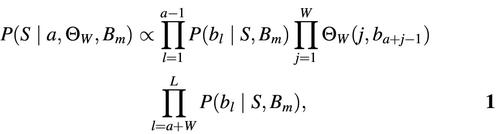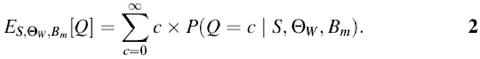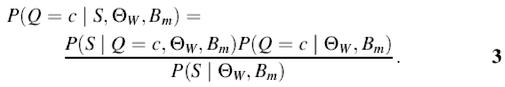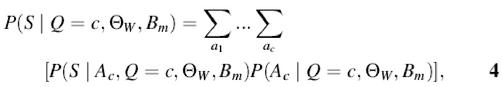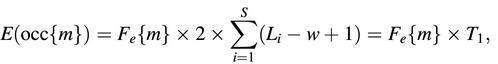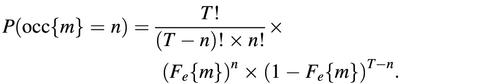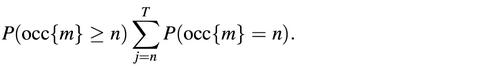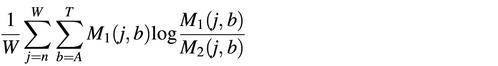Toucan: deciphering the cis-regulatory logic of coregulated genes (original) (raw)
Abstract
TOUCAN is a Java application for the rapid discovery of significant _cis_-regulatory elements from sets of coexpressed or coregulated genes. Biologists can automatically (i) retrieve genes and intergenic regions, (ii) identify putative regulatory regions, (iii) score sequences for known transcription factor binding sites, (iv) identify candidate motifs for unknown binding sites, and (v) detect those statistically over-represented sites that are characteristic for a gene set. Genes or intergenic regions are retrieved from Ensembl or EMBL, together with orthologs and supporting information. Orthologs are aligned and syntenic regions are selected as candidate regulatory regions. Putative sites for known transcription factors are detected using our MotifScanner, which scores position weight matrices using a probabilistic model. New motifs are detected using our MotifSampler based on Gibbs sampling. Binding sites characteristic for a gene set—and thus statistically over-represented with respect to a reference sequence set—are found using a binomial test. We have validated Toucan by analyzing muscle-specific genes, liver-specific genes and E2F target genes; we have easily detected many known binding sites within intergenic DNA and identified new biologically plausible sites for known and unknown transcription factors. Software available at http://www.esat.kuleuven.ac.be/∼dna/BioI/Software.html.
INTRODUCTION
Genomes contain vast amounts of _cis_-regulatory DNA responsible for directing spatial and temporal patterns of gene expression in response to metabolic requirements, developmental programs and a plethora of external stimuli (reviewed in 1). Genes of multicellular organisms contain both proximal modules (with the promoter-proximal elements) and distal modules (enhancers and silencers), as well as a basal transcription apparatus (i.e., a TATA box or another element to position RNA polymerase II). Distal modules can lie many kilobases on either side of a coding region or within an intron. Each module may contain multiple binding sites that interact with a specific combination of transcription factors (2). The characterization of such regions is a fundamental step toward understanding the largely unexplored networks of gene regulation.
DNA microarrays and other functional genomics technologies are frequently used to yield sets of coregulated genes to find common regulatory modules. Several tools and algorithms already exist for comparative sequence analysis, for scoring known transcription factor binding motifs (TFBMs) using position weight matrices (PWMs), for detecting new patterns using Gibbs sampling techniques (3,4), and for clustering binding sites to find regions where their local density is high (5,6). In most cases, however, a combination of approaches is required to minimize false positives (7,8). This research domain is extensive, and since not every biologist has access to the bioinformatics expertise to integrate several tools and web applications, biologists will benefit from an efficient tool to perform their regulatory sequence analysis. Further more, if such analyses are carried out on a large scale, the efficient retrieval of promoter sequences is essential. This task is now becoming more straightforward for organisms with fully sequenced genomes. By querying genomic databases like Ensembl (9) for a gene and walking up- or downstream from it, the intergenic regions can be retrieved.
Toucan allows the user to construct a gene set by importing or retrieving sequences from local or online sources, to visualize, manipulate, cut and export sequences, to select putative regulatory regions, to score and annotate sequences with putative binding sites, to find new motifs, and to perform a statistical analysis to select over-represented sites. Toucan was designed with the analysis of regulation in gene sets of higher eukaryotes as its primary goal. It is in this setting that it provides the user with the most added value as compared to existing tools. The web-based Regulatory Sequence Analysis Tools (10) was designed for the analysis of prokaryotic and yeast sequences. Sequences of such organisms with compact genomes can also be analyzed with Toucan but the benefits of Toucan will be more limited (e.g., Ensembl access is not available and the current comparative sequence analysis methodology is not well suited to these cases). Tools like rVISTA (7) and TraFaC (11) work on higher eukaryotic sequences and also use comparative sequence analysis to detect transcription factor binding sites in a sequence, but they work with single genes and have a less integrated scope. The Genomatix software (http://www.genomatix.de) shares the integration aspect with Toucan, but does not use comparative sequence analysis to detect distal regulatory regions (PromoterInspector predicts only proximal promoters). In fact most of the existing tools for regulatory sequence analysis are compatible with Toucan as long as their output is formatted as, or can be converted to, Sanger’s General Feature Format (GFF, see http://www.sanger.ac.uk/Software/formats/GFF). Toucan has both the advantages of a local installation (e.g., a higher user interactivity) and the advantages of distributed computing by using new technologies like web services (12). The BioJava library (http://www.biojava.org) is used as the back-end for most of the biological sequence manipulations.
METHODS
Java and BioJava
Toucan was developed using the Java 2 SDK version 1.4.1 (Sun Microsystems). It has been tested under the Windows, Linux and MacOS operating systems. The application can either be run directly from our website using Java Web Start or it can be downloaded from http://www.esat.kuleuven.ac.be/∼dna/BioI/Software.html. The BioJava library was used and can be downloaded from http://www.biojava.org.
Web services
The MotifScanner, MotifSampler and AVID/VISTA programs are used through web services using the Apache implementation of Simple Object Access Protocol, version 2.3. The services, which reside on a Tomcat server, start the actual programs on a Linux cluster with 10 nodes using Java Remote Method Invocation.
Online sequence and information retrieval
Ensembl gene identifiers and the identifiers of orthologous genes are retrieved from other database identifiers directly from the Ensembl MySQL database at kaka.sanger.ac.uk. Sequences and flanking regions are retrieved from Ensembl using HTTP access on the Export functionality of Ensembl. Using HTTP queries and XEMBL access, sequences are also retrieved from the EMBL nucleotide sequence database.
MotifScanner
To search for instances of a known motif we have built a new algorithm from the core modules of the MotifSampler (4,13). The basic sequence model assumes that binding sites are hidden in a noisy background sequence. The model of the binding sites is based on a frequency residue model and is represented by a position probability matrix Θ_W_ of length W. The background model is represented by the transition matrix B m of a _m_th-order Markov model. The usage of a higher-order background model allows to better distinguish between motifs that occur frequently throughout the genome and the ones that are specific to a certain region. If the start position of the motif instance is known and indicated by a, then the probability that the sequence is generated given the model parameters is
where L is the length of the sequence, b l is the nucleotide at position l in the sequence S, Θ_W_(j, b a + j – 1) is the probability of finding b at position j in the motif model, and P(b l|S, B m) is the probability of finding b l in the sequence according to the background model. This formula is easily extended to multiple instances of the motif model by adding more motif terms to equation 1.
Estimating number of motif instances. The aim of the algorithm is to find the number of instances of a known motif model in the input sequence. To solve this problem we should compute the expected number of instances Q as
To compute equation 2 we need to estimate the probability P(Q = c|S, Θ_W_, B m) of finding c instances of the motif in the noisy background sequence. Applying Bayes’ rule to this probability leads to
We can distinguish three different parts in equation 3. The denominator P(S|Θ_W_, B m) serves as the normalization factor. The first term P(S|Q = c, Θ_W_, B m) of the numerator is the probability that the sequence is generated by the motif model Θ_W_, the background model B m, and contains c motif instances. This probability can be calculated by summing over all possible non-overlapping combinations of c motifs in sequence S.
with A c the set of c start positions a_1,…,a c. By applying equation 1, this equation can be efficiently computed in linear time. Assuming that each position is equally probable, the factor P(A c|Q = c, Θ_W, B m) is replaced by a constant inversely proportional to the number of possible combinations of c motif instances in a sequence of length L. Within this model, we see the motif instances in the context of the noisy background sequence. This implies that the longer the sequence is, the harder it is to find an instance within this noise. Therefore, in a long sequence only those instances that have a very high score with the motif model rise above the noise level and can be selected. The second term P(Q = c|Θ_W_, B m) in the numerator is the prior probability of finding c instances given the motif model and the background model. Let us define P(Q = c|Θ_W_, B m) as γ0(c). Since the complete prior distribution is not known, we propose one. There are two conditions to construct this distribution: (i) Σ∞c = 0γ0(c) should be equal to 1; (ii) for all c > 1, γ0(c + 1) is smaller than γ0(c). The user should define only γ0(1), a value between 0 and 1, as the probability of finding 1 instance. Initially γ0(0) is set to 1 – γ0(1) and the remainder of the distribution γ0(c) is set to κγ0(c – 1) and the distribution is then normalized. We use κ = 0.25. The effect of lowering the prior is that _E_[Q_] decreases and that less instances will be selected. Normally, we should compute the sum in equation 2 for c from 0 to ∞. Since this is unpractical, we propose to compute the next term in the distribution P(Q = c|S, Θ_W, B m) as long as the previous value is larger than a predefined small value ε (e.g., 0.0001).
Algorithm. The previous defined formulas can be combined to create the following algorithm. For each sequence S in the data set do: (i) score sequence S with motif model Θ_W_; (ii) score sequence S with background model B m; (iii) initialize P(Q = 0|S, Θ_W_, B m) and P(Q = 1|S, Θ_W_, B m); (iv) while P(Q = i|S, Θ_W_, B m) > ε augment i and update P(Q = c|S, Θ_W_, B m) for c = 0…i using equation 3; (v) compute the expected number of copies E S,θ,_Bm_[_Q_] with equation 2; and (vi) select the Q best scoring positions as motif instances.
MotifScanner is implemented in C++ and in addition to its use in Toucan through a web service we also provide a web-based version and a standalone version at our website: http://www.esat.kuleuven.ac.be/dna/BioI/Software.html. Further details about the influence of the prior probability and the length of the sequence on the detection rate of the MotifScanner can also be found at the MotifScanner website.
Statistical analysis
The calculation of a p value and a significance score for each motif was done as described in van Helden et al. (14), where it was developed to detect over-represented hexanucleotides within the upstream regions of families of coregulated genes in yeast. The frequency of binding sites observed throughout large reference sets like all the promoters in the Eukaryotic Promoter Database (EPD) or a large set of randomly selected putative regulatory regions from Ensembl are used to estimate the expected frequency for each motif m, F e{m}. These expected frequencies are used to calculate the number of expected occurrences for each motif in the set of regulatory regions under analysis:
where T is (by definition) the number of possible start positions, L i is the length of the _i_th sequence and w is the length of the motif. The probability to observe exactly n occurrences of the motif m is estimated by the binomial formula:
The probability to observe n or more occurrences of the motif m is:
A significance coefficient sig is used to select the most over-represented patterns:
sig = – log10[P(occ{m} ≥ n) × _D_]
where D is the number of distinct motifs that are used.
Matrix similarity
The similarity between two motifs, _M_1 and _M_2, is measured with the mutual information or Kullback–Leiber distance (15). The mutual information is computed as
where _M_1(j, b) is the probability of finding base b at position j in Motif 1. Since this equation is asymmetric, we take the average between the distance from _M_1 to _M_2 and from _M_2 to _M_1.
RESULTS
Toucan Overview
An analysis in Toucan starts with the creation of a set of sequences that might share a common _cis_-regulatory element or modules consisting of multiple elements. The goal is to identify significant elements and to visually inspect them. Possible sequence sets could be derived from gene clusters from microarray data analysis, from known target genes of a transcription factor, from putative target genes of a gene regulatory network, or genes involved in the same biological pathway, process or tissue (16,17).
Constructing sets of regulatory sequences
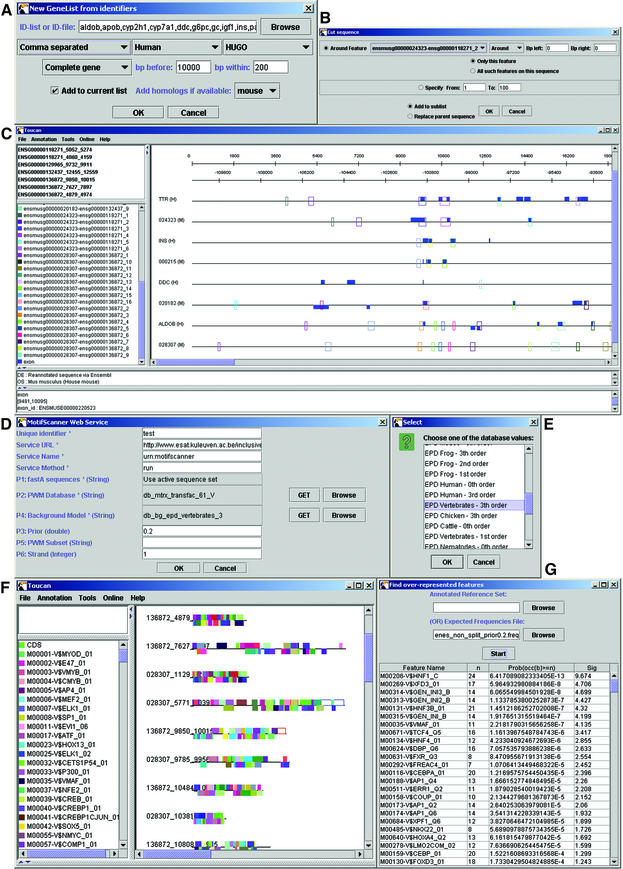
There are three different ways to import DNA sequences into Toucan. First, local sequence files in fastA, EMBL or GenBank format can be imported, either as separate sequences or as multiple sequences in one file. Secondly, sequences can be retrieved automatically from the EMBL nucleotide sequence database using a list of accession numbers. Thirdly, complete genes with their flanking regions (or optionally only the flanking regions) can be retrieved from the Ensembl genome database for all available organisms. In that case any identifier available in the Ensembl database can be used. For human genes these identifiers currently are Ensembl gene, SWISSPROT, EMBL accession number, protein_id, MIM, HUGO, Gene Ontology, PDB, RefSeq, LocusLink, SPTREMBL and Interpro. Sequences of orthologous genes can be retrieved simultaneously for the genes that have an ortholog correspondence in the Ensembl database.
Next, we wanted to give a solution for the detection of both proximal promoter regions and distal regulatory regions within the large sequences that are retrieved in this way. The presence of a promoter within the 5′ flanking region of a gene can be predicted by annotating CpG islands (18), which is included in Toucan. Promoter predictions using more sophisticated algorithms (e.g., PromoterInspector; 19) can be annotated on the sequences if the output of such external tools can be converted to GFF. Another approach is to predict the transcription start site (TSS) itself (e.g., by using Eponine; 20). Eponine outputs its predictions in GFF by default so they can be directly applied on the active gene set. In all these cases, however, the coverage of annotated promoters is limited to 50–60% at most (19).
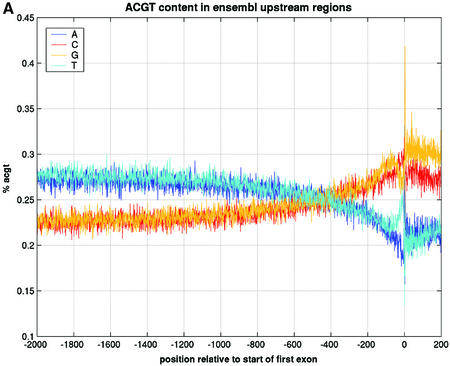
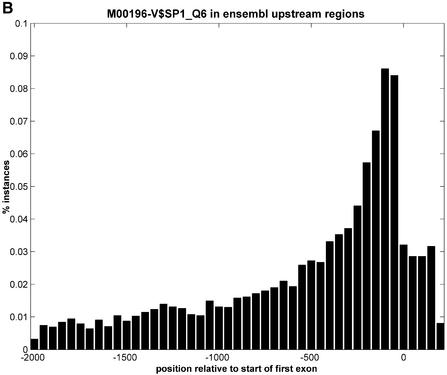
We therefore investigated whether genome annotation databases like Ensembl contain enough information (e.g., in the form of known transcripts mapped to the genomic sequence) to extract the location of the TSS. We believe that in most cases the start of the Exon 1 annotation in Ensembl, which lies generally further upstream than the ATG start codon, coincides with the TSS of the gene. If that is true, then sequences directly upstream of Exon 1 would contain the promoter-proximal sequences that we are interested in. It would also imply that we would not need to use promoter prediction tools and thus we would not be restricted to the limitations described above. To prove this statement, we have retrieved 2000 bp upstream of Exon 1 for 4000 randomly selected genes from the human genome. First, we calculated the percentages of A, C, G and T at each position in this stretch of DNA (Fig. 1A). The G/C content rises when approaching position 1 of Exon 1 and drops again after this position. We cannot think of any other DNA signal with such impact on the GCAT content than the TSS. A similar finding can be observed in the regions upstream of the ATG start codon in yeast (21). A second proof of this statement is the rise in the number of putative SP1 binding sites that occur within these 4000 regions (Fig. 1B). Since SP1 is known to be a proximal _cis_-acting factor (22), this analysis shows that is it likely that the first 500 bp upstream of the TSS are predominantly promoters. Because the goal of the analysis is to find over-represented motifs in sets of genes, and not in individual genes, it is still acceptable that for some genes in the set we would not have the correct promoter-proximal sequences if for these the start of Exon 1 would not be the TSS (e.g., if longer and yet unknown transcripts exist).
Figure 1.
Representation of the genomic region 2000 bp upstream of Exon 1 annotation in Ensembl and 200 bp after the start of Exon 1, taken from 4000 randomly selected genes from the human genome (homo_sapiens_8_30a database at kaka.sanger.ac.uk). The relative position of 0 on the _x_-axis is the start of Exon 1. (A) Percentages of A, C, G and T at each position. (B) Number of instances of a SP1 binding site at each position.
To predict other putative regulatory regions that lie more distal from the TSS, genomic sequences of the genes and their flanking regions can be aligned with the same regions of orthologous genes. For this purpose, highly specialized alignment algorithms exist like AVID (together with its visualization tool VISTA) (23,24), Bayes Aligner (25), DNA Block Aligner (26) or PipMaker (27). The AVID/VISTA tools can be used transparently through a web service. For the other tools, we provide an online GFF toolbox at our website to convert their alignment outputs to GFF. After annotation of the latter, the similar parts in the upstream sequences of the orthologs can be selected to construct a new sequence set for the analysis of regulatory elements. Alternative approaches could be (i) to use Alfresco (28) (which is a visualization tool that integrates multiple tools for comparative sequence analysis) or related tools, save interesting regions as GFF, and import these into Toucan; or (ii) to retrieve syntenic regions directly from specialized databases like CORG (29). Figure 2C shows an example of a sequence set.
Figure 2.
(Previous page) Screenshots of Toucan during the analysis of liver-specific genes. (A) Dialog where all gene names (HUGO symbols) are entered as a comma separated list. In the second drop-down box ‘Human’ is selected to search for and retrieve human genes. All organisms that are available in Ensembl (see http://www.ensembl.org) can be chosen from this list, and in the ‘Preferences’ menu the user can update these settings if Ensembl were to add new organisms. Depending on which organism is chosen, the third drop-down box shows all available external database identifiers that can be mapped to a stable Ensembl gene. The fourth drop-down box allows to choose between ‘complete gene’, ‘upstream of CDS’ and ‘upstream of Exon 1’. The latter corresponds in most cases to the region upstream of the TSS. The text boxes labeled with ‘bp before’ and ‘bp within’ state how many base pairs should be retrieved as flanking sequence upstream or around the specified region. In the last drop-down menu ‘mouse’ is selected to retrieve also the mouse orthologous sequences for each human gene in the list. (B) Every region that seems likely to contain putative regulatory modules (e.g., because it is conserved between species or because it contains a CpG island) can be selected and added to a sequence sublist. (C) Feature map. All open boxes represent regions that are at least 75% similar with their respective orthologous region, resulting from the AVID/VISTA web service. (D) Matrices, background model, and all other parameters are set in the dialog box of the MotifScanner. (E) Dialog showing the background models on our server. The values are retrieved transparently through the web service when the user presses the ‘GET’ button. (F) The results of the MotifScanner can either be saved or can be automatically added as features on the currently active sequence set. (G) Results of using the binomial formula to detect over-represented motifs. n is the number of occurrences of a binding site within this set, the third column is the p value for this motif, the fourth column the sig value (see Methods). The top scoring motifs for the human–mouse conserved regions in 10 kb upstream sequence of liver-specific genes are shown.
Scoring transcription factor binding sites
In Toucan, a set of sequences can be annotated with IUPAC consensus sequences. A string containing IUPAC symbols is translated into a regular expression, which is used to find matching positions on both sequence strands (e.g., WWC{2,3}AA becomes [at][at]c{2,3}aa internally). More refined methods are based on scoring sequences at each nucleotide with PWMs (e.g., MatInspector; 30,31). PWMs provide a quantitative rating (score) suggesting likelihood of protein binding to the site analyzed. A selection of positive hits is then made by imposing a threshold on the normalized scores. Here we introduce a new algorithm for scoring sequences with PWMs, MotifScanner. The method is based on a sequence model which states that the binding sites are hidden in a noisy background sequence. We use this probabilistic model to estimate the number of instances of a motif in a specific sequence, given the background model and the motif model, instead of using a predefined threshold that is independent of the sequence being scored. The advantages of this method can be summarized as follows. First, by choosing an appropriate background model for the sequences to be scored we can reduce the number of false positive hits (32). For example, when scoring human promoter sequences using a 3rd-order background model that is calculated from a large set of human promoters, a putative motif instance would need a higher resemblance to the PWM to be a positive hit than when using a background model of mouse sequences to score the same human promoters. Another example can be given by the fact that a A/T rich motif scores higher with MotifScanner in a G/C rich context than in a A/T rich context. Because the presence of a motif in a dissimilar context can imply a functional conservation we believe that its detection should indeed be promoted. Secondly, by estimating the number of motif instances instead of using a threshold, only the best matching instances are considered as hits instead of all the instances that score above a certain threshold. This approach reflects the situation in a cell where a transcription factor is bound more often to the stronger sites than to the weaker sites. However, if one also wishes to select the weaker sites then the prior parameter can be increased, the sequences trimmed (to remove noise), or a matrix scoring program can be used. Because this algorithm has not been described before, we elaborate on it in the Methods section.
The user can transparently use this algorithm as a web service from within Toucan and the resulting GFF formatted output can be applied to the active sequence set. By clicking on a feature of a sequence (a colored box) the information on this feature is shown in the bottom window of the application, in this case it is the name of the transcription factor, the actual sequence instance and the matching score. This score is the absolute value of the ratio between the probability of the site being generated by the motif model and the probability of the site being generated by the background model. The PWM databases and background models that are used by the service reside on the server and can be selected within a dialog window in Toucan (see Fig. 2D, E and F).
A binomial distribution model is used to correlate all annotated features with a p value and a significance score based on their occurrence in the sequence set and relative to their expected frequency (see Methods and Fig. 2G). The expected frequency of a feature can be approximated by calculating the respective actual frequencies, expressed as occurence per base pair, either from another sequence set or from a general (genome-wide) reference set (e.g., all promoters in the EPD). At our website we provide several files with expected frequencies calculated from the EPD or from the upstream regions of random gene subsets from complete genomes.
Detecting new patterns using Gibbs sampling
Unfortunately, some factors are not sufficiently well described to allow the construction of reliable weight matrices. For this reason, current PWM databases (like TRANSFAC; 33) and the supporting scoring algorithms can only characterize part of the logic of _cis_-regulatory regions. It is therefore useful to search directly for statistically over-represented motifs (which may possibly correspond to yet unknown TFBMs) in the DNA of a regulatory sequence set. To find such motifs, our MotifSampler (4) (which is a probabilistic method for motif finding based on Gibbs sampling) can be accessed through a web service. This algorithm uses the same probabilistic framework as in the MotifScanner to estimate the expected number of copies of a motif in a sequence set and also uses a higher-order background model based on a Markov chain. The web service returns, besides its GFF output, also the position probability matrices of the motifs which can afterwards be used to scan related sequence sets using the MotifScanner service.
Case studies
We have performed several analyses on human gene sets. They serve both as a biological validation of Toucan and as examples for the user. In the first example DNA regions of 500 bp directly upstream of the start of Exon 1 (as annotated in the Ensembl database) of eight known E2F target genes are investigated. Table 1A lists all factors with a sig factor ≥2. In the second example we investigated a set of liver specific genes and a set of muscle specific genes. We tested whether certain binding sites of known transcription factors would be over-represented in either known regulatory regions, promoter-proximal regions or mouse-syntenic regions of these genes. It is shown that the retrieval of orthologous sequences (human and mouse) enables the selection of putative regulatory regions through comparative sequence analysis. Starting from upstream sequences tens of kilobases long, this selection narrows down the search region for regulatory modules to a couple of hundred base pairs—this length restriction is essential for the detection of over-represented motifs (if not, the over-representation statistic is buried by the sequence noise). In Table 1B and C we report all factors with a sig factor >2 in at least one of the three analyses performed (also see the Methods section). Lastly, in the third example we use the mouse syntenic regions of muscle and liver genes again for the detection of over-represented DNA words using the MotifSampler, and we show that the results are comparable to those of the second example, and that they can contain extra information.
Table 1. Statistically over-represented TFBMs in several gene sets.
| A E2F targets | B Muscle-specific | C Liver-specific | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 Proximal | B1 Exp. | B2 Proximal | B3 Syntenic | C1 Exp. | C2 Proximal | C3 Syntenic | |||
| E2F | ***** | SRFa | ***** | ***** | ***** | HNF1a | ***** | ** | ***** |
| ETF | ***** | Myogenina | ***** | ***** | ***** | HNF3a | * | **** | |
| SP1 | ** | MAZ | ***** | * | ** | HNF4a | * | ** | |
| MYODa | ***** | IPF | **** | ||||||
| HEB | ***** | *** | AP1 | **** | ** | ||||
| LBP1 | ***** | * | **** | C/EBPa | *** | ** | |||
| MEF2a | ***** | ** | COUP | ** | ** | ||||
| SP1a | ***** | * | FOX | * | ** | ||||
| MZF | ***** | * | * | VMAF | **** | ||||
| MINI | **** | TCF4 | *** | ||||||
| LMO2COM | **** | DBP | ** | ||||||
| MAZR | **** | *** | FXR | ** | |||||
| E12 | **** | ERR | ** | ||||||
| MEF3 | **** | ||||||||
| RREB | ***** | ||||||||
| ZIC | ** | ** | |||||||
| LYF | ** | ||||||||
| VDR | ** | ||||||||
| GC | ** | ||||||||
| TFIII | ** | ||||||||
| TEFa | * | * | |||||||
| AP4 | *** | *** | |||||||
| RSRFC4 | **** | ||||||||
| STAT6 | **** | ||||||||
| CP2 | ** | ||||||||
| PAX4 | ** |
E2F target genes
In this example, we investigated eight human genes of which the E2F complex is a known regulating transcription factor: CAV1, CDC6, MYC, DHFR, E2F2, RBL1, TK1 and RB1. Since E2F mostly binds to the proximal promoter of its target genes, a region of 500 bp upstream of the putative TSS (start of Exon 1) was obtained from Ensembl.
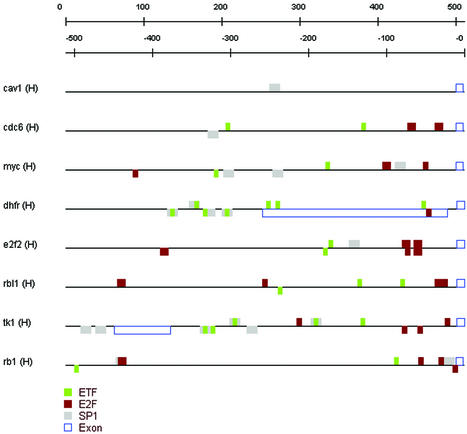
All retrieved sequences are visualized in a sequence feature map (Fig. 3). Next we have scored these sequences with PWMs that reside on our server by using the MotifScanner web service. The matrices, background model and all other parameters are set in a dialog box (see Fig. 2D and E). The results of the MotifScanner can be automatically added as features on the currently active sequence set. Although a low prior (0.2) was used, most of the sequences are packed with putative binding sites. Running the binomial analysis we could select the significantly over-represented motifs (see Table 1A). The expected frequencies needed for this statistic were calculated by scoring the same matrices on all human sequences in the EPD (see http://www.epd.isb-sib.ch/) (see Methods). The presence of E2F, ETF and SP1 was significant (sig ≥ 2). Figure 3 shows the sequence set with the instances of these motifs annotated. The presence of two to three putative ETF binding sites in almost all E2F target genes is interesting since this is also the case in the mouse p53 promoter, which is bound by E2F and ETF upon adenovirus infection in the presence of the Early 1a protein (34).
Figure 3.
Promoter regions of eight E2F target genes with the over-represented TFBMs. The sequences were retrieved from Ensembl starting from a comma separated list of HUGO symbols and choosing ‘upstream of Exon 1’, 500 ‘bp before’ and 10 ‘bp within’.
Muscle and liver-specific genes using TRANSFAC
Wasserman and Fickett (17) and Krivan and Wasserman (16) have compiled and analyzed respectively muscle-specific and liver-specific regulatory regions that are experimentally verified. They found a significant occurrence of specific binding site clusters within these regions. We have done a regulatory analysis on their training set sequences using Toucan. The fastA formatted sequence files were downloaded from http://bio.cse.psu.edu/mousegroup/Reg_annotations/ and loaded straight into Toucan (after removing blanks within the sequences). We used the MotifScanner with the TRANSFAC collection of vertebrate matrices, a prior of 0.2, and a background model of vertebrate sequences of EPD. The result of the statistics applied afterwards are represented in Figure 2F and Table 1, columns B1 and C1. Some of the motifs are known to be muscle specific: SRF, myogenin, MYOD, MEF-2, MZF, MINI and MEF-3; so their presence in these sequences is not surprising. The only muscle-specific factor that was used in Wasserman and Fickett (17) that we could not confirm with sig ≥ 2 is TEF. Some others can interact with muscle-specific factors: E12 (dimerizes with MYOD and myogenin of the Myf family) and HEB (interacts with E12 and myogenin). The finding that their actual binding sites are significantly present in these sequences is new. The detection of SP1 is not surprising since it is a general promoter element. Some of the remaining factors may not be muscle specific but they may play a role in transcriptional regulation in certain circumstances. Vitamin D receptor (VDR) for example is involved in the genomic response of avian embryonic skeletal muscle cells to vitamin D3 (35), LMO2 (LIM-only protein) may play a role in differentiation and myofibrillogenesis of heart (36) and LBP-1 (UBP-1) binds at the promoter of skeletal troponin I (37). For the remaining factors we could not find any references that point to regulation of muscle genes. These are MAZ (Pur-1, Zif87), MAZ related factor (MAZR), ZIC and Ras-responsive element binding protein (RREB).
An analogous analysis on the set of liver-specific regions shows similar results, although fewer factors have over-represented sites. Of the factors having sig ≥ 2, HNF and C/EBP were also used by Krivan and Wasserman (16) and are known to be liver specific. Other significant factors include COUP, which may antagonize with HNF-4 (38), and Insulin Promoter Factor (IPF). Mutations in IPF or HNF both result in a common progression of maturity-onset diabetes of the young (MODY) (39). Their involvement in _cis_-regulation may therefore be an interesting hypothesis. The last one is AP1, a general regulatory factor.
Although these analysis give remarkably good results, starting from experimentally determined regulatory regions considerably facilitates the analysis—while obtaining such regions experimentally is difficult. Therefore, we tested another approach starting only from approved HUGO symbols, and retrieving the sequences automatically from the Ensembl database. We used the same genes that were represented in the set of known regulatory sequences used above: for the muscle set these are CHRM2, CHRM3, ACTC, CKM, DES, MYF6, MYOG, MYL1, MYLA, TNNI3, MYHCA, ACTA1, DMD, ANF and ALDOA; and for the liver set these are ALDOB, APOB, CYP2H1, CYP7A1, DDC, G6PC, GC, IGF1, INS, PAH, PROC, SLCA2, SULT2A2, SULT2A1, TTR and UGT1A1.
When using only 400 bp upstream of Exon 1 like in the E2F analysis, fewer elements were detected both for the muscle and for the liver genes (see Table 1, columns B2 and C2). For muscle, the highly significant elements are SRF and MAZR, and for liver HNF-1 and FOX (previously called HNF-3/forkhead transcription factors).
If we look at the location of the known regulatory regions relative to the TSS, we see that most of the regions are actually enhancers that lie further upstream, or even downstream of the TSS. We therefore retrieved, in a new analysis, 10 kb of sequence upstream of the translation start (start of CDS annotation) together with the same part of the mouse ortholog when such a correspondence was available. The different steps of the analysis for the liver genes are summarized in Figure 2. For each pair of orthologous sequences we used AVID and VISTA to detect regions having minimal 75% of similarity in a sliding window of 100 bp. The regions located 5′ upstream of the TSS or in the 5′ UTR were selected and scored with the TRANSFAC collection of vertebrate matrices using the same parameter settings as before. The results of the statistical analysis performed thereafter are shown in Table 1, columns A3 and B3. Both for the liver set and the muscle set, the presence of the same elements as in the experimental regions is more pronounced than for the proximal regions, for example HEB, LBP and MEF-2 in the muscle regions and HNF-3, HNF-4, C/EBP, COUP and AP1 in the liver regions. These are probably factors that bind to sites in distal modules rather than in the region just upstream of the TSS. There are also factors that were present in neither of the two other analyses: for muscle RSRFC4 (SRF-related), STAT6 (involved in hypercontractility of smooth muscle cells) and others without established muscle relatedness (see Table 1); for liver TCF4 (tumors arising in the liver can be caused by a complex of TCF4 and mutated beta-catenin), DBP (a member of the C/EBP family that is enriched in liver) and others without established liver relatedness (see Table 1). This shows that putative regulatory motifs can be detected computationally that have not been detected experimentally yet, which might be due to the difficulty of mimicking every developmental and metabolic condition in the cell. The presence of factors without a direct link with the experimental setup can sometimes be due to the fact that they recognize sequences which are related to the sites of other factors. This is probably the case for v-MAF which binds to AP-1 sites since v-MAF forms heterodimers with Fos and Jun (the consensus binding site of v-MAF is TGCTGACTCAGCA and the consensus site of AP-1 is GVTGACTCA so they are very similar).
