 14).
Figure 1.
Differential sensing of cytosolic PAMP by CARD15. Briefly, a minimal amount (30 ng) of CARD15- or CARD15ΔLRR-encoding vector was used for reliable detection of the NF-κB activation by the WT allele. Values are noted in relative luciferase units (RLU) after standardization with the pnull-Renilla system (Promega). Means and SEM are shown for at least three independent experiments, each performed in duplicate. For positive control stimulation, HEK 293 cells were treated with 10 ng/ml IL-1 for 4 h. After transfection, HEK 293 cells were either not exposed (−) or exposed to 10 μg/ml of each of the following PAMP: the most active tested commercial LPS (from serotype O111:B4 E. coli, labeled cLPS E. coli), hexaacylated protein-free purified from E. coli F515 (pLPS E. coli), lipoteichoic acid from B. subtilis (LTA B. s.), lipid A from E. coli (LipidA E. coli), PGN-associated lipoprotein from E. coli (PAL E. coli), PAM3-CysOH synthetic bacterial lipopeptides (Lipop. Synt.) purchased from Novabiochem, and PGN from S. aureus _(_PGN S. a.). Cells were also exposed to 5 μM CpG oligodeoxyribonucleic acids (CpG ODN) and non-CpG oligodeoxyribonucleic acids (non-CpG ODN). In addition to PGN or IL-1, vector-transfected cells were exposed to all tested PAMPs, and the NF-κB pathway was shown to be nonactivated (data not shown).
Mutations Causing BS Are Associated with Increased Basal NF-κB Activity.
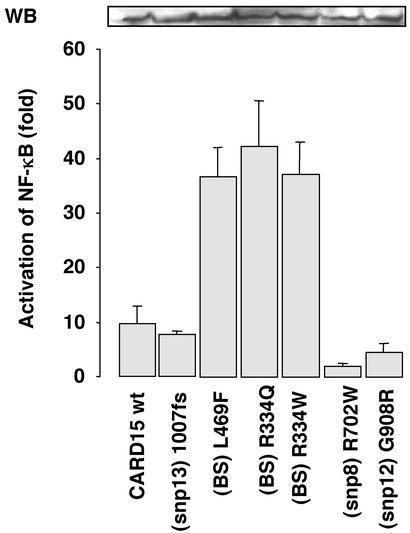
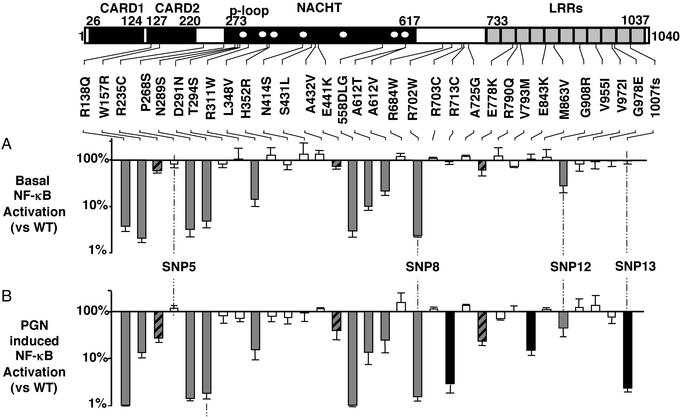
At equivalent expression levels, the three BS-causing mutations revealed an ≈4-fold increase in basal activity, compared with the WT allele, indicating that in BS, the NF-κB CARD15-dependent activity may be constitutive (Fig. 2). In contrast, the CD-associated CARD15 variants suggested decreased basal activity of NF-κB (Fig. 2). To assess this decrease more adequately, transfection experiments were performed with ≈20-fold higher amounts of CARD15. These experiments demonstrated that the basal activity of SNP8 and SNP12, but not that of SNP13, were constitutively impaired (Fig. 3A).
Figure 2.
