Microbial Characterization of Biofilms in Domestic Drains and the Establishment of Stable Biofilm Microcosms (original) (raw)
Abstract
We have used heterotrophic plate counts, together with live-dead direct staining and denaturing gradient gel electrophoresis (DGGE), to characterize the eubacterial communities that had formed as biofilms within domestic sink drain outlets. Laboratory microcosms of these environments were established using excised biofilms from two separate drain biofilm samples to inoculate constant-depth film fermentors (CDFFs). Drain biofilms harbored 9.8 to 11.3 log10 cells of viable enteric species and pseudomonads/g, while CDFF-grown biofilms harbored 10.6 to 11.4 log10 cells/g. Since live-dead direct staining revealed various efficiencies of recovery by culture, samples were analyzed by DGGE, utilizing primers specific for the V2-V3 region of eubacterial 16S rDNA. These analyses showed that the major PCR amplicons from in situ material were represented in the microcosms and maintained there over extended periods. Sequencing of amplicons resolved by DGGE revealed that the biofilms were dominated by a small number of genera, which were also isolated by culture. One drain sample harbored the protozoan Colpoda maupasi, together with rhabtidid nematodes and bdelloid rotifers. The microcosm enables the maintenance of stable drain-type bacterial communities and represents a useful tool for the modeling of this ecosystem.
Clinical epidemiologists have long recognized the potential of sink drains in hospital wards to harbor pathogens. Several studies have identified sink drains within medical-surgical intensive-care wards (19, 22, 30, 35) and cystic fibrosis units (24) as possible sources of infection. Despite the increased information relating to the occurrence of bacterial biofilms and their reported involvement in the biofouling of domestic drains (7), there are few reports in the literature concerning the ecology and microbiology of this environment. The persistence (1) and significance (10) of biofilms in virtually all environments is widely acknowledged. Studies in the home (14, 34) have identified the potential health risks of microbial contamination. Scott et al. (34) identified possible pathogens in the kitchen, toilet, and bathrooms in >200 homes in the United Kingdom. More recent studies (3, 8, 9) have demonstrated that homes represent an environment into which bacterial, viral, and fungal pathogens are continuously introduced in association with food, people, and pets. Studies by Cogan et al. (8, 9) showed that detergent-based cleaning was relatively ineffective in controlling the spread of salmonella and campylobacter to kitchen surfaces during the preparation of contaminated poultry. Despite such concerns, there have been few investigations into the bacterial composition of biofilms within domestic sink drains. As with hospital drains, the pipe work presents a variety of solid surfaces that are suitable substrates for biofilm formation (7, 26). Biofilm has been implicated in a high proportion of slow-running drains in the United States (7). Domestic drains are subject to intermittent wetting, periodic feeding with a plethora of nutrients, hydrodynamic stresses of various intensities, and frequent subeffective antimicrobial treatments. Importantly, the open nature of drains means that they are continuously challenged by a wide range of microbes, which vary depending on the site of the drain. In the kitchen, a variety of normal domestic wastes will pass through the drain conduit, especially where waste disposal units are fitted, along with residues from high-risk infectious materials, such as uncooked meat (12) and vegetable waste (15). Additionally, potential pathogens, including pseudomonads and Legionella pneumophila, may also enter the drain through tap water (6, 20, 27). Domestic sink drain biofilms therefore represent a largely unstudied nidus for potentially pathogenic bacteria, situated close to food preparation areas. The persistence and maturity of drain biofilms demonstrates the general ineffectiveness of chemical control agents (1).
The aim of this study was to develop and evaluate an in vitro system for the establishment and maintenance of a domestic-drain biofilm community. Constant-depth film fermentors (CDFFs) were used to simulate this environment, using drain biofilms as inocula. This culture apparatus has previously been used to model complex (45) and defined (43) oral bacterial communities. In order to obtain baseline data for drain biofilm composition, the eubacterial compositions of domestic-drain biofilms from four houses were characterized using heterotrophic plate counting and culture-independent methods (denaturing gradient gel electrophoresis [DGGE]) (33) in conjunction with sequencing and phylogenetic analysis. The developed model broadly reproduced the physicochemical and substrate environment of a kitchen sink drain biofilm. Potential applications of the system include studying the effects of long-term biocide exposure on drain biofilm communities and investigating the maintenance of pathogens or the transfer of resistance plasmids within a simple laboratory simulator of a drain-type ecosystem.
MATERIALS AND METHODS
Biofilm samples.
Material was obtained from the horizontal pipe sections of PVC kitchen drain outlets from four houses situated in Greater Manchester, United Kingdom. The biofilm samples, designated B1 to B4, were taken from conventional sink drains and had been in situ for at least 5 years. Sample B4 was removed from a drain outlet from a sink with an attached waste disposal unit, which had been in situ for >12 years. These households did not use biocidal detergent products other than bleach. In all cases, the pipe joints were separated and biofilm was excised using a sterile scalpel. The samples were then transported to the laboratory for processing within 2 h. A further sample of B4 material was removed 2 years later to study possible long-term changes in in situ communities.
Domestic drain microcosms.
Drain biofilm (2.5 g) was macerated using a sterile mortar and pestle and diluted 1:10 in sodium phosphate buffer (22.5 ml; 0.1 M; pH 6.5) containing 0.45% (wt/vol) NaCl which had been prereduced (boiled for 5 min and cooled under a constant stream of anaerobic gas [5:95 CO2-N2]). The samples were homogenized for 1 min in a flask shaker (Griffin, London, United Kingdom) in the presence of approximately five glass beads (3.5- to 5.5-mm diameter; BDH, Poole, United Kingdom). Two portions (1.5 ml) of each slurry were removed for immediate bacteriological analysis, and thin wet preparations were examined by differential interference contrast (DIC) microscopy using a Zeiss Axioskop 2 microscope. The remaining 19 ml of diluted material from samples B1 and B4 was used to inoculate CDFFs, designated microcosms M1 and M4, respectively. The fermentors were further inoculated 7 days later, using an additional sample of homogenized but undiluted drain biofilm. Anaerobiosis was maintained for the first 48 h by continuous gassing with oxygen-free gas (5:95 CO2-N2) at 1 liter/h; the temperature was uncontrolled (the ambient laboratory temperature ranged from 18 to 24°C). The biofilms were shielded from light by covering the CDFFs with aluminum foil shrouds. Growth medium was added to the fermentors by a peristaltic pump (Gilson, Villers le Bel, France). Throughout, the microcosms were maintained on a feast-famine regimen (four times daily; 20-min perfusion with 0.5 ml of synthetic dishwater/min) as follows (in grams/liter in tap water): starch, 1.0; peptone, 0.5; tryptone, 0.5; yeast extract, 0.5; NaCl, 1.0; margarine (Flora; Van den Berg Foods, Ltd., Crawley, United Kingdom), 0.05; domestic detergent (Fairy Original; Procter and Gamble, Newcastle Upon Tyne, United Kingdom), 0.05; hemin, 0.001; tomato ketchup (Heinz, Uxbridge, United Kingdom), 0.05. Discontinuous feeding regimens were controlled using programmable electronic timers (Micromark, London, United Kingdom). In order to model more accurately the open nature of a domestic sink drain, the fermentor pans were continuously wetted with untreated tap water (1 ml/h). CDFFs allow the continuous culture of biofilms at accurately set depth (31). In all cases, the biofilm (Teflon plug) depth was set at 5.0 mm. The developed communities were characterized periodically over the course of the investigation.
