Breast cancer stem cells revealed (original) (raw)
A fundamental problem in cancer research is identification of the cell type capable of sustaining the growth of the neoplastic clone. There is overwhelming evidence that virtually all cancers are clonal and represent the progeny of a single cell. What is less clear for most cancers is which cells within the tumor clone possess tumor-initiating cell (T-IC) function and are capable of maintaining tumor growth.
A remarkable series of transplant experiments from the 1950s, which reflect the state of human experimentation of a bygone era, demonstrated that autologous injection of tumor cells into the thigh could only be initiated if >106 tumor cells were injected (1). Subcutaneous injection of human tumor samples into mice gave similar results. Two theories were developed to explain why not every cell within a tumor was capable of regenerating the tumor (2). The stochastic theory predicts that every cell within the tumor is potentially tumor-initiating but entry into the cell cycle is governed by low probability stochastic events. By contrast, the hierarchy theory proposes that the tumor is functionally heterogeneous and only a limited number of cells are capable of initiating the tumor. However, cells of this class all initiate tumors at high frequency. Although each theory predicts that a limited number of cells within a tumor will initiate tumorigenic growth, the underlying biological mechanisms are radically different.
The stochastic model predicts that the tumor is relatively homogeneous and the tumorigenic mechanisms (pathways, genetic programs) that underlie the malignancy are operative in all cells. Thus, studying the bulk of the cells that make up the tumor mass can identify the key properties of the tumor. The hierarchy model predicts functional heterogeneity among the cells that comprise the tumor and that the rare T-IC are different from the vast majority of the cells that make up the tumor. Therefore, tumorigenic pathways may function differentially in distinct tumor subpopulations. This model also predicts that although eradication of the non-T-IC cells may result in a remission, the disease will relapse if the T-IC cells are not eliminated. Resolution of the T-IC problem requires both purification of tumor cells into subfractions and a functional assay to detect cells with the capacity to initiate tumor growth in vivo. The stochastic model suggests that it will be impossible to predict which kind of cells become T-IC and that stochastic events will cause T-IC cells to be found in any two sorted cell fractions with equal probabilities. By contrast, the hierarchy model predicts that it should be possible to separate T-IC from non-T-IC.
Research on T-IC is most advanced for the hematological malignancies. Key to these studies is the depth of understanding of normal hematopoietic development that has been gained in the past four decades. Functional in vitro and in vivo assays are available for all stem and progenitor cell types ranging from the primitive pluripotential stem cells to multipotential and unipotential progenitors (3). In addition, a rich collection of cell surface differentiation markers enable detailed characterization of normal hematopoietic development, as well as providing insight into how normal differentiation becomes disrupted in human leukemia. It is clear that leukemic tissues, although abnormal, still retain remnants of normal differentiation and developmental programs (4). Unfortunately, purification of solid tumor T-IC has been difficult because of the paucity of cell surface markers that enable cell sorting. Moreover, T-IC xenograft assays for primary human solid tumor tissue have traditionally been carried out in nude mice. These mice still possess significant residual immune function causing variability in the rate of xenograft initiation and it is possible that host resistance mechanisms will not permit single T-IC to be detected.
For most cancers it is less clear which cells within the tumor clone possess tumor-initiating cell function.
The paper by Al-Hajj et al. (5) in this issue of PNAS provides a major step forward with the identification of human breast cancer initiating cells (BrCa-IC). First a reliable xenograft assay was developed that enabled the injection of single cell suspensions, not just fragments, of human breast cancer tissue into the mammary fat pad of immune-deficient NOD/SCID mice which are more immune-deficient than nude mice. Nine of nine samples, including metastatic and primary breast tumors were able to grow in NOD/SCID mice. Thus, this system provides a reliable and sensitive in vivo T-IC assay for human breast cancer. Al-Hajj et al. then used four cell surface markers (adhesion molecules CD44 and CD24, a breast/ovarian cancer specific marker B38.1, and the epithelial specific antigen, ESA), whose expression is heterogeneous in breast cancer tissue, to sort into expressing and nonexpressing fractions, alone or in combination. The BrCa-IC capacity of each cell population was determined by using the NOD/SCID xenotransplant assay. Because breast tumor tissue can contain hematopoietic, endothelial, mesothelial, and fibroblast cells, lineage markers for all of these tissues (Lin+) were used to show that only the breast cancer tissue (Lin−) contained BrCa-IC. All mice injected with either CD44+, B38.1+, or CD24−/low generated palpable tumors in 12 weeks, whereas none of the CD44−, B38.1− injections caused tumors. For three samples, the Lin−CD44+CD24−/low fractions were subdivided by expression of ESA: Lin−ESA+CD44+CD24−/low cells generated breast tumors in NOD/SCID mice, whereas ESA−CD44+CD24−/low cells did not. The Lin−ESA+CD44+CD24−/low cells represented a minor subpopulation (2%) of the unfractionated breast cancer cells and semiquantitative limiting dilution analysis suggested that BrCa-IC activity was enriched by 50-fold in this fraction. This study demonstrates the crucial importance of combining cell sorting with functional assays because there were no morphological differences between the two fractions. Collectively, these results conclusively demonstrate that breast cancer is functionally heterogeneous and that a rare BrCa-IC is the only cell type capable of establishing human breast cancer after transplant into NOD/SCID mice. Moreover, the BrCa-IC express unique cell surface markers that enable prospective isolation strongly supporting the hierarchy model of cancer.
Examination of tumor tissues that were generated from the injection of ESA+CD44+CD24−/low cells revealed another important property of BrCa-IC: the ability to reestablish tumor heterogeneity after transplantation. Tumors in NOD/SCID mice contained the same complex mixture of ESA−, CD44−, and CD24+ cells as tumors from the original donor. Importantly, secondary transplant experiments indicated that ESA+CD44+CD24−/low cells were the only fraction with the capacity for tumor initiation on serial transplant. These data provide strong support for the idea that on cell division the BrCa-IC can self-renew (i.e., make other BrCa-IC) as well as make progeny that acquire maturation markers and lose the ability to initiate tumor growth. This process is very similar to the process whereby normal stem cells can both self-renew to maintain the stem cell pool and also produce differentiated progeny to create the mature cell types specific to that organ. Thus, like leukemic cells, breast cancer cells retain remnants of normal developmental programs.
The study of normal and leukemic human hematopoiesis provides a roadmap of what may be possible for breast cancer. Progress to study normal and leukemic human stem cells has been greatly aided by the NOD/SCID xenotransplant system in which human stem cells repopulate the hematopoietic tissues of this profoundly immunodeficient recipient (6). Transplantation of human leukemia into NOD/SCID mice generates a disease in the mice that recapitulates the human disease. In acute myeloid leukemia (AML), an impaired differentiation program results in the excess production of leukemic blasts, of which the vast majority have limited proliferative capacity. Morphologically, AML reflects abnormal development in one of the major blood lineages, but the blasts from different patients are heterogeneous with respect to the lineage antigens they express. The human AML-initiating cell, termed the SCID Leukemia-Initiating Cell (SL-IC), was identified and purified by transplantation into NOD/SCID mice (7). The CD34+CD38− cell fraction that represents from 0.1% to 1% of the AML cell population contained all of the SL-IC. The CD34− and CD34+CD38+ cell fractions that comprised the bulk of the cells contained no SL-IC, although they did contain clonogenic leukemic progenitors capable of in vitro growth. Thus, these studies conclusively showed that the leukemic clone is organized as a hierarchy in which only a rare subset of leukemic cells possesses the ability to initiate the clone on transplantation and that these SL-IC are distinct from the clonogenic progenitors that can be assayed in vitro and the nonclonogenic blast population that makes up the majority of the leukemic clone. Moreover, the cell surface phenotype of the SL-IC is similar to that of the normal human stem cell (8). These data support a model in which the cellular targets for transformation reside in the primitive stem cell compartment, not in the lineage-committed clonogenic progenitors (reviewed in refs. 6 and 9). According to this model, the heterogeneity observed within the leukemic clone results from the variable ability of these primitive leukemic stem cells to differentiate or acquire lineage markers, depending on the repertoire and direct influence of specific transformation or progression-related gene(s) and not on the degree of lineage commitment of the target cell. This model suggests that the cellular milieu of the stem cell itself plays an important role in the leukemogenic process. It is likely that solid tumors such as breast cancer may also follow this paradigm.
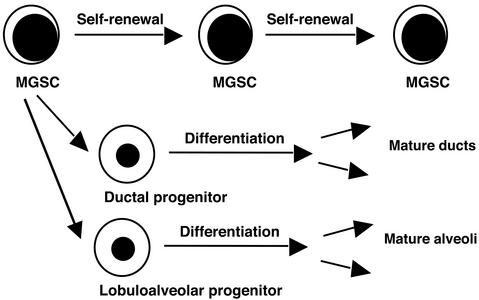
Significant progress has already been made in the identification of the stem cell responsible for the generation of the entire mammary gland of the mouse (Fig. 1, ref. 10), though relatively little work has been carried out in humans. Clonal tracking of retroviral integration sites has shown that the entire mouse mammary gland can be regenerated from a single cell after implantation into recipient mice (11). Other studies have suggested that there are also separate progenitor cells restricted to ductal or lobuloalveolar lineages. Taking a page from the hematopoietic literature, mammary gland stem cells (MGSC) capable of generating the entire ductal and lobuloalveolar tissues in the mouse were enriched by using cell sorting strategies that were developed for hematopoietic stem cell (HSC) purification. HSC express the membrane transporter, ABCG2, whereas most non-stem cells do not. ABCG2 enables efflux of the DNA dye Hoechst 33342 providing a means to isolate stem cells (termed SP cells) without the need for cell surface marker analysis (12, 13). In breast, SP cells comprise 0.2–2% of the epithelial tissue. On injection into the mammary fat pad, SP cells generated both ductal and lobuloalveolar tissues. Breast SP cells were enriched for expression of the HSC marker Sca-1. Injection of only 1000 sorted Sca-1+ cells could initiate mammary gland regeneration, whereas 10,000 Sca-1− did not contain any MGSC (12). These studies give hope that the development of an assay for the human MGSC will be readily accessible by combining cell sorting with the NOD/SCID xenotransplant assay after the paradigm established by Al-Hajj et al. Availability of additional cell surface markers and functional assays for human MGSC will allow precise definition and comparison of the hierarchical organization of normal and malignant breast tissue. If experience from the hematopoietic field is a guide, it will become possible to gain insight into the nature of the normal mammary cell type that is the target for the initiating tumorigenic events. Moreover, it should also be possible to model the earliest steps in the subversion of the normal developmental program of the human MGSC into a tumorigenic one (14).