PGN-independent NF-κB activation potential of CARD15 variants associated with CD and BS. The WT reference CARD15 construct and six derived constructs, each carrying a single mutation associated with either BS (L469F, R334Q, and R334W) or CD susceptibility (R702W-SNP8, G908R-SNP12, and 1007fs-SNP13), were transiently cotransfected in HEK 293 cells with a minimal amount of DNA (30 ng). (Upper) A typical CARD15 immunoblot (WB) of the transfected HEK 293 cells revealing comparable levels of exogenous protein expression. Means and SEM of at least five independent duplicate experiments are shown.
Figure 3.
In vitro functional analysis of CARD15_variants. From its N terminus to its C terminus, CARD15 is composed of two caspase recruitment domains (CARD), a centrally placed NACHT domain with seven highly conserved regions (white dots), including a_p loop (9), and 11 LRR (10). Locations of the 32 tested amino acid changes/deletions observed in CD patients are shown. In a test using minimal amounts of DNA, such as the one reported in Fig. 2, none of these mutations demonstrated increased basal activity comparable to that observed for the BS mutants. (A) Ratio of the basal NF-κB activity measured after transient transfection of 500 ng of CARD15-variant expression vector DNA in HEK 293 cells over that observed with the WT construct (log scale). The latter was 80-fold that observed in mock-transfected cells. (B) Ratio of the NF-κB activity measured after transient transfection of 30 ng of CARD15-variant expression vector DNA in HEK 293 cells and after a 24-h incubation in the presence of PGN (10 μg/ml) over that observed with the WT construct under the same conditions (log scale). The latter was 110-fold that observed in mock-transfected cells. White, gray, and black bars correspond to prototypes I, II, and III, respectively, as described in_Results_. Hatched bars correspond to the variants with borderline behavior between prototypes II and III.
Functional Characterization of CARD15 CD-Associated Variants and 29 Rare CARD15 Variants Observed in CD Patients.
The potential of PGN to activate the NF-κB pathway with CD-associated CARD15 variants was then addressed. SNP8 and SNP13 were essentially unresponsive to PGN, whereas the SNP12 response was half of the WT (Fig. 3B). To probe the influence of the frequent P268S polymorphism (SNP5) on SNP8, SNP12, and SNP13 alleles, double-mutant constructs were obtained. Double and single mutants displayed comparable basal and induced responses, indicating that the SNP5 polymorphism has no influence on the functions of the CD-associated variants of CARD15 (data not shown).
Together with the 3 CD-associated variants, 29 rare CARD15 variants observed in CD patients were characterized with respect to both their basal activity and their ability to respond to PGN. This investigation identified three prototypes among the total of 32 _CARD15_alleles studied (Fig. 3; refs. 6, 8, and 15). (I) Sixteen variants were indistinguishable from WT: R235C, T294S, R311W, H352R, N414S, S431L, A432V, E441K, R684W, R703C, A725G, V793M, R790Q, M836V, V955I, and V972I. (II) SNP8, SNP12, and eight additional variants (two in the CARD domain, R138Q and W157R, and six within the NACHT domain: N289S, D291N, L348V, 558delLG, A612T, and A612V) were characterized by at least a 2-fold reduction in both basal activity and PGN-induced response, indicating a constitutive defect of CARD15 function. (III) SNP13 and two other mutations (R713C and E843K), located close or within the LRR domain, showed an almost unchanged basal activity (at least 80% residual activity), but a major impairment of the PGN-response (<20% that of WT), thus demonstrating a specific defect in PGN-sensing caused by alteration in the LRR domain. R235C, E441K, E778K demonstrated borderline properties among prototypes II and III.
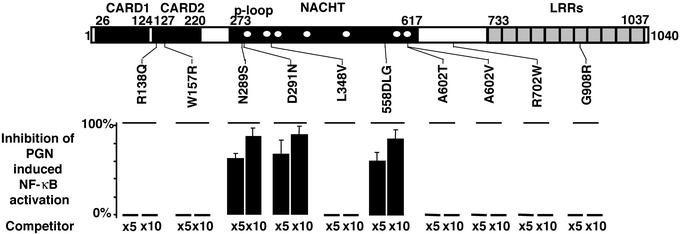
We hypothesized that among the type-II prototype variants, dominant-negative mutants may occur, as shown in plants (16). Comparable to the dominant-negative phytopathogen-resistant gene mutants, cotransfection of 5× or 10× molar excess of 3 of the 10 type-II variants together with the WT construct caused a reduction in the WT-CARD15 response to PGN, thus revealing their _in vitro_dominant-negative properties (Fig. 4). Under similar conditions, the three CD-predisposing CARD15 mutations failed to demonstrate dominant-negative properties with themselves and with the WT-allele (data not shown).
Figure 4.
Search for in vitro dominant-negative properties among the prototype II CARD15 variants observed in inflammatory bowel disorder patients. Inhibition (%) of the CARD15 response to PGN (10 μg/ml) in cotransfection experiments using the WT allele (30 ng) and 5× or 10× molar excess of the mutant construct (competitor) is reported. In all cases, means ± SEM of at least three independent duplicate experiments are reported.
Genotype–Phenotype Relationship: A Codominant Interaction.
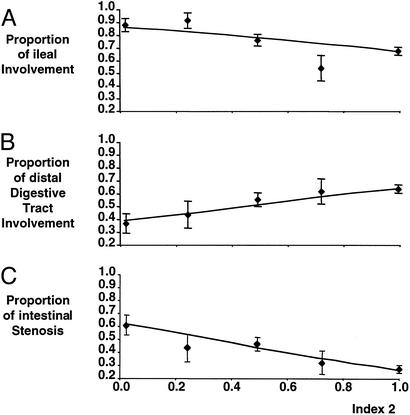
By using the data of Lesage et al. (15), the clinical consequences of the spectrum of CARD15 functional alterations were investigated for CD patients not carrying an in vitro_dominant-negative allele. For each patient, an index, Ind1, was defined as the mean of the basal_in vitro activities (non-PGN induced) associated with his/her CARD15 alleles, as reported in Fig. 3B. Linear standard or regressions of Ind1 on each of 26 clinical items (15) did not reveal a correlation, thus failing to support a role for the intrinsic functional deficiency of CARD15 variants in CD manifestations. A second index, Ind2, was defined as the mean of the PGN-induced in vitro activities associated with the patient's CARD15 alleles (Fig.3B). In contrast to the analysis with Ind1, 4 of 26 statistical tests performed with Ind2 were significant, after correction for multiple testing (Table 1). Ind2 was negatively associated with ileal inflammation at presentation (_P_corr = 0.01). It was positively associated with involvement of transverse colon (_P_corr = 0.05) or of the most distal part of the digestive tract (i.e., from transverse colon to rectum, _P_corr = 0.01) at presentation. Most conspicuously, Ind2 was negatively associated with subsequent intestinal stenosis (_P_corr = 0.00006), a known consequence of long-term bacterial-induced chronic inflammation (Fig.5; refs. 17 and 18).
Table 1.
Clinical correlation with the _in vitro_PGN-dependent activity deduced from the _CARD15_genotype
| No. of patients | Significance of regression | Logit model | |
|---|---|---|---|
| Characteristics | |||
| Sex (ratio M/F = 0.55) | NS | ||
| Age at onset (mean = 18.9; SD = 9.6) | NS | ||
| Age at diagnosis (mean = 20.5; SD = 10.0) | NS | ||
| Age at inclusion in study (mean = 31.8; SD = 12.7) | NS | ||
| Familial/sporadic (ratio F/S = 0.63) | NS | ||
| Smoking status (mean = 1.22; SD = 0.88) | NS | ||
| Disease location at onset | |||
| Esophagus | 14 | NS | |
| Stomach | 46 | NS | |
| Duodenum | 52 | NS | |
| Jejunum | 33 | NS | |
| Upper digestive tract* | 99 | NS | |
| Ileum | 291 | 0.01 | logit(P) = −1.2 Ind2 + 1.9 |
| Ascending colon | 225 | NS | |
| Transverse colon | 163 | 0.05 | logit(P) = 0.3 Ind2 − 1.1 |
| Descending colon and sigmoid | 204 | NS | |
| Distal digestive tract† | 238 | 0.01 | logit(P) = 1.0 Ind2 − 0.4 |
| Rectum | 114 | NS | |
| Perineum | 125 | NS | |
| Pathology at recruitment | |||
| Transmural involvement | 169 | NS | |
| Stenosis | 153 | 6·10−5 | logit(P) = −1.4 Ind2 + 0.4 |
| Granuloma | 215 | NS | |
| Extradigestive symptoms‡ | 143 | NS | |
| Medical management | |||
| Steroids | 357 | NS | |
| Azathioprine/6-mercaptopurine | 163 | NS | |
| Nutritional support | 150 | NS | |
| Surgery | 227 | NS |
Figure 5.
Clinical consequences of the CARD15 deficiency in PGN-induced NF-κB activation. The data of the series described by Lesage et al. (15) were used. For each CD patient not carrying an_in vitro_ dominant-negative allele, a parameter Ind2 reflecting the CARD15 ability of PGN sensing was defined as the average of the relative _in vitro_activities of each of his/her two CARD15 alleles compared with the WT allele. The relative activities associated with each allele are taken from Fig. 3B [e.g., Ind2 for a patient who is a compound heterozygote (SNP12/SNP13) is computed as the mean of the residual PGN-dependent activity of SNP12, i.e., 0.45, and that of SNP13, 0.02, resulting in Ind2 = 0.235]. Patients were classified into five groups according to their Ind2 indexes: group 1 (Ind2 < 0.2, 41 patients), group 2 (0.2–0.4, 23 patients), group 3 (0.4–0.6, 87 patients), group 4 (0.6–0.8, 26 patients), and group 5 (Ind2 > 0.8; 222 patients). The proportion of patients and its estimated SE (binomial distribution) with a given clinical manifestation are plotted as a function of the average Ind2 in each group. The line shows the fit of the ungrouped clinical data to the linear regression model. (A) Proportion of patients with ileal involvement at presentation. (B) Proportion of patients with distal digestive tract (from transverse colon to rectum) involvement at presentation. (C) Proportion of patients with intestinal stenosis.
In contrast with other CD patients, the unrelated CD patients heterozygous for in vitro dominant-negative mutations, presented at disease-onset very frequent distal digestive tract involvement (11 of 12 vs. 230 of 399, P < 0.02, by Fisher's exact test). Two first-degree relatives of CD-patients who inherited such mutations were affected by an inflammatory bowel disorder (IBD) diagnosed as ulcerative colitis, a disease strictly limited to the colorectum.
Discussion
CARD15 is constitutively expressed in phagocytic cells (10) and may be induced in epithelial cells upon exposure to proinflammatory cytokines (19, 20). The most prominent phenotype associated with CARD15 deficiency is observed in the ileum, the site in the gastrointestinal tract with the highest density of lymphoid nodules, which are major location sites for bacteria before their engulfment by phagocytic cells. The CARD15-dependent sensing of PGN in phagocytic cells may be critical in a balanced inflammatory host response to intracellular bacterial invasion (21). It has recently been suggested that CARD15 activates NF-κB pathway in response to PGN through the specific recognition of bacterial muramyl dipeptide (22,23), a very efficient immunogenic antigen commonly used in vaccine preparation and not present in the structure of LPS. Impairment in this defense process may lead to a persistent triggering of alternate stress routes, for example, through the surface TLRs, subsequently leading to T cell NF-κB activation and to abundant local secretions of proinflammatory cytokines (e.g., TNF-α; refs. 24–26). Alternatively, an NF-κB-dependent secretion of down-regulatory factors, such as transforming growth factor-β, may be impaired (27, 28). Consistent with this mechanism, PGN induces granulomatous enterocolitis, which resembles CD in genetically susceptible rats (29), and _nf-κb1_-deficient mice exhibit chronic inflammation (30).