Muscle and liver-specific genes without TRANSFAC
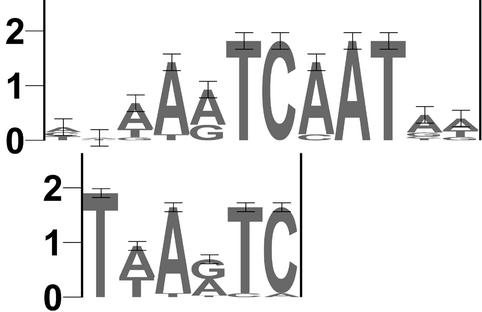
So far, we have only used a collection of PWMs of known transcription factors. Because some of these matrices may be of inferior quality, and also because there must exist other transcription factors with yet unknown binding sites, we have used the MotifSampler to detect over-represented DNA motifs in the liver and muscle sets of human–mouse syntenic regions. We have used the sampler five times, each time with a different length of the motif to be found (6, 7, 8, 10 and 12 bp). The number of different motifs to be found was always set to 4, the prior to 0.2, and the number of runs to 10. This way, a total of 4 × 10 = 40 motifs were found for each motif length. These were ranked by their log likelihood score (4) and the top five motifs were selected. All the top motifs were taken together (5 × 5 = 25 in total), ranked again, and similar motifs were grouped together (see Methods). This resulted in 18 distinct motifs for the liver set and 15 for the muscle set (see Table 2). On these sets we performed two kinds of validation. First, we calculated a significance score similar to the one used after the MotifScanner. We scored a set of putative regulatory regions selected by aligning 3500 randomly selected orthologous gene pairs of human and mouse with all the newly found motifs and we calculated the respective expected frequencies. These were used in Toucan to calculate p values and sig scores (see the Methods section) in the sets of coregulated genes. As can be seen in Table 2, many of the motifs are significantly over-represented compared to their expected frequency (sig ≥ 2). As a second validation we compared each newly found motif matrix with the position weight matrices of TRANSFAC professional version 6.1 (see Table 2). Most of the motifs have a good similarity (see Methods for the calculation of the similarity measure) with one or more known matrices that were detected in the previous analysis using the MotifScanner. By comparing the sequence logo of the matrix found by the MotifSampler with the best matching matrix of TRANSFAC (e.g., HNF-6 in Fig. 4), one might detect gene set or process specific sequence preferences, like the extra T in the new motif at the 5′ end that is not present in the TRANSFAC matrix. A few motifs seem to appear for the first time (they were not over-represented in the MotifScanner approach), like HEN (or STAF) and HSF for the muscle group and PPARA, SRY, MIF1, BRACH and NFY (or AP4) for the liver group. Finally, there are several motifs that do not match a known PWM. These could be binding sites of unknown transcription factors.
Table 2. Statistically over-represented DNA motifs.
| A Muscle-specific | B Liver-specific | ||||
|---|---|---|---|---|---|
| Motif consensus | sig | PWM similar to | Motif consensus | sig | PWM similar to |
| AsCTGGTGwk | ***** | - | TTkGmTnAry | ***** | - |
| nGCCyGGkyC | ***** | - | wrkkkAmTwA | ***** | - |
| GGGrCnGGks | ***** | - | nTkATTGAnnwA | ***** | HNF1 |
| CnyCTCyCTC | ***** | MAZ | nCnwAGkTmA | ***** | PPARA |
| rGGGnwGGGGC | ***** | MZF | kAwGwGTyTG | ***** | SRY |
| AAGCCT | **** | HSF | nTTTGmTywr | ***** | DBP, HNF3 |
| rCCTGGk | ** | - | wGTyAwT | ***** | HNF1, AFP, IPF, FXR, MEF2, AMEF2, COMP1 |
| GsAGGGG | ** | - | CTwnGTmn | ***** | MIF1, PPARA |
| rCCCAGs | ** | HEN1, STAF | ACyTAsn | ***** | BRACH |
| ACCCAG | ** | CP2, RORA2, STAF, PPARG | TTwwTsmTTnrC | ***** | HNF1 |
| GGGCwG | * | SP1 | nATTnGCT | ***** | DBP, HNF3 |
| CCTGCT | * | HEB, MYOGENIN, E12, E47 | TTGAyTwwnrGw | ***** | - |
| GGGrmAGG | - | ynnGAsyTTnnn | ***** | - | |
| CCTGGsnCnGG | - | CTAnGTm | ***** | - | |
| GCTGCCC | - | AAkywAAT | ***** | HNF3, HNF1 | |
| TwArTC | ***** | HNF6 | |||
| CTGrTT | *** | NFY, AP4 | |||
| CTkTGA | TCF4, ATF, ER, PPARG, GFI |
Figure 4.
Sequence logos (40) of a pair of similar motifs (see Table 2), one motif derived from the scoring matrix M00639 (HNF-6, upper logo) of the TRANSFAC database and one motif found by the MotifSampler (lower logo). The first is based on 13 binding sites in TRANSFAC, the second is based on 16 motif instances in our liver regulatory dataset. Positions 2–6 of the new motif match perfectly with the known motif. Position 1 of the new motif is certainly a T while the known motif has no information at that position.
CONCLUSION
In summary, Toucan provides an efficient and integrated environment for gene regulation bioinformatics. Starting only from gene identifiers, it can retrieve, visualize, annotate and analyze proximal and distal regulatory sequences of coregulated genes. Because we use web services, we can add more services that work with fastA formatted sequence files and we will be able to link with bioinformatics service registries in the future. This flexibility will help to improve the interoperability among visualization tools, algorithms and data providers for gene regulation bioinformatics (12).
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank M. Dabrowski and B. De Strooper from the Center for Human Genetics, VIB4, Leuven, Belgium for helpful discussions. S.A. is a research assistant of the Katholieke Universiteit (K.U.)Leuven. G.T. is a research assisant of IWT. Y.M. is a post-doctoral researcher with the FWO-Vlaanderen and an assistant professor at the K.U.Leuven. B.D.M. is a full professor at the K.U.Leuven, Belgium. Research supported by Research Council KUL [GOA-Mefisto 666, IDO (IOTA Oncology, Genetic networks), several PhD/postdoc and fellow grants]; Flemish Government: FWO [PhD/postdoc grants, projects G.0115.01 (microarrays/oncology), G.0240.99 (multilinear algebra), G.0407.02 (support vector machines), G.0413.03 (inference in bioi), G.0388.03 (microarrays for clinical use), G.0229.03 (ontologies in bioi), research communities (ICCoS, ANMMM)]; AWI [Bil. Int. Collaboration Hungary/Poland]; IWT [PhD Grants, STWW-Genprom (gene promotor prediction), GBOU-McKnow (Knowledge management algorithms), GBOU-SQUAD (quorum sensing), GBOU-ANA (biosensors)]; Belgian Federal Government: DWTC [IUAP IV-02 (1996–2001) and IUAP V-22 (2002–2006)]; EU [CAGE; ERNSI]; Contract Research/agreements [Data4s, Electrabel, Elia, LMS, IPCOS, VIB].
REFERENCES
- 1.Lemon B. and Tjian,R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- 2.Davidson E.H. (2001) Genomic Regulatory Systems. Development and Evolution. Academic Press, San Diego, CA.
- 3.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of _cis_-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 4.Thijs G., Marchal,K., Lescot,M., Rombouts,S., De Moor,B., Rouze,P. and Moreau,Y. (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol., 9, 447–464. [DOI] [PubMed] [Google Scholar]
- 5.Berman B.P., Nibu,Y., Pfeiffer,B.D., Tomancak,P., Celniker,S.E., Levine,M., Rubin,G.M. and Eisen,M.B. (2002) Exploiting transcription factor binding site clustering to identify _cis_-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA, 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markstein M., Markstein,P., Markstein,V. and Levine,M.S. (2002) Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 99, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loots G.G., Ovcharenko,I., Pachter,L., Dubchak,I. and Rubin,E.M. (2002) rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res., 12, 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halfon M.S., Grad,Y., Church,G.M. and Michelson,A.M. (2002) Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res., 12, 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down, T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Helden J., Andre,B. and Collado-Vides,J. (2000) A web site for the computational analysis of yeast regulatory sequences. Yeast, 16, 177–187. [DOI] [PubMed] [Google Scholar]
- 11.Jegga A.G., Sherwood,S.P., Carman,J.W., Pinski,A.T., Phillips,J.L., Pestian,J.P. and Aronow,B.J. (2002) Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res., 12, 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein L. (2002) Creating a bioinformatics nation. Nature, 417, 119–120. [DOI] [PubMed] [Google Scholar]
- 13.Thijs G., Lescot,M., Marchal,K., Rombauts,S., De Moor.,B., Rouze,P. and Moreau,Y. (2001) A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics, 17, 1113–1122. [DOI] [PubMed] [Google Scholar]
- 14.van Helden J., Andre,B. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 15.Kullback S. (1959) Information Theory and Statistics. John Wiley and Sons, New York, NY.
- 16.Krivan W. and Wasserman,W.W. (2001) A predictive model for regulatory sequences directing liver-specific transcription. Genome Res., 11, 1559–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wasserman W.W. and Fickett,J.W. (1998) Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol., 278, 167–181. [DOI] [PubMed] [Google Scholar]
- 18.Ioshikhes I.P. and Zhang,M.Q. (2000) Large-scale human promoter mapping using CpG islands. Nature Genet., 26, 61–63. [DOI] [PubMed] [Google Scholar]
- 19.Scherf M., Klingenhoff,A. and Werner,T. (2000) Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J. Mol. Biol., 297, 599–606. [DOI] [PubMed] [Google Scholar]
- 20.Down T.A. and Hubbard,T.J.P. (2002) Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res., 12, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazma A., Jonassen,I., Vilo,J. and Ukkonen,E. (1998) Predicting gene regulatory elements in silico on a genomic scale. Genome Res., 8, 1202–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook T., Gebelein,B. and Urrutia,R. (1999) Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci., 880, 94–102. [DOI] [PubMed] [Google Scholar]
- 23.Dubchak I., Brudno,M., Loots,G.G., Pachter,L., Mayor,C., Rubin,E.M. and Frazer,K.A. (2000) Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res., 10, 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayor C., Brudno,M., Schwartz,J.R., Poliakov,A., Rubin,E.M., Frazer,K.A., Pachter,L.S. and Dubchak,I. (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, 16, 1046–1047. [DOI] [PubMed] [Google Scholar]
- 25.Wasserman W.W., Palumbo,M., Thompson,W., Fickett,J.W. and Lawrence,C.E. (2000) Human–mouse genome comparisons to locate regulatory sites. Nature Genet., 26, 225–228. [DOI] [PubMed] [Google Scholar]
- 26.Jareborg N., Birney,E. and Durbin,R. (1999) Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res., 9, 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jareborg N. and Durbin,R. (2000) Alfresco—a workbench for comparative genomic sequence analysis. Genome Res., 10, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dieterich C., Wang,H., Rateitschak,K., Luz,H. and Vingron,M. (2003) CORG: a database for COmparative Regulatory Genomics. Nucleic Acids Res., 31, 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frech K., Quandt,K. and Werner,T. (1997) Finding protein–binding sites in DNA sequences: the next generation. Trends Biochem. Sci., 22, 103–104. [DOI] [PubMed] [Google Scholar]
- 32.Marchal K., Thijs,G., De Keersmaecker,S., Monsieur,P., De Moor,B. and Vanderleyden,J. (2003) Genome-specific higher-order background models to improve motif detection. Trends Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- 33.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Pruss,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hale T.K. and Braithwaite,A.W. (1999) The adenovirus oncoprotein E1a stimulates binding of transcription factor ETF to transcriptionally activate the p53 gene. J. Biol. Chem., 274, 23777–23786. [DOI] [PubMed] [Google Scholar]
- 35.Capiati D., Benassati,S. and Boland,R.L. (2002) 1,25(OH)2-vitamin D3 induces translocation of the vitamin D receptor (VDR) to the plasma membrane in skeletal muscle cells. J. Cell. Biochem., 86, 128–135. [DOI] [PubMed] [Google Scholar]
- 36.Li H.Y., Kotaka,M., Kostin,S., Lee,S.M., Kok,L.D., Chan,K.K., Tsui,S.K., Schaper,J., Zimmermann,R., Lee,C.Y., Fung,K.P. and Waye,M.M. (2001) Translocation of a human focal adhesion LIM-only protein, FHL2, during myofibrillogenesis and identification of LIM2 as the principal determinants of FHL2 focal adhesion localization. Cell Motil. Cytoskeleton, 48, 11–23. [DOI] [PubMed] [Google Scholar]
- 37.Nikovits W. Jr, Mars,J.H. and Ordahl,C.P. (1990) Muscle-specific activity of the skeletal troponin I promoter requires interaction between upstream regulatory sequences and elements contained within the first transcribed exon. Mol. Cell. Biol., 10, 3468–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mietus-Snyder M., Sladek,F.M., Ginsburg,G.S., Kuo,C.F., Ladias,J.A., Darnell,J.E.J. and Karathanasis,S.K. (1992) Antagonism between apolipoprotein AI regulatory protein 1, Ear3/COUP-TF and hepatocyte nuclear factor 4 modulates apolipoprotein CIII gene expression in liver and intestinal cells. Mol. Cell. Biol., 12, 1708–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stride A. and Hattersley,A.T. (2002) Different genes, different diabetes: lessons from maturity-onset diabetes of the young. Ann. Med., 34, 207–216. [PubMed] [Google Scholar]
- 40.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]