Figure 1.
Schematic representation of the organization of the mammary gland. This model proposes that only mammary gland stem cells possess high self-renewal capacity and the developmental capacity to produce lineage-committed progenitors. The two major lineage-restricted progenitors have lost self-renewal capacity but still possess extensive proliferation and differentiation capacity. MGSC and progenitors possess the capacity for mammary gland regeneration after in vivo transplantation.
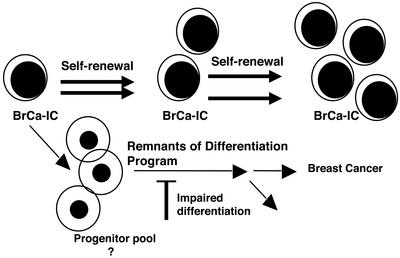
The identification of a breast cancer stem cell represents a major step forward in the elucidation of the breast cancer tumor hierarchy and signals the beginning of a new era of breast cancer research. The BrCa-IC (Fig. 2) hold the key to understanding the origin and maintenance of breast cancer. With this knowledge, it should be possible to design therapies targeted to the unique properties of the tumor stem cell to enable selective killing. New therapeutics will need to take into account the existence of BrCa-IC and ensure that they are targeted for eradication. Indeed, all of the rich information that has been gained on tumorigenic cellular pathways in breast cancer needs to be reevaluated in light of the functional heterogeneity that exists in the tumor clone. For example, the biological consequence of a particular signaling pathway might be different in the rare BrCa-IC compared with the more numerous non-BrCa-IC cells. Perhaps the most important outcome of this study extends beyond the breast cancer field to cancer research in general. Although the cancer stem cell hypothesis is well established, much modern cancer research still treats tumors as homogeneous collections of cells that can be simply disrupted for biochemistry studies or for gene expression profiling. The focus of future studies in cell signaling, molecular and cellular comparisons of normal and tumorigenic pathways, gene expression profiling, and drug development must include the cancer stem cell. This will require the same approaches used here for breast cancer: the use of primary tissue and not cell lines, functional transplantation assays for the cancer stem cell, and cell purification using cell surface or metabolic properties to isolate enriched populations.
Figure 2.
A model depicting the hierarchical organization of the breast cancer tumor. A rare and primitive breast cancer-initiating cell (BrCa-IC) maintains the breast cancer tumor. BrCa-IC possess high self-renewal capacity; however, they can also mature or differentiate into breast cancer cells that have lost the ability to sustain the tumor. Some of these cells may still possess extensive proliferative capacity indicative of a progenitor cell, but have lost the ability to sustain the tumor after transplantation. The breast cancer cells that comprise the bulk of the tumor tissue retain remnants of normal differentiation genetic programs.
Footnotes
See companion article on page 3983.
References
- 1.Southam C, Brunschwig A. Cancer. 1960;14:971–978. [Google Scholar]
- 2.Reya T, Morrison S J, Clarke M F, Weissman I L. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Weissman I L. Science. 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha M S, Benito-Hernandez A, Morrison S J, Clarke M F. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J C, Dorrell C, Ito C Y, Inamitsu T, Guenechea G, Gan O I, Dick J E. In: Hematopoiesis: A Developmental Approach. Zon L I, editor. New York: Oxford Univ. Press; 2001. pp. 99–118. [Google Scholar]
- 7.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri M, Dick J E. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia M, Wang J C Y, Kapp U, Bonnet D, Dick J E. Proc Natl Acad Sci USA. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet D, Dick J E. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 10.Smith G H, Chepko G. Microsc Res Tech. 2001;52:190–203. doi: 10.1002/1097-0029(20010115)52:2<190::AID-JEMT1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 11.Kordon E C, Smith G H. Development (Cambridge, UK) 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 12.Welm B E, Tepera S B, Venezia T, Graubert T A, Rosen J M, Goodell M A. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 13.Alvi A, Clayton H, Joshi C, Enver T, Ashworth A, Vivanco M, Dale T, Smalley M. Breast Cancer Res. 2003;5:10.1186. doi: 10.1186/bcr547. /bcr/547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira D S, Dorrell C, Ito C Y, Gan O I, Murdoch B, Rao V N, Zou J P, Reddy E S P, Dick J E. Proc Natl Acad Sci USA. 1998;95:8239–8244. doi: 10.1073/pnas.95.14.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]