In the colorectum, where the highest concentration of luminal bacteria is present, adequate control of bacterial proliferation may require epithelial up-regulation of CARD15. For WT and most_CARD15_ variants, the enhanced expression in the colorectum mucosae would tend to clear inflammation by increasing NF-κB activation (27, 31). For patients with a dominant-negative allele, an up-regulation of CARD15 expression would have the opposite effect.
In summary, two distinct groups of functional alterations of CARD15 are involved in different granulomatous inflammatory diseases. For BS, the observed gain-of-function mutations of CARD15 explain dominant inheritance. Furthermore, the PGN-independent NF-κB activation observed for these mutations accounts for the inflammatory response in aseptic sites, which is characteristic of this disease. Similar functional alterations may be caused by the NACHT-domain mutations of_CIAS1_/Cryopyrin/NALP3/Pypaf1_observed for three other rare dominant autoinflammatory, extradigestive disorders (32, 33). In contrast, conditional on the presence of specific environmental factors, the CARD15 genotype is shown to contribute in an additive manner to the disease phenotype, thus explaining the multifactorial nature and the variable expressivity of this complex disease. This work highlights the relevance of a cytosolic surveillance pathway mediated by CARD15 that detects specifically PGN. This finding strikingly parallels the recent discoveries in_Drosophila that have shown the role of PGN-binding proteins in the detection of bacterial infection (34). Furthermore, our studies describe the functional implication of high allelic heterogeneity that may exist at a single locus involved in predisposition to rare monogenic as well as common multifactorial disorders.
Acknowledgments
We thank Howard Cann for his careful and critical reading of the manuscript. We thank the staff of the sequencing and bioinformatics facility of the Fondation Jean Dausset for invaluable assistance. We thank Dr. Roland Lloubes (Institut de Biologie Structurale et Microbiologie, Centre National de la Recherche Scientifique, Marseille, France), who provided recombinant PGN-associated lipoprotein. This project received financial support from European Economic Community Contract Biomed 2 no. BMH4-97-2098, the Institut National de la Santé et de la Recherche Médicale, the Direction Générale de la Santé (convention no. 4T004C), and the Association François Aupetit. M.C. and S.E.G. are supported by grants from the Ministère Français de l'Education Nationale, de la Recherche et de la Technologie and Danone Vitapole, respectively. P.J.S. is a Howard Hughes Medical Institute Scholar.
Abbreviations
CD
Crohn disease
BS
Blau's syndrome
SNP
single nucleotide polymorphism
LPS
lipopolysaccharides
LRR
leucine-rich repeats
HEK
human embryonic kidney
PAMP
pathogen-associated molecular pattern
PGN
peptidoglycan
References
- 1.Shanahan F. Lancet. 2002;359:62–69. doi: 10.1016/S0140-6736(02)07284-7. [DOI] [PubMed] [Google Scholar]
- 2.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 3.Schultsz C, Van Den Berg F M, Ten Kate F W, Tytgat G N, Dankert J. Gastroenterology. 1999;117:1089–1097. doi: 10.1016/s0016-5085(99)70393-8. [DOI] [PubMed] [Google Scholar]
- 4.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel J F. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, van Kruiningen H J, West A B, Cartun R W, Cortot A, Colombel J F. Gastroenterology. 1995;108:1396–1404. doi: 10.1016/0016-5085(95)90687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugot J P, Chamaillard M, Zouali H, Lesage S, Cezard J P, Belaiche J, Almer S, Tysk C, O'Morain C A, Gassull M, et al. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y, Bonen D K, Inohara N, Nicolae D L, Chen F F, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr R H, et al. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 8.Miceli-Richard C, Lesage S, Rybojad M, Prieur A M, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot J P. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E V, Aravind L. Trends Biochem Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 10.Kobe B, Kajava A V. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 11.Ogura Y, Inohara N, Benito A, Chen F F, Yamaoka S, Nuñez G. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 12.Inohara N, Ogura Y, Chen F F, Muto A, Nuñez G. J Biol Chem. 2000;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 13.Kirschning C J, Wesche H, Merrill A T, Rothe M. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardin S E, Tournebize R, Mavris M, Page A L, Li X, Stark G R, Bertin J, DiStefano P S, Yaniv M, Sansonetti P J, Philpott D J. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesage S, Zouali H, Cezard J P, Colombel J F, Belaiche J, Almer S, Tysk C, O'Morain C, Gassull M, Binder V, et al. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao Y, Yuan F, Leister R T, Ausubel F M, Katagiri F. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mourelle M, Salas A, Guarner F, Crespo E, Garcia-Lafuente A, Malageada J-R. Gastroenterology. 1998;114:519–526. doi: 10.1016/s0016-5085(98)70535-9. [DOI] [PubMed] [Google Scholar]
- 18.van Tol E A, Holt L, Li F L, Kong F M, Rippe R, Yamauchi M, Pucilowska J, Lund P K, Sartor R B. Am J Physiol. 1999;277:G245–G255. doi: 10.1152/ajpgi.1999.277.1.G245. [DOI] [PubMed] [Google Scholar]
- Berrebi, D., Maudinas, R., Hugot, J.-P., Chamaillard, M., Chareyre, F., De Lagausie, P., Yang, C., Desreumaux, P., Giovannini, M., Cézard, J.-P., et al. (2003) Gut, in press. [DOI] [PMC free article] [PubMed]
- 20.Gutierrez O, Pipaon C, Inohara N, Fontalba A, Ogura Y, Prosper F, Nuñez G, Fernandez-Luna J L. J Biol Chem. 2002;277:41701–41705. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 21.O'Riordan M, Yi C H, Gonzales R, Lee K-D, Portnoy D A. Proc Natl Acad Sci USA. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, et al. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 24.Beutler B. Immunity. 2001;15:5–14. doi: 10.1016/s1074-7613(01)00176-5. [DOI] [PubMed] [Google Scholar]
- 25.Kosiewicz M M, Nast C C, Krishnan A, Rivera-Nieves J, Moskaluk C A, Matsumoto S, Kozaiwa K, Cominelli F. J Clin Invest. 2001;107:695–702. doi: 10.1172/JCI10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kourilsky P, Truffa-Bachi P. Trends Immunol. 2001;22:502–509. doi: 10.1016/s1471-4906(01)02012-9. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence T, Gilroy D W, Colville-Nash P R, Willoughby D A. Nat Med. 2001;7:1291–1297. doi: 10.1038/nm1201-1291. [DOI] [PubMed] [Google Scholar]
- 28.Fadok V A, Bratton D L, Konowal A, Freed P W, Westcott J Y, Henson P M. J Clin Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartor R B, Cromartie W J, Powell D W, Schwab J H. Gastroenterology. 1985;89:587–595. doi: 10.1016/0016-5085(85)90455-x. [DOI] [PubMed] [Google Scholar]
- 30.Erdman S, Fox J G, Dangler C A, Feldman D, Horwitz B H. J Immunol. 2001;166:1443–1447. doi: 10.4049/jimmunol.166.3.1443. [DOI] [PubMed] [Google Scholar]
- 31.Baer M, Dillner A, Schwartz R C, Sedon C, Nedospasov S, Johnson P F. Mol Cell Biol. 1998;18:5678–5689. doi: 10.1128/mcb.18.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman H M, Mueller J L, Broide D H, Wanderer A A, Kolodner R D. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldmann J, Prieur A M, Quartier P, Berquin P, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khush R S, Leulier F, Lemaitre B. Science. 2002;296:273–275. doi: 10.1126/science.1071208. [DOI] [PubMed] [Google Scholar]