Direct bacterial-cell counts.
The proportion of the viable bacterial communities that could be cultured by the methods employed was estimated by comparison using vital staining and direct microscopy. A subsample (100 μl) of the 10−2 or 10−3 dilution (prepared for viable counting) was stained using a live-dead bacterial-viability kit (BacLight; Molecular Probes, Leiden, The Netherlands) and counted using an improved Neubauer counting chamber in conjunction with fluorescence microscopy using a 100-W mercury vapor lamp. Live (green fluorescent) and dead (red fluorescent) cells were visualized separately using fluorescein and Texas red bandpass filters, respectively, according to the manufacturer's instructions.
Bacterial characterization by culture.
Drain biofilm material (1.0 g) or CDFF biofilm associated with two plugs was macerated using a sterile mortar and pestle, homogenized, and diluted 1:10 as described above. For enumeration, dilutions of macerated drain or model biofilm (1:10) were serially diluted using prereduced half-strength peptone water. In order to minimize variation due to the sampling of immature biofilms, only those CDFF pans that had been in situ for at least 10 days were removed for analysis. Aliquots (0.1 ml) of appropriate dilutions were plated in triplicate onto a variety of selective and nonselective media (Oxoid, Basingstoke, United Kingdom) as follows: Wilkins-Chalgren agar (for anaerobic and facultative heterotrophic counts and gram-positive cocci), R2A (for aerobic and facultative heterotrophic counts), pseudomonas agar with C-N selective supplements (for Pseudomonas aeruginosa), mannitol salts agar (for gram-positive cocci), and MacConkey agar no. 3 (for enteric organisms). The plates were incubated for up to 5 days both aerobically and in an anaerobic chamber (atmosphere: H2, 10%; CO2, 10%; N2, 80%) (Don Whitley Scientific, Shipley, United Kingdom). The criteria used for selecting bacterial populations for use as markers of population change in the biofilm communities included numerical importance and ease of selective cultivation and identification. In order to compare DGGE community characterization with that of culture, morphologically distinct colonies were subcultured from B1, M1, B2, B3, and M4; archived (at −60°C); and identified on the basis of morphology, an oxidase test, Gram reaction, and ribosomal DNA (rDNA) sequencing.
Community DNA extraction.
Archived biofilm material (0.2 to 0.5 g) was mixed with 1 ml of sodium phosphate buffer (0.12 M; pH 8.0), vortex mixed, and subjected to two cycles of freeze-heating (−60°C for 10 min and 60°C for 2 min). Samples were then transferred to a bead beater vial containing 0.3 g of sterile zirconia beads (0.1-mm diameter). Tris-equilibrated phenol (pH 8.0; 150 μl) was added, and the suspension was shaken three times for 80 s each time at maximum speed (Mini-Bead-Beater; Biospec Products, Bartlesville, Okla.). After 10 min of centrifugation at 13, 000 × g, the supernatant was extracted three times with an equal volume of phenol-chloroform and once with chloroform-isoamyl alcohol (24:1 [vol/vol]). The DNA was precipitated from the aqueous phase with 3 volumes of ethanol, air dried, and resuspended in 100 μl of deionized water. The amount and quality of DNA extracted was estimated by electrophoresis of 5-μl aliquots on a 0.8% agarose gel and comparison to a molecular weight standard (stained with ethidium bromide). The DNA extracts were stored at −60°C prior to analysis.
PCR amplification for DGGE analysis.
The V2-V3 region of the 16S rRNA genes (16S rDNA) (corresponding to positions 339 to 539 of Escherichia coli) was amplified with the eubacterium-specific primers HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) as used by Walter et al. (44). The reactions were performed in 0.2-ml tubes using a Perkin-Elmer (Cambridge, United Kingdom) DNA thermal cycler (model 480). In all cases, reactions were carried out using Red Taq DNA polymerase ready mix (25 μl; Sigma, Dorset, United Kingdom), HDA primers (2 μl each; 5 mM), nanopure water (16 μl), and extracted community DNA (5 μl). Optimization studies, as described by Muyzer et al. (25), showed that extracted community DNA required a minimum of a 1:10 dilution to ensure reliable PCRs. The thermal program was as follows: 94°C (4 min) followed by 30 thermal cycles of 94°C (30 s), 56oC (30 s), and 68oC (60 s). The final cycle incorporated a 7-min chain elongation step (68°C).
DGGE analysis.
Biofilm samples were analyzed by DGGE using a D-Code universal mutation detection system (Bio-Rad, Hemel Hempstead, United Kingdom). Polyacrylamide (8%) gels (16 by 16 cm; 1 mm deep) were run with 1× TAE buffer diluted from 50× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, and 1 mM EDTA). Initially, separation parameters were optimized by running PCR products from selected pure cultures of drain bacteria and PCR amplicons from extracted drain DNA on gels with a 0-to-100% denaturation gradient, perpendicular to the direction of electrophoresis (100% denaturing solution contained 40% [vol/vol] formamide and 7.0 M urea). Denaturing gradients were formed with two 8% acrylamide (acrylamide-bisacrylamide, 37.5:1) stock solutions (Sigma). On this basis, a denaturation gradient for parallel DGGE analysis ranging from 20 to 60% was selected, and PCR amplicons from the isolates Pseudomonas sp. strain MBRG 4.7 and Stenotrophomonas maltophilia MBRG 4.17, together with the type strains P. aeruginosa PAO1 (ATCC 15692) and Pseudomonas fluorescens B52 (Tatua Dairy Company, Morrinsville, New Zealand). For community analyses, the gels also contained a 20 to 60% denaturing gradient. Electrophoresis was carried out at 150 V and 60°C for approximately 4.5 h. All gels were stained using SYBR Gold stain [diluted to 10−4 in 1× TAE [Molecular Probes (Europe), Leiden, The Netherlands] for 30 min. The gels were viewed and images were documented using a BioDocit system (UVP, Upland, Calif.).
Partial 16S rDNA gene sequencing of bacterial isolates and excised gel bands.
Strains were subcultured on R2A agar until pure cultures were obtained, and then bacterial colonies (two to three) were aseptically removed from the surface of the plate and homogenized in a reaction tube containing nanopure water (100 μl). The bacterial suspension was heated to 100°C in a boiling-water bath for 10 min and then centrifuged for 10 min (10,000 × g). The supernatant was used as a template for PCR. Partial 16S rRNA gene sequences were amplified using the primers 8FPL1 (5′-GAG TTT GAT CCT GGC TCA G-3′) and 806R (5′-GGA CTA CCA GGG TAT CTA AT-3′) at 5 μM each. Each PCR mixture consisted of Red Taq DNA polymerase ready mix (25 μl), forward and reverse primers (2 μl each; 5 μM), nanopure water (16 μl), and template DNA (5 μl). A Perkin-Elmer thermal DNA cycler, model 480, was used to run 35 thermal cycles as follows: 94°C (1 min), 53°C (1 min), and 72°C (1 min). The final cycle incorporated a 15-min chain elongation step. For analysis of the major resolved DGGE amplicons, selected resolved bands were cut out of the polyacrylamide gels under UV illumination and incubated at 4°C for 20 h together with 20 μl of nanopure water in nuclease-free universal bottles. Portions (5 μl) were then removed and used as templates for a PCR identical to that outlined in “PCR amplification for DGGE analysis” above, although only the reverse (non-GC clamp) primer (HDA2) was used. The amplified products were purified using Qiaquick PCR purification kits (Qiagen Ltd., West Sussex, United Kingdom) and sequenced using the primers described above. DNA sequences were compiled using GENETOOL LITE 1.0 (http://www.biotools.com) to obtain consensus sequences or to check and edit unidirectional sequences. For excised DGGE band PCRs, the presence of a GC clamp upon sequence analyses confirmed that the correct target had been reamplified rather than an extraneous contaminant.
Identification of strains by comparative methods.
Both FASTA3 (http://www.ebi.ac.uk/FASTA3/) and BLAST (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST) searches were performed with each compiled sequence against those in the EMBL prokaryote database.
Construction of neighbor-joining tree.
The closest relative species were assigned based on compiled partial 16S rRDA gene sequence comparisons with FASTA3 and BLAST against sequences in the EMBL database. Unambiguous positions of representative sequences of closely related strains were then aligned by using CLUSTALX version 1.64b. Neighbor-joining analysis was conducted with the correction of Jukes and Cantor (18) using TREECON 1.3b (42) with Methanobacterium thermoautotrophicum as the outgroup and showing bootstrap values as percentages of 100 replications.
Microscopic enumeration of metazoa.
Nematode worms and rotifers were enumerated by direct counting, using a Sedgewick Rafter Counting Chamber, combined with DIC microscopy (37).
MPN estimation of protozoa.
Protozoan numbers were estimated using a most-probable number (MPN) method (11, 36, 37). Briefly, biofilm samples (0.4 g of drain material or a single CDFF plug) were diluted 1:10 in Neff's amoebal saline and macerated using a pestle and mortar in the presence of 0.1 g of sterile sand. Fifteen twofold dilutions were made using amoebal saline (n = 8) in sterile, 96-well, flat-bottom microtiter plates (Corning Glass Works, Corning, N.Y.) (final volume, 100 μl). Prey bacteria comprised E. coli K-12, which was grown overnight in Luria-Bertani broth at 37°C (approximate final density, 5.0 × 108 CFU/ml). The cells were washed once in Neff's amoebal saline, and 100 μl was added to each well. The MPN plates were incubated in darkness at 23°C for 3 weeks, and the wells were checked for the presence of protozoa and metazoa after 5, 7, 14, and 21 days using an inverted microscope. Number estimates were calculated using Microsoft Excel (4).
Identification of protozoa and metazoa.
Provisional identification of protozoa was done by G. Esteban, Institute of Freshwater Ecology, Cumbria, United Kingdom. The identification was confirmed by staining the cells with dry nigrosin, dried silver staining, and the Feulgen reaction (5) and comparing the morphology with published images (28). Nematodes were kindly identified by D. Hunt, CABI Biosciences, Surrey, United Kingdom. Rotifers were identified by key using DIC morphology (13).
Chemicals.
Unless otherwise stated, chemicals were obtained from Sigma. Formulated bacteriological media were supplied by Oxoid.
RESULTS AND DISCUSSION
DGGE optimization.
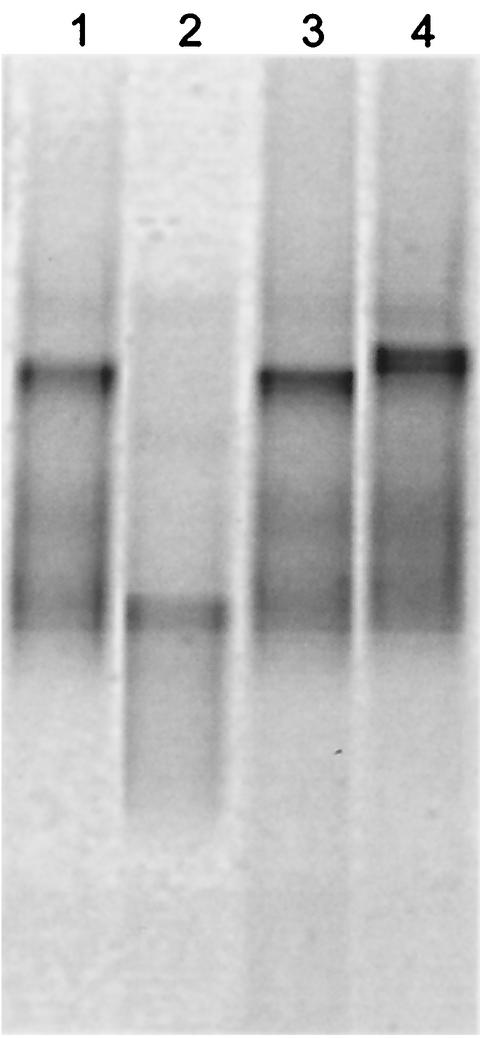
Fingerprinting techniques such as DGGE allow reproducible comparisons of DNA profiles obtained from microbial communities to be made (25). As such, an additional advantage of DGGE is that selected bands can be sequenced and the presence of a particular bacterium can be monitored. In the present study, DGGE of 16S rDNA was optimized for a number of selected drain isolates. Perpendicular DGGE optimization gels showed that a denaturing gradient ranging from 20 to 60% gave good separation of PCR products (data not shown). Parallel DGGE using these parameters to run products from pure cultures showed that the drain isolate S. maltophilia MBRG 4.17 and P. aeruginosa PAO1 could be readily resolved, while PAO1 and P. fluorescens B52 were only poorly resolved. PAO1 and the drain isolate Pseudomonas sp. strain 4.7 had very similar migration properties and could not be resolved (Fig. 1) due to close phylogenetic relatedness. DGGE analysis, however, proved sufficiently robust to provide a means of analysis of drain biofilm and microcosm samples.
FIG. 1.
Negative image of a parallel DGGE gel showing separation between selected drain-isolated bacteria and reference strains. Lane 1, Pseudomonas sp. strain MBRG 4.7; lane 2, S. maltophilia strain MBRG 4.17; lane 3, P. aeruginosa PAO1; lane 4, P. fluorescens B52.
Bacterial analysis.
Direct total- and viable-cell counting, selective isolation and enumeration of “marker bacteria,” 16S rDNA sequencing of “cell clones,” and PCR-DGGE community fingerprinting, combined with sequence analysis of the dominant amplicons, were used to characterize excised drain biofilms and microcosms. Table 1 shows that the excised materials, together with their cultured microcosms, harbored large numbers of bacteria. Total cell counts ranged from 9.9 to 11.4 log10 cells/g. Based on live-dead direct viability counts, the majority of these cells appeared to be viable. Differential plate counting showed that oxidase-negative gram-negative bacteria were the major class of bacteria isolated on MacConkey agar no. 3; pseudomonads were not isolated on this medium. Pseudomonas agar (plus C-N selective supplement) was highly selective for P. aeruginosa, and enteric species did not form colonies. Mannitol salts agar was less selective and grew various pseudomonads, as well as staphylococci and enterococci. This analysis showed that all drain biofilms harbored large numbers of enteric species (range, 7.9 to 10.6 log10 CFU/g). Pseudomonads also colonized these biofilms, with counts ranging from <4.0 to 9.5 log10 CFU/g. Differential counts for gram-positive cocci showed all samples, with the exception of B3, to harbor these bacteria. Direct viable-cell counts were marginally higher than the inoculum counts for M1 and lower for M4. Overall, bacterial plate counts were higher in the microcosms than in the excised material (Table 1). Colony-counting procedures indicated a deficit in the viable bacterial counts compared to vital staining in all samples tested. The percentage efficiency of plate counting ranged from 4 to 68% for in situ material and from 45 to 68% for the corresponding microcosms.
TABLE 1.
Microflora and microfauna within excised kitchen drain biofilm and mean composition after growth in drain simulator
| Organism | No.a | |||||
|---|---|---|---|---|---|---|
| Drain 1 | Drain 2 (sink drain biofilm) | Drain 3 (sink drain biofilm) | Drain 4 | |||
| Sink drain biofilm | Microcosm biofilm | Sink drain biofilm | Microcosm biofilm | |||
| Total direct cell count | 11.24 ± 0.06 | 11.40 ± 0.03 | 11.16 ± 0.06 | 9.86 ± 0.04 | 11.35 ± 0.05 | 10.63 ± 0.01 |
| BacLight viable-cell count | 11.07 ± 0.09 | 11.39 ± 0.04 | 11.06 ± 0.01 | 9.80 ± 0.04 | 11.26 ± 0.03 | 10.58 ± 0.02 |
| Total culturable heterotrophsb | 10.90 ± 0.04 | 11.22 ± 0.12 | 10.20 ± 0.03 | 9.20 ± 0.01 | 9.85 ± 0.07 | 10.23 ± 0.04 |
| Percentage culturable heterotrophsc | 68 | 68 | 14 | 25 | 4 | 45 |
| Enteric species | 10.63 ± 0.04 | 10.84 ± 0.21 | 9.97 ± 0.07 | 9.16 ± 0.05 | 7.92 ± 0.00 | 8.77 ± 0.01 |
| P. aeruginosa | 9.52 ± 0.07 | 8.37 ± 0.17 | 7.80 ± 0.05 | <4.0 | 6.00 ± 0.05 | 7.04 ± 0.02 |
| Gram-positive cocci | 9.76 ± 0.03 | 8.79 ± 0.49 | 6.78 ± 0.07 | <4.0 | 9.65 ± 0.07 | 7.60 ± 0.29 |
Culture-independent analysis.
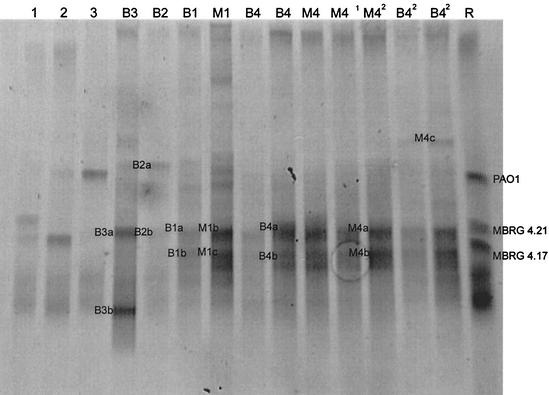
Table 1 shows significant differences between the viable- and total cell counts and indicates either the presence of unculturable bacteria or highly variable plate count efficiencies. Giovannoni et al. (16) discussed various aspects of the “great plate count anomaly” by proposing two hypotheses (39). Based on this, the worst-case scenario when analyzing a community by culture is that the community is composed mainly of unknown species that cannot be grown on common microbiological media (17). The alternative hypothesis is that a community is composed of known species that are capable of forming colonies but do so with various efficiencies. DGGE was therefore carried out in order to provide phylogenetic information about the predominant PCR amplicons derived from extracted community DNA. PCR-DGGE using eubacterium-specific primers (the 16S rRNA gene) showed that all drain biofilms were dominated by only a few species, which were also dominant in the corresponding microcosms (Fig. 2). Table 2 shows the major biofilm phylotypes, based on PCR of isolated cell clones. Table 3 shows dominant DGGE sequences, as derived from excised DGGE bands. These combined data show that both model systems maintained the dominant phylotypes, although one band (M1a) that was not detected in B1 was resolved in the model biofilm M1. Based on sequence homology, this phylotype was related to Sphingobacterium multivorum (Table 3). Overall, the species identified by DGGE as being dominant (Table 3) were putatively culturable (Table 2). The dominant resolvable PCR amplicons in each biofilm were as follows: B1 and M1, bacteria related to Klebsiella oxytoca; B2, bacterium related to Moraxella osloensis; and B3, bacterium related to E. coli. The dominant phylotype from B4 and M4 was related to Chryseobacterium sp_._
FIG. 2.
Negative image of a parallel DGGE gel showing community fingerprints for biofilm samples B3, B2, B1, M1, B4 (in duplicate), and three time intervals of M4 and duplicate samplings of B42 (B4 drain biofilm, excised 2 years after the original B4 sampling). Individual reference stains are included (lane 1, B. cereus MBRG 4.21; lane 2, S. maltophilia MBRG 4.17; lane 3, P. aeruginosa PAO1), together with a reference mix (lane R).
TABLE 2.
Sequencing of PCR amplicons derived from isolated cell clonesa
| Sample | Cell clone | Sequence length (bp) | Ambiguity | Most closely related species (% sequence similarity)c | |
|---|---|---|---|---|---|
| bp | % | ||||
| B1, M1b | MBRG 1.5 | 750 | 0 | 0 | B. subtilis AJ276351 (99) |
| B1, M1 | MBRG 1.6 | 696 | 8 | 1.1 | E. asburiae AB004744 (99) |
| B1, M1 | MBRG 1.2 | 742 | 7 | 0.9 | H. alvei M59155 (99%) |
| B1, M1 | MBRG 1.5 | 757 | 16 | 2.1 | K. oxytoca AB053117 (99) |
| B1, M1 | MBRG 1.4 | 798 | 15 | 1.9 | L. paracasei AF243147 (97) |
| B1, M1, B2 | MBRG 1.7 | 756 | 2 | 0.3 | Pseudomonas sp. strain P400Y AB076857 (99) |
| B1, M1 | MBRG 1.1 | 772 | 6 | 0.8 | K. planticola AF129443 (99) |
| B2 | MBRG 2.4 | 760 | 2 | 0.3 | B. subtilis Z99104 (99) |
| B2 | MBRG 2.3 | 744 | 0 | 0 | M. osloensis X95304 (99) |
| B2 | MBRG 2.1 | 771 | 5 | 0.6 | E. coli U18997 (99) |
| B2 | MBRG 2.5 | 785 | 4 | 0.5 | S. epidermidis AF269927 (99) |
| B3 | MBRG 3.1 | 739 | 4 | 0.5 | E. coli AE000406 (98) |
TABLE 3.
Sequencing of PCR amplicons (dominant phylotypes) derived from DGGE gelsa
| Sample | Cell clone | Sequence length (bp) | Ambiguity | Most closely related species (% sequence similarity)c | |
|---|---|---|---|---|---|
| bp | % | ||||
| B1 | B1a/M1b | 181 | 19 | 10.5 | K. oxytoca AB004754 (80) |
| B1, M1b | B1b/M1c | 183 | 37 | 20.2 | K. oxytoca AB004754 (72) |
| M1 | M1a | 180 | 31 | 17.2 | S. multivorum AB020205 (72) |
| B2 | B2a | 182 | 10 | 5.5 | M. osloensis AF00519016S (94) |
| B2 | B2b | 185 | 12 | 6.5 | M. osloensis AJ269515 (89) |
| B3 | B3a | 183 | 13 | 7.1 | E. coli AF076037 (92) |
| B3 | B3b | 181 | 10 | 5.5 | E. coli AF076037 (91) |
| B4, M4 | B4a/M4b | 184 | 42 | 22.9 | Chryseobacterium sp. strain AJ309324 (62) |
| B4, M4 | B4b/M4b | 177 | 38 | 21.5 | Chryseobacterium defluvium AJ309324 (65) |
| M4 | M4c | 180 | 23 | 12.8 | P. dentalis X81876 (81) |
Comparison of DGGE community fingerprints from original in situ biofilm samples from B4, with biofilm removed from the same drain twice with a 2-year interval, revealed that the major bacteria remained dominant. However, a new band was resolved in the latter sample (M4c) which was related to the anaerobic oral bacterium Prevotella dentalis (Table 3). These analyses suggest that the major species detected by DGGE were also isolated on formulated media (Tables 2 and 3). Interestingly, conventional culture characterized major portions of the community from B4 which were not detected on DGGE gels. This observation is significant for such ecological studies and suggests that bacteria were present which could be isolated but which were below the detection threshold of the DGGE process. In this respect, PCR-based methods may be biased in that the relative concentrations of PCR products do not properly reflect the composition of the in situ community (38). Sequences obtained from DGGE bands are short and of variable quality. For example, the data in Table 2 show that the mean percent ambiguity for sequenced cell clone PCR products was 0.75 compared to 12.9 for DGGE-derived sequences (Table 3). Such ambiguities for directly sequenced PCR amplification products probably arise from amplification of different phylotypes with similar or identical electrophoretic mobilities. This is especially a problem where resolution of bands is relatively poor. This is likely to be most pronounced where a community is composed of multiple species of a given genus. The relatively short sequences derived from DGGE also reduce the refinement of phylogenetic determination. These issues do not prohibit identification with BLAST, although they do reduce the confidence of identifications.
Stability of microcosms.
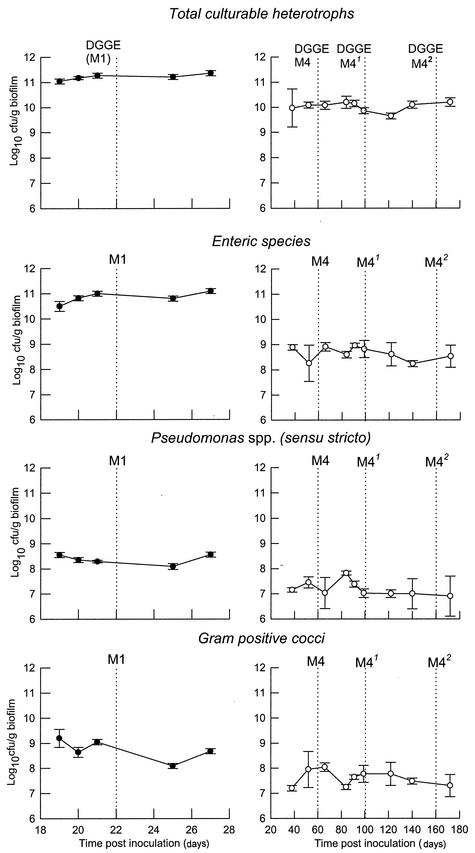
The data in Fig. 3 show that microcosm communities achieved dynamic stability with respect to culturable heterotrophic populations of marker bacteria (total heterotrophs, enteric species, pseudomonads, and gram-positive cocci) between 10 days and 1 month following inoculation. Steady states could be maintained for prolonged periods (M1, 28 days; M4, >160 days). Comparative analysis based on DGGE fingerprints (Fig. 2) showed that the microcosm communities M1 and M4 were highly similar to those detected in the corresponding excised in situ biofilms. DGGE analyses of microcosm samples from M4 taken on three separate occasions showed that the major amplicons were maintained over extended periods (Fig. 2).
FIG. 3.
Stabilities of selected bacteria in domestic-drain biofilm microcosms. Solid symbols, M1; open symbols, M4. The data are means ± standard deviations from two bioreactors (M4) or two separate sample pans (M1) (three separate analyses per replicate sample for each point). The units are log10 CFU per gram (wet weight) of biofilm. The dashed vertical lines indicate the times of DGGE samplings.
Phylogenetic characterization of biofilms by culture and DGGE.
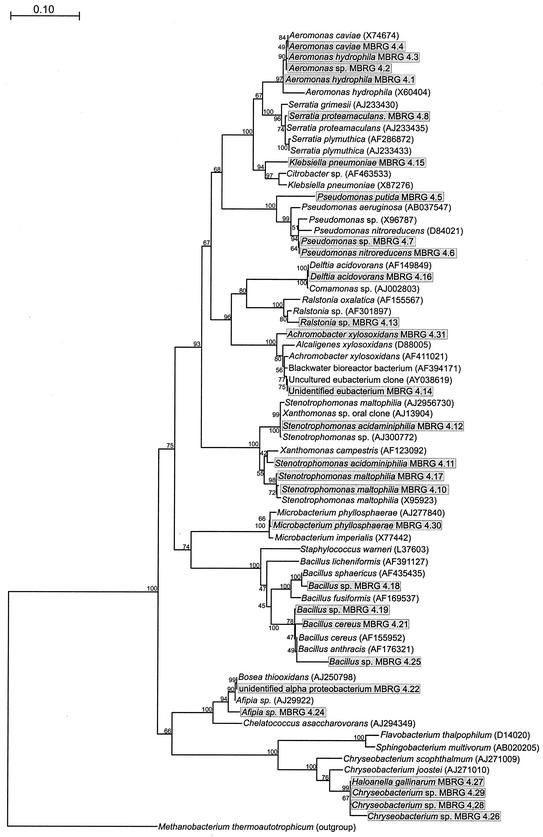
The isolation and identification of distinct morphotypes from the isolation media showed that bacterial diversity in different drain biofilms varied considerably (Tables 2 and 3 and Fig. 2 and 4). Drains B1 and B2, for example, harbored four and six distinct morphotypes, respectively, corresponding to bacterial species belonging to the families Enterobacteriaceae, Pseudomonadaceae, and Bacillaceae and the Xanthomonas group. Biofilm removed from B3, however, was dominated by E. coli. M4 harbored by far the most complex community (Fig. 4) and was derived from B4, the only sink outlet sampled that had a waste disposal unit attached. This might account for the much greater diversity in this biofilm, since there would have been greater substrate input than in the other sink drains examined in this study. Since it was not possible to unambiguously identify two of the isolates from M4, a phylogenetic tree was constructed for this ecosystem (Fig. 4). The unidentified eubacterium strain MBRG 4.14 was most closely related to a black water bioreactor bacterium (AY394171) and an uncultured eubacterium clone (AY038619), while the unidentified alpha proteobacterium strain MBRG 4.22 was related to Achromobacter xylosoxidans (AF411021) and Bosea thiooxidans (AJ250798).
FIG. 4.
Phylogenetic tree showing bacteria isolated from M4 (boxed and shaded) together with selected reference strains based on 540-bp-long sequences of 16S rRNA genes. The tree was constructed using the neighbor-joining method of Jukes and Cantor (18). The scale bar indicates 0.10 estimated substitution per nucleotide. TREECON for Windows (42) was used to construct the tree. The bootstrap values indicate confidence limits of the phylogenies, based on percentages of 100 replications. M. thermoautotrophicum was used to root the tree.
Protozoa and metazoa.
Light microscopy (DIC) and MPN analysis, respectively, revealed the presence of considerable numbers of rhabtidid nematode worms (ca_._ 4.6 log10 cells/g) and protozoa (Colpoda maupasi) (ca_._ 4.6 log10 cells/g) in drain B4. These organisms were also present in the corresponding microcosm (M4) at 3.9 and 6.2 log10 cells/g for nematodes and protozoa, respectively, together with bdelloid rotifers (genus Habrotrocha) at 3.9 log10 cells/g, which became established within the microcosms but were not detected in the original excised material. These were presumably either present but undetectable in the original samples or introduced in the untreated potable-water feed.
Nucleotide sequence accession numbers.
The following sequences for isolated cell clones were deposited in the EMBL sequence database (the accession numbers are given in parentheses): Klebsiella planticola MBRG 1.1 (AJ508364), Hafnia alvei MBRG 1.2 (AJ508360), K. oxytoca MBRG 1.3 (AJ508361), Lactobacillus paracasei MBRG 1.4 (AJ508362), Bacillus subtilis MBRG 1.5 (AJ508358), Enterobacter asburiae MBRG 1.6 (AJ508359), Pseudomonas sp. strain MBRG 1.7 (AJ508363), E. coli MBRG 2.1 (AJ508367), M. osloensis MBRG 2.3 (AJ508366), B. subtilis MBRG 2.4 (AJ508365), Staphylococcus epidermidis MBRG 2.5 (AJ508368), E. coli MBRG 3.1 (AJ508369), Aeromonas hydrophila MBRG 4.1 (AJ508693), Aeromonas sp. strain MBRG 4.2 (AJ508692), A. hydrophila MBRG 4.3 (AJ508690), Aeromonas caviae MBRG 4.4 (AJ508691), Pseudomonas putida MBRG 4.5 (AJ508696), Pseudomonas nitroreducens MBRG 4.6 (AJ508698), Pseudomonas sp. strain MBRG 4.7 (AJ508697), Serratia proteamaculans MBRG 4.8 (AJ508694), S. maltophilia MBRG 4.10 (AJ508703), Stenotrophomonas acidaminiphila MBRG 4.11 (AJ508701), S. acidaminiphila MBRG 4.12 (AJ508700), Ralstonia sp. strain MBRG 4.13 (AJ508607), unidentified eubacterium strain MBRG 4.14 (AJ508699), Klebsiella pneumoniae MBRG 4.15 (AJ508695), Delftia acidovorans MBRG 4.16 (AJ508611), S. maltophilia MBRG 4.17 (AJ508702), Bacillus cereus MBRG 4.19 (AJ508707), B. cereus MBRG 4.21 (AJ508706), unidentified alpha proteobacterium strain MBRG 4.22 (AJ508610), unidentified alpha proteobacterium strain MBRG 4.24 (AJ508612), B. cereus MBRG 4.25 (AJ508705), Flavobacterium sp. strain MBRG 4.26 (AJ508710), Haloanella gallinarum MBRG 4.27 (AJ508708), Flavobacterium sp. strain MBRG 4.28 (AJ508709), H. gallinarum MBRG 4.29 (AJ508711), Microbacterium phyllosphaerae MBRG 4.30 (AJ508704), and A. xylosoxidans MBRG 4.31 (AJ508608).
Conclusions
In these investigations, we have shown that large sessile populations of bacteria colonize domestic drain conduits and that the species composition varies widely between individual locations. The presence of certain species of bacteria appeared to be specific to the drain tested. For example, L. paracasei was detected in B1, M. osloensis was detected in B2, and E. coli was detected in B2 and B3.
While biofilm formation within water treatment and distribution systems has been studied in considerable detail, the composition of biofilms within domestic drains has been unknown. As well as considerable numbers of opportunistic pathogens, such as aeromonads and pseudomonads (29), biofilms formed within potable-water systems contain bacterial pathogens such as L. pneumophila (40) and coliforms of intestinal and nonintestinal origin (46). Furthermore, protozoa are commonly found within water distribution systems (32, 41) and have been associated with the persistence and invasiveness of L. pneumophila (41). The formation of thick biofilm within a sink drain is not surprising in view of the increased substrate availability (food residues, etc.) in this environment compared to oligotrophic potable-water systems.
The model described here has been developed as a paradigm of domestic-drain biofilm communities for the long-term maintenance and laboratory analysis of this ecosystem, which is a major site of bacterial colonization in the home.
There is considerable concern that the overuse of biocides such as Triclosan within the home could select for reduced susceptibility within target bacteria, which may have clinical implications (21, 23). The effect of biocides on community composition and resistance properties is being studied in such models, together with the biodegradation of biocides and the maintenance of pathogens.
Acknowledgments
We are grateful to Procter and Gamble USA for funding the program; Walid Naser, Department of Microbiology, University of New Hampshire, Durham, for advice concerning the DGGE analyses; G. Esteban for help with identification of protozoa; and D. Hunt for nematode identification.
REFERENCES
- 1.Allison, D. G., A. J. McBain, and P. Gilbert. 2000. Microbial biofilms: problems of control, p. 310-327. In H. Lappin-Scott, P. Gilbert, M. Wilson, and D. Roberts (ed.), Community structure and cooperation in biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 2.Barker, J., D. Stevens, and S. F. Bloomfield. 2001. Spread and prevention of virus infections in community settings and domestic homes. J. Appl. Microbiol. 91**:**7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloomfield, S. F. 2001. Gastrointestinal disease in the domestic setting. What are the issues? J. Infect. 43**:**23-29. [DOI] [PubMed] [Google Scholar]
- 4.Briones, A., and W. Reichardt. 1999. Estimating microbial population counts by “most probable number” using Microsoft Excel. J. Microbiol. Methods 35**:**157-161. [DOI] [PubMed] [Google Scholar]
- 5.Burt, R. 1940. Specific analysis of the genus Colpoda with special reference to the standardization of experimental material. Trans. Am. Soc. Microbiol. 59**:**414-432. [Google Scholar]
- 6.Camper, A., M. Burr, B. Ellis. P. Butterfield., and C. Abernathy. 1999. Development and structure of drinking water biofilms and techniques for their study. J. Appl. Microbiol. **35:**1S-12S. [DOI] [PubMed]
- 7.Charaf, U. K. 1997. Biofilms in our drains, p. 175-181. In J. Wimpenny, P. Handley, P. Gilbert, H. Lappin-Scott, and M. Jones (ed.), Biofilms: community interactions and control. Bioline Press, Cardiff, United Kingdom.
- 8.Cogan, T. A., J. Slader, S. F. Bloomfield, and T. J. Humphrey. 2002. Achieving hygiene in the domestic kitchen: the effectiveness of commonly used cleaning products. J. Appl. Microbiol. 92**:**885-892. [DOI] [PubMed] [Google Scholar]
- 9.Cogan, T. A., S. F. Blomfield, and T. J. Humphrey. 1999. The effectiveness of hygiene procedures for the prevention of cross contamination from chicken carcasses in the domestic kitchen. Lett. Appl. Microbiol. 29**:**354-358. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., K. Cheng, G. Geesey, T. Ladd, J. Nickel, and M. Dasgupta. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41**:**435-464. [DOI] [PubMed] [Google Scholar]
- 11.Cutler, D. 1920. A method for estimating the number of active protozoa in the soil. J. Agric. Sci. 10**:**135-143. [Google Scholar]
- 12.Davidson, C. M., J. Y. Daoust, and W. Allewell. 1985. Steam decontamination of whole and cut-up raw chicken. Poultry Sci. 64**:**765-767. [Google Scholar]
- 13.Donner, J. 1966. Rotifers. Frederick Warne and Co. Ltd., London, United Kingdom.
- 14.Finch, J. E., J. Prince, and M. Hawkworth. 1978. A bacteriological survey of the domestic environment. J. Appl. Microbiol. 45**:**357-364. [DOI] [PubMed] [Google Scholar]
- 15.Francis, G. A., C. Thomas, and D. O'Beirne. 1999. The microbiological safety of minimally processed vegetables. Int. J. Food Microbiol. 34**:**1-22. [Google Scholar]
- 16.Giovannoni, S. J., T. D. Mullins, and G. Field. 1995. Microbial diversity in oceanic systems: rRNA approaches to the study of unculturable microbes, p. 217-248. In I. Joint (ed.), Molecular ecology of aquatic microbes. Springer, Berlin, Germany.
- 17.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33**:**1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jukes, T. H., and C. P. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 19.Levin, M. H., B. Olson, C. Nathan, S. A. Kabins, and R. A. Weinstein. 1984. Pseudomonas in the sinks in an intensive care unit: relation to patients. J. Clin. Pathol. 37**:**424-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lye, D., G. S. Fout, S. R. Crout, R. Danielson, C. L. Thio, and C. M. Paszko-Kolva. 1997. Survey of ground, surface, and potable waters for the presence of Legionella species by EnviroAmp(R) PCR Legionella kit, culture, and immunofluorescent staining. Water Res. 31**:**287-293. [Google Scholar]
- 21.McBain, A. J., and P. Gilbert. 2001. Biocide tolerance and the harbingers of doom. Int. Biodeterior. Biodegradation 47**:**55-61. [Google Scholar]
- 22.McGeer, A., D. E. Low, J. Penner, J. Ng, C. Goldman, and A. E. Simor. 1990. Use of molecular typing methods to study the epidemiology of Serratia marcescens. J. Clin. Microbiol. 28**:**55-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Triclosan targets lipid synthesis. Nature 394**:**531-532. [DOI] [PubMed] [Google Scholar]
- 24.Moore, J. E., I. Thompson, M. Crowe, J. Xu, A. Shaw, B. C. Millar, A. O. B. Redmond, A. J. M. Reid, C. Clarke, and J. S. Elborn. 2002. Burkholderia cepacia from a sink drain. J. Hosp. Infect. 50**:**235-237. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 26.Niquette, P., P. Servais, and R. Savoir. 2000. Impacts of pipe materials on densities of fixed bacterial biomass in a drinking water distribution system. Water Res. 43**:**1952-1956. [DOI] [PubMed] [Google Scholar]
- 27.Ollos, P., R. Slawson, and P. Huck. 1998. Bench scale investigations of bacterial re-growth in drinking water distribution systems. Water Sci. Technol. 38**:**275-282. [Google Scholar]
- 28.Patterson, D. J. 1998. Free-living freshwater protozoa: a colour guide. Mason Publishing Ltd., London, United Kingdom.
- 29.Payment, P., F. Gamache, and G. Paquette. 1988. Microbiological and virological analysis of water from two water filtration plants and their distribution systems. Can. J. Microbiol. 34**:**1304-1309. [DOI] [PubMed] [Google Scholar]
- 30.Perryman, F. A., and D. J. Flournoy. 1980. Prevalence of gentamycin- and amikacin-resistant bacteria in sink drains. J. Clin. Microbiol. 12**:**79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters, A., and J. Wimpenny. 1988. A constant-depth laboratory model film fermenter. Biotech. Bioeng. 32**:**263-270. [DOI] [PubMed] [Google Scholar]
- 32.Rogers, J., A. B. Dowsett, P. J. Dennis, J. V. Lee, and C. W. Keevil. 1994. Influence of plumbing materials on biofilm formation and growth of Legionella pneumophila in potable water systems. Appl. Environ. Microbiol. 60**:**1842-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67**:**504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, E., S. F. Bloomfield, and C. G. Barlow. 1982. An investigation of microbial contamination in the home. J. Hyg. 89**:**279-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shlaes, D. M., C. C. Currie-McCumber, M. Eanes, G. Rotter, and R. Floyd. 1986. Gentamycin-resistance plasmids in an intensive care unit. Infect. Control 7**:**355-361. [DOI] [PubMed] [Google Scholar]
- 36.Singh, B. 1946. A method of estimating numbers of soil protozoa, especially amoebae, based on their differential feeding on bacteria. Ann. Appl. Biol. 33**:**112-120. [DOI] [PubMed] [Google Scholar]
- 37.Stevik, T., J. Hanssen, and P. Jenssen. 1998. A comparison between DAPI direct counts (DDC) and most probable number (MPN) to quantify protozoa in filtration systems. J. Microbiol. Methods 33**:**13-21. [Google Scholar]
- 38.Suzuki, M. T., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64**:**4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, M. T., M. S. Rappe, Z. W. Haimberger, H. Winfield, N. Adair, J. Strobel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63**:**983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiefenbrunner, F., A. Arnold, M. Dierichi, and K. Emde. 1993. Occurrence and distribution of Legionella pneumophila in water systems of Central European private homes, p. 235-238. In J. Barbaree, R. Brieman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 41.Tyndall, R. L., and E. L Domingue. 1982. Co-cultivation of Legionella pneumophila and free-living amoebas. Appl. Environ. Microbiol. 44**:**954-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van de Peer, Y., and R. De Wachter. 1997. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput. Appl. Biosci. 13**:**227-230. [DOI] [PubMed] [Google Scholar]
- 43.Vroom, J. M., K. J. De Grauw, H. C. Gerritsen, D. J. Bradshaw, P. D. Marsh, G. K. Watson, J. J. Birmingham, and C. Allison. 1999. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl. Environ. Microbiol. 65**:**3502-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66**:**297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, M., H. Patel, and J. H. Noar. 1998. Effect of chlorhexidine on multi-species biofilms. Curr. Microbiol. 36**:**13-18. [DOI] [PubMed] [Google Scholar]
- 46.World Health Organization. 1993. Guidelines for drinking water quality, 2nd ed., vol. 1. World Health Organization, Geneva, Switzerland.