A Genome-Wide Survey of R Gene Polymorphisms in Arabidopsis (original) (raw)
Abstract
We used polymorphism analysis to study the evolutionary dynamics of 27 disease resistance (R) genes by resequencing the leucine-rich repeat (LRR) region in 96 Arabidopsis thaliana accessions. We compared single nucleotide polymorphisms (SNPs) in these R genes to an empirical distribution of SNP in the same sample based on 876 fragments selected to sample the entire genome. LRR regions are highly polymorphic for protein variants but not for synonymous changes, suggesting that they generate many alleles maintained for short time periods. Recombination is also relatively common and important for generating protein variants. Although none of the genes is nearly as polymorphic as RPP13, a locus previously shown to have strong signatures of balancing selection, seven genes show weaker indications of balancing selection. Five R genes are relatively invariant, indicating young alleles, but all contain segregating protein variants. Polymorphism analysis in neighboring fragments yielded inconclusive evidence for recent selective sweeps at these loci. In addition, few alleles are candidates for rapid increases in frequency expected under directional selection. Haplotype sharing analysis revealed significant underrepresentation of R gene alleles with extended haplotypes compared with 1102 random genomic fragments. Lack of convincing evidence for directional selection or selective sweeps argues against an arms race driving R gene evolution. Instead, the data support transient or frequency-dependent selection maintaining protein variants at a locus for variable time periods.
INTRODUCTION
Plant disease resistance (R) genes are abundant in every plant species (Michelmore and Meyers, 1998). Annotation of the Arabidopsis thaliana genomic sequence (Arabidopsis Genome Initiative, 2000) recognizes 207 genes with coding domains characteristic of plant resistance proteins, of which 149 belong to the largest class of nucleotide binding site plus leucine-rich repeat (NBS-LRR) genes (Meyers et al., 2003). NBS-LRR R genes can be further subdivided based on their N-terminal structural features into TIR-NBS-LRR (TNL), which have homology to the Drosophila Toll and mammalian interleukin-1 receptors and CC-NBS-LRR (CNL), which contain a putative coiled-coil motif (Richly et al., 2002).
R genes are characterized by a gene-for-gene interaction (Flor, 1956) in which a specific allele of a disease resistance gene recognizes an avirulence protein encoded by the pathogen, leading to a hypersensitive response. This specificity is encoded, at least in part, in a relatively fast-evolving LRR region, which consists of a varying number of LRR modules. Inappropriate activation of at least some NBS-LRR proteins are negatively regulated by trans partners, as has been shown for RPM1 and RPS2 (Mackey et al., 2003).
In the Columbia-0 (Col-0) ecotype, the majority of the 149 NBS-LRR genes are part of clusters of two to nine loci, the remaining 40 are single-copy loci. Clusters and single-copy loci are organized in superclusters (Arabidopsis Genome Initiative, 2000; Richly et al., 2002). Phylogenetic analysis shows that clusters are the result of both old segmental duplications and recent gene rearrangements (Michelmore and Meyers, 1998; Richly et al., 2002).
Only a small number of the NBS-LRR class R genes have been functionally characterized, and of the subset subjected to molecular population genetic analysis, balancing selection appears to be the predominant force acting to maintain both resistance and susceptibility alleles (RPP1, Botella et al., 1998; RPS2, Caicedo et al., 1999; RPP5, Noël et al., 1999; RPM1, Stahl et al., 1999; RPS5, Tian et al., 2002). These loci share distinctive signatures of molecular variation between resistance and susceptibility alleles that include highly diverged alleles at intermediate frequencies, a high level of silent polymorphism, and/or a relative excess of amino acid substitutions. Several authors have attributed the observed polymorphism patterns to frequency dependent selection, where a cost of resistance or allele-specific pathovar recognition couples R allele frequency with pathogen density (Ellis et al., 1999; Stahl et al., 1999; Tian et al., 2003).
The coevolutionary arms-race dynamic in a gene-for-gene interaction (Flor, 1956) is expected to drive a high rate of turnover for R gene alleles and thus generate loci with few, relatively young alleles with extended haplotypes. Unexpectedly, there is only one record of an Arabidopsis R gene that exhibits characteristics of a recent selective sweep (RPS4; Bergelson et al., 2001). However, whether balancing selection is more prevalent than directional selection in the evolution of R genes is debatable because the R genes investigated to date were initially identified in mapping studies of loci segregating for resistance and susceptibility alleles; R gene loci harboring little genetic variation would thus not be identified in such a screen.
Tests to detect selection have traditionally used predictions from the theory of neutrally evolving sites as a null hypothesis. Departures from equilibrium expectations can indicate the presence of natural selection acting either at one or more of the sites under investigation or at a tightly linked site (Kreitman, 2000). However, recent surveys of genome-wide polymorphism in Arabidopsis show that the data do not fit standard neutral models in several ways (Nordborg et al., 2005; Schmid et al., 2005). For example, there is a systematic shift across the genome toward lower than expected allele frequencies, perhaps an indication of recent range expansion (Innan et al., 1997). But this skew is much greater for nonsynonymous than for synonymous polymorphisms, suggesting at least some role of selection (Nordborg et al., 2005). Similarly, the distribution of polymorphism levels is broader than expected, with relatively too many loci with unusually low or unusually high levels of polymorphism. These observations suggest possible variation in mutation rates across loci, or alternatively directional and balancing selection, respectively. The inability to model these observations (i.e., generate a plausible null hypothesis) argues for substituting an empirical distribution of genome-wide polymorphism for a theoretical distribution as baseline against which to identify unusual features of R gene polymorphism, as has been suggested by Kreitman (2000) and Nordborg et al. (2005).
This empirical modeling approach was recently used by Akey et al. (2004) to identify eight disease genes under selection in humans, where polymorphism patterns obtained for 132 genes were used to distinguish between the effects of demographic history and selection. However, an additional 14 genes that were previously found to be significant under a standard neutral model did not fall into the tails of the empirical distribution, indicating that studies that have used departures from equilibrium models to infer selection are probably less robust than previously assumed. In another study, Akey et al. (2002) used an empirical approach without relying on any models, where Fst values of individual single nucleotide polymorphisms (SNPs) were contrasted to the empirical genome-wide distribution of Fst. Although 174 candidate genes were identified to have been targets of selection based on this approach, this study was limited by a relatively small sample size representing only a few populations. Therefore, our analysis of 27 NBS-LRR R genes in Arabidopsis relies on empirical distributions obtained from genome-wide polymorphism patterns based on a worldwide sampling of this species (Nordborg et al., 2005).
The objective of this study is to explore the prevalence of directional and balancing selection in a class of genes that are expected to be under continual selection for alleles that allow the plant to defend against pathogen attack. To achieve this goal, we investigated polymorphism in 27 of the 149 known members of the NBS-LRR R gene class, including 19 single-copy and eight multiple-copy loci. We minimized sampling artifacts by resequencing the same set of 96 accessions that were used to establish the empirical distribution of polymorphism in 876 randomly distributed genomic regions, which will be referred to as the empirical distribution (Nordborg et al., 2005). Polymorphism patterns exhibited by these R genes can be viewed as many independent snapshots of possible states of the evolutionary process and are expected to reveal features of the breadth and distribution of evolutionary outcomes for this group of loci.
RESULTS
Overview of Polymorphism Patterns in R Genes
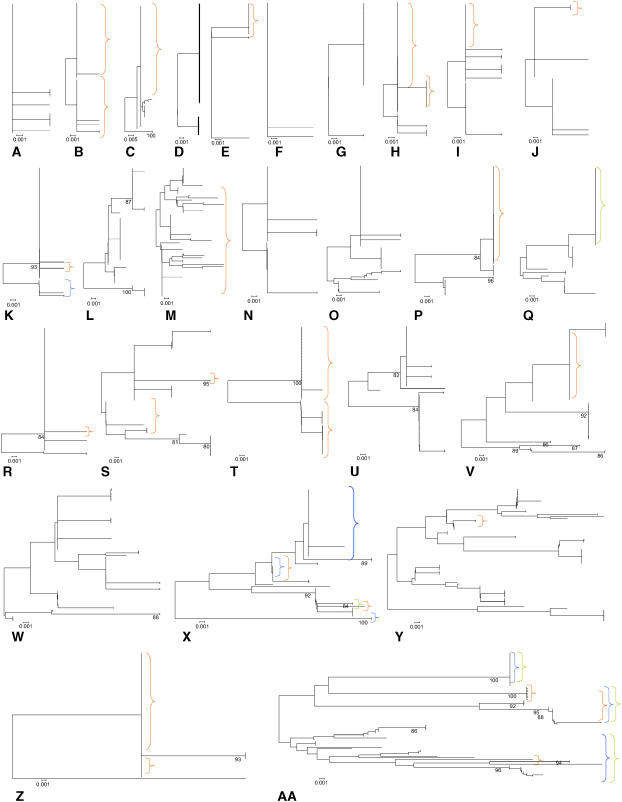
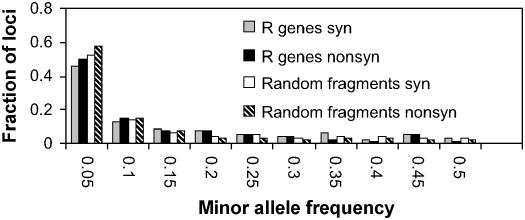
An overview of R gene evolution was visualized by plotting neighbor-joining trees based on Jukes-Cantor corrected silent sites (Nei and Gojobori, 1986) for all 27 R genes using MEGA version 2.1 (Kumar et al., 2001; Figure 1). This figure reveals substantial variation in tree shape and depth, indicative of a wide range of evolutionary histories. The majority of loci possess many low-frequency variants, whereas only a small number of them contain multiple alleles at intermediate frequencies (Figure 2). Nevertheless, comparison with the allele frequency distribution of the set of 876 random genomic fragments shows that the R genes tend to have a higher minor allele frequency for both nonsynonymous and synonymous SNPs (Mann-Whitney U-test, P < 0.00001 and P < 0.005, respectively), which is a possible indication of positive selection acting on these R genes.
Figure 1.
Neighbor-Joining Trees Based on Jukes-Cantor Corrected Silent Sites (Nei and Gojobori, 1986) with 100 Bootstraps, Sorted Based on the Level of Allelic Divergence.
At1g12290 (A), At1g17600 (B), At1g27170 (C), At4g33300 (D), At1g63730 (E), At1g64070 (F), At5g04720 (G), At1g53350 (H), At1g63740 (I), At1g65850 (J), At5g11250 (K), At5g17680 (L), At1g59620 (M), At1g33560 (N), At1g56540 (O), At4g26090 (P), At5g44870 (Q), At5g38850 (R), At4g14370 (S), At5g63020 (T), At3g50950 (U), At5g58120 (V), At1g63750 (W), At1g59780 (X), At4g14610 (Y), At2g16870 (Z), and At3g46530 (AA). Bar beneath each tree reflects 0.001 substitutions per site. Clades highlighted in orange contain at least one nonfunctional allele due to mutations resulting in a frameshift or premature stop codon. Clades highlighted in blue and green contain at least one allele with one or more amino acid insertion or deletion, respectively.
Figure 2.
The Allele Frequency Distribution for Synonymous and Nonsynonymous SNPs Observed for the Set of 27 R Genes and the Set of 876 Random Genomic Fragments.
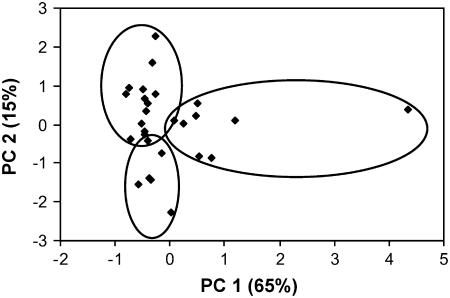
Principal component analysis (PCA) based on eight summary statistics (KSmax, π, S, Tajima's D, Rh, Fst, number of protein variants for the full sequence, and number of protein variants for the xxLxLxx motif) shows that the 27 R genes fall into three overlapping clusters based on breaks in the scatter of two principal components (Figure 3). The first principal component (PC1; 65% of total variation) differentiates the set of R genes based on Ksmax (maximum interallelic synonymous divergence, a measure of the maximum depth of a gene tree), π (nucleotide diversity), S (number of segregating sites), and number of protein variants. R genes with high PC1 values have relatively more highly diverged alleles, an indicator of balancing selection. The second principal component (PC2; 15% of total variation) differentiates the set of R genes based on Tajima's D (test for selection) and Fst (population differentiation). R genes with low PC2 values are characterized by a small number of alleles at intermediate frequency, another indicator of balancing selection (Table 1).
Figure 3.
PCA for 27 R Genes Based on the Correlation Matrix of Eight Summary Statistics.
Summary statistics are KSmax, π, S, Tajima's D, Rh, Fst, number of protein variants for the full sequence, and number of protein variants for the xxLxLxx motif.
Table 1.
Loadings for the Summary Statistics That Constituted the First and Second Principal Components in a PCA of the Set of 27 R Genes
| Summary Statistic | PC1 | PC2 |
|---|---|---|
| KSmax | 0.164 | 0.112 |
| π | 0.186 | −0.005 |
| S | 0.187 | 0.086 |
| Tajima's D | 0.073 | −0.631 |
| Rh | 0.183 | 0.043 |
| Fst | −0.084 | 0.600 |
| Protein variants LRR | 0.141 | 0.086 |
| Protein variants xxLxLxx | 0.175 | 0.243 |
We could divide the R genes into three overlapping clusters. The clusters overlap such that two R genes (At1g63750 and At4g26090) are shared by two clusters (Table 2). R genes located in cluster I tend to have high levels of polymorphism and more diverged alleles. R genes located in cluster II have fewer and less diverged alleles compared with the loci in cluster I, but the alleles tend to be at more intermediate frequencies (i.e., more positive Tajima's D values). R genes located in cluster III tend to have low levels of polymorphism, and their alleles are little diverged (Table 2). This group contains the best candidates for directional selection or selective sweeps. Since outliers can have an effect on PCA results, we also conducted this PCA excluding the major outlier RPP13. Although clustering was less apparent, we could still identify a grouping corresponding with the three previously defined clusters.
Table 2.
Population Genetic Summary Statistics for the Set of 27 R Genes
| R Genea | Cb | Classc | PCAd | KSmaxe | πf | Sg | Dh | Rhi | Fstj | P1k | P2l | Fun.m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At1g12290 | 1 | CNL-B5 | 3 | 0.0106 | 0.0010 | 0.0125 | −1.6058 | 0.0000 | 0.3309 | 6 | 3 | 0.00 |
| At1g17600 | 2 | TNL-D | 3 | 0.0109 | 0.0013 | 0.0124 | −1.3671 | 0.0007 | 0.3624 | 10 | 3 | 14.92 |
| At1g27170* | 3 | TNL-C | 3 | 0.0479 | 0.0023 | 0.0290 | −1.8802 | 0.0018 | 0.3678 | 18 | 5 | 1.513 |
| At1g33560* | S | CNL-A | 3 | 0.0222 | 0.0018 | 0.0151 | −1.0061 | 0.0000 | 0.2384 | 5 | 1 | 0.00 |
| At1g53350* | S | CNL-D | 3 | 0.0204 | 0.0028 | 0.0243 | −1.1962 | 0.0012 | 0.2948 | 15 | 5 | 3.491 |
| At1g56540* | 4 | TNL-H | 1 | 0.0315 | 0.0140 | 0.0582 | 0.7111 | 0.0145 | 0.3149 | 16 | 9 | 0.00 |
| At1g59620 | S | CNL-D | 1 | 0.0417 | 0.0062 | 0.0483 | −1.0861 | 0.0251 | 0.2160 | 33 | 6 | 3.292 |
| At1g59780 | S | CNL-D | 1 | 0.0792 | 0.0280 | 0.1651 | −0.4290 | 0.0125 | 0.1936 | 26 | 10 | 6.331 |
| At1g63730* | 10 | TNL-H | 3 | 0.0303 | 0.0042 | 0.0327 | −1.0167 | 0.0014 | 0.2333 | 8 | 2 | 1.163 |
| At1g63740* | 10 | TNL-H | 3 | 0.0222 | 0.0040 | 0.0242 | −0.4846 | 0.0000 | 0.3248 | 11 | 5 | 1.233 |
| At1g63750 | 10 | TNL-H | 1, 3 | 0.0546 | 0.0056 | 0.0419 | −0.9304 | 0.0164 | 0.2387 | 17 | 4 | 0.00 |
| At1g64070* | S | TNL-H | 3 | 0.0162 | 0.0014 | 0.0252 | −1.9570 | 0.0018 | 0.1806 | 15 | 2 | 0.00 |
| At1g65850 | S | TNL-G3 | 3 | 0.0254 | 0.0021 | 0.0324 | −1.9306 | 0.0000 | 0.2704 | 12 | 4 | 5.191 |
| At2g16870* | S | TNL-H | 3 | 0.1036 | 0.0025 | 0.0437 | −2.1191 | 0.0000 | 0.4215 | 5 | 6 | 2.533 |
| At3g46530 | S | CNL-C2 | 1 | 0.1529 | 0.0968 | 0.3586 | 1.2938 | 0.0931 | 0.1620 | 33 | 24 | 12.31 |
| At3g50950 | S | CNL-C2 | 2 | 0.0472 | 0.0056 | 0.0179 | 1.7060 | 0.0044 | 0.2710 | 6 | 1 | 0.00 |
| At4g14370 | S | TNL-H | 1 | 0.0747 | 0.0125 | 0.0671 | −0.1403 | 0.0128 | 0.1557 | 23 | 4 | 7.321 |
| At4g14610 | S | CNL-B5 | 1 | 0.0802 | 0.0130 | 0.0559 | 0.6065 | 0.0116 | 0.1757 | 29 | 7 | 2.413 |
| At4g26090* | S | CNL-B1 | 2, 3 | 0.0357 | 0.0035 | 0.0227 | −0.6030 | 0.0013 | 0.2300 | 8 | 3 | 1.153 |
| At4g33300 | S | CNL-A | 2 | 0.0378 | 0.0045 | 0.0179 | 0.7928 | 0.0000 | 0.2126 | 5 | 2 | 0.00 |
| At5g04720 | S | CNL-A | 2 | 0.0127 | 0.0018 | 0.0069 | 0.7997 | 0.0000 | 0.2046 | 4 | 2 | 0.00 |
| At5g11250 | S | TNL-G3 | 3 | 0.0277 | 0.0052 | 0.0305 | −0.4149 | 0.0032 | 0.3636 | 17 | 6 | 1.563 |
| At5g17680 | S | TNL-D | 3 | 0.0287 | 0.0048 | 0.0274 | −0.3467 | 0.0000 | 0.4025 | 13 | 4 | 0.00 |
| At5g38850 | S | TNL-H | 3 | 0.0258 | 0.0037 | 0.0347 | −1.3290 | 0.0018 | 0.2540 | 13 | 1 | 1.183 |
| At5g44870* | 36 | TNL-B2 | 2 | 0.0340 | 0.0055 | 0.0264 | 0.1746 | 0.0044 | 0.2234 | 12 | 5 | 0.00 |
| At5g58120 | S | TNL-H | 1 | 0.0705 | 0.0166 | 0.0929 | −0.2703 | 0.0109 | 0.2888 | 12 | 11 | 8.861 |
| At5g63020 | S | CNL-B5 | 2 | 0.0429 | 0.0144 | 0.0390 | 2.7922 | 0.0011 | 0.2190 | 11 | 2 | 17.91 |
| 5%n | 0.0000 | 0.0003 | 0.0052 | −2.0959 | 0.0000 | 0.0922 | 1 | NA | NA | |||
| 95% | 0.1148 | 0.0165 | 0.0824 | 1.3715 | 0.0076 | 0.4593 | 13 | NA | NA |
Pseudogenes and Nonfunctional Alleles
None of the 27 R genes appear to be pseudogenes in the sense that all of the alleles are rendered nonfunctional by frameshifts or premature stop codons. Seventeen of the loci, however, possessed nonfunctional alleles, the occurrence of which ranged from 1.15 to 32.8% due to frameshifts (including indels up to 55 bp in length) or premature stop codons (Table 2, Figure 1). This is most likely an underestimate of the percentage of accessions containing nonfunctional alleles since missense mutations, which are abundant, can also render an allele nonfunctional.
Information about the resistance and susceptibility phenotype of individual alleles is available for only a small number of R genes. However, alleles rendered nonfunctional by frameshifts or premature stop codons can be assumed to produce a susceptible phenotype in the accessions in which they reside. Under this assumption, we can draw inferences about the occurrence of susceptible alleles and their evolutionary history. The set of 17 R genes for which we observed nonfunctional alleles fall into three categories based on the distribution of frameshift mutations across the R gene trees. First, there is a group of seven R genes with either an entire clade composed of accessions with nonfunctional alleles or, alternatively, with one or more clades containing accessions differing by the presence/absence of a frameshift mutation. The second category, represented by two R genes, has many independent nonfunctional alleles scattered throughout a gene tree. The third category, consisting of eight R genes, has a nonfunctional allele represented in one or two accessions (Table 2).
Four R genes (At1g59780, At3g46530, At5g11250, and At5g44870) have in-frame indels (multiples of three bases), ranging between one and four codons, which are mostly restricted to relatively deep clades (Figure 1). These clades in At1g59780 and At3g46530 also contain alleles rendered nonfunctional by frameshifts or premature stop codons. Whereas for At1g59780, At5g11250, and At5g44870 in-frame indels are located outside the xxLxLxx motif, in-frame indels occur inside this motif in At3g46530 and might affect the β-sheet structure, which is assumed the primary target of pathogen recognition. For none of the 27 R gene loci did we observe major structural rearrangements, a feature present in flax L and M locus alleles (Anderson et al., 1997; Ellis et al., 1999).
Population Structure
Genes under strong selection, especially ones involved in defense against pathogens, might be expected to show geographic patterns of genetic subdivision different from genome-wide patterns, which are generally assumed to reflect demographic history. To investigate this possibility, we measured differentiation for the R genes across eight groups of accessions, which were previously identified as distinguishable clusters from the Structure software (Falush et al., 2003; Nordborg et al., 2005) (see Supplemental Table 1 online). These eight groups roughly followed plausible geographic boundaries. Population differentiation (Fst) was calculated using alleles identified based on both nucleotide substitutions and indels. Although no significant average difference was observed between R genes and the genome-wide Fst distributions (Mann-Whitney U-test, P = 0.36), none of the R gene Fst values were in the lower or upper 5% tails of the random fragment distribution. Fst values for all 27 R genes ranged between 0.1620 and 0.4215, whereas Fst values for the empirical distribution ranged between 0 and 0.8654. Despite this apparent difference in the range of Fst values, the variance in Fst for the set of 27 R genes (0.0051) was not significantly different from the empirical distribution (0.0125). These results argue against widespread regional selection for particular disease resistance alleles.
Balancing Selection
Large (positive) values of the summary statistics KSmax (allelic divergence), π (nucleotide diversity), S (number of segregating sites), Tajima's D (test for selection), number of protein variants, and interallelic Ka:Ks ratios are possible indicators of a locus being under the influence of balancing selection. Interallelic Ka:Ks ratios for the solvent-exposed amino acids in the xxLxLxx motif were significantly higher than what would be expected under the neutral model for three R genes (At1g56540, At3g46530, and At5g58120) (P < 0.05, after Bonferroni correction; Table 3). These three genes, plus one other (At1g59780), also revealed significantly high Ka:Ks ratios between alleles for the left and/or right domains flanking the xxLxLxx motif within each LRR module (P < 0.05, after Bonferroni correction; Table 3), indicative of balancing selection. In no case did we observe a significant excess of amino acid replacements in the solvent-exposed residues in the xxLxLxx motif relative to the rest of the LRR region. Three R genes (At1g33560, At3g50950, and At5g38850) possessed no amino acid substitutions in the sequenced xxLxLxx motifs, although all three have experienced amino acid replacements in other parts of the LRR region. The absence of amino acid replacements in the xxLxLxx motif of the entire LRR is a certainty for At3g50950; for the other two R genes, we sequenced only eight out of a total of 11 to 12 LRR modules and therefore cannot rule out the possibility of amino acid replacements in LRR modules for which no sequence was available.
Table 3.
Interallelic Maximum Ka:Ks Ratios for the xxLxLxx Motif and Its Two Flanking Domains within the LRR Modules
| Left Domain | xxLxLxx Motif | Right Domain | |||||||
|---|---|---|---|---|---|---|---|---|---|
| R Gene | Ka | Ks | Ka:Ks | Ka | Ks | Ka:Ks | Ka | Ks | Ka:Ks |
| At1g56540 | 0.054 | 0.019 | 2.80** | 0.159 | 0.028 | 5.66**** | 0.018 | 0.013 | 0.97 |
| At1g59780 | 0.055 | 0.013 | 4.07**** | 0.202 | 0.082 | 2.47 | 0.088 | 0.031 | 2.83 |
| At3g46530 | 0.070 | 0.020 | 3.55**** | 0.205 | 0.029 | 7.04**** | 0.200 | 0.005 | 43.39**** |
| At5g58120 | 0.009 | 0.026 | 0.33 | 0.159 | 0.029 | 5.42**** | 0.046 | 0.014 | 3.26**** |
We also compared the 27 R genes with the empirical distribution for a number of summary statistics. R genes in general have higher values for these summary statistics when compared with the corresponding empirical distributions. Nucleotide diversity (π) ranged between 0.00095 and 0.0968 (average = 0.0098), and the number of segregating sites per base pair (S) ranged between 0.0125 and 0.3586 (average = 0.0505; Table 2). Both π and S were significantly higher for the set of R genes than for the empirical distribution (average = 0.0047 and 0.0314, respectively; Mann-Whitney U-test, P < 0.05), showing that R genes are, in fact, slightly more variable than a typical locus. However, the number of R genes located in the upper 5% tail (three for both π and S) of the empirical distribution was not significantly different from the 1.35 expected by chance (P = 0.31). Nevertheless, π and S values for RPP13 (At3g46530) and S value for At1g59780 were higher than the 99th percentile of the empirical distribution and thus stand out with regard to both high levels of nucleotide diversity and numbers of segregating sites.
Despite having greater segregating variation than genome-wide tendencies, R genes do not have unusually old (or deep) genealogies. KSmax, a measure of the maximum number of synonymous substitutions between any pair of accessions at a locus, provides a gauge of the time to the common ancestry of the sample. Large values indicate the presence of long-lived alleles at a locus, whereas small values imply a recent common ancestry of alleles. Compared with the empirical distribution, the most diverged alleles at an R locus do not tend to be unusually old (Mann-Whitney U-test, P = 0.28). Again, one possible exception is RPP13 (At3g46530), where KSmax = 0.153, which is located above the 99th percentile of the empirical distribution. Maximum allele ages (average over fragment length) were calculated based on the KSmax values for the LRR region using a mutation rate of 1.5 × 10−8 (Koch et al., 2000) and were found to fall within a range of 0.35 to 5.1 million years.
The LRR regions of our 27 R genes encode a large number of protein variants ranging from 4 to 33 (average = 14.2; Table 2). As expected for a protein domain characterized by its rapid rate of evolution, as evidenced by generally high Ka:Ks ratios, the number of R gene protein variants was significantly higher than the number of protein variants encoded by the set of random coding fragments (average = 5.4; Mann-Whitney U-test, P < 0.0001). Eleven R genes were located in the upper 5% tail of the random fragment distribution, far exceeding the genome-wide expectation (Table 2).
Tajima's D distributions for R genes and random fragments are both skewed toward negative values, indicating a relative excess of low frequency variants compared with expectations under a stationary neutral model. R genes, however, have more positive Tajima's D values than the random fragments (Mann-Whitney U-test, P = 0.053). In addition, R genes have more positive Tajima's D values for nonsynonymous changes than for synonymous changes, whereas the reverse is true for the random fragments. The surprising finding that nonsynonymous sites have slightly more positive values than synonymous sites may be an indication of positive rather than relaxed selection acting on segregating nonsynonymous changes in the R genes.
Selective Sweep, Directional Selection, and Adaptive Evolution
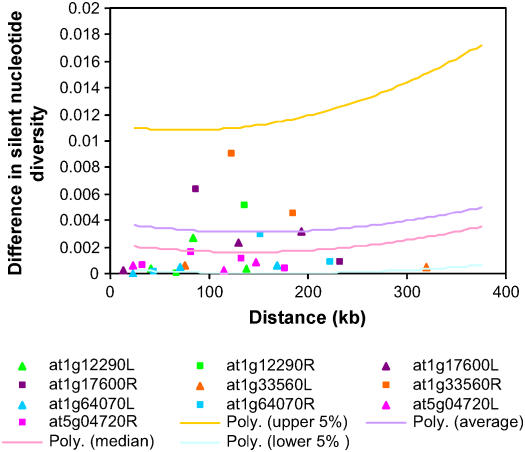
We investigated the possibility of a recent selective sweep in five R genes with the lowest nucleotide diversity among the set of 27 (At1g12290, At1g17600, At1g33560, At1g64070, and At5g04720). None of the five genes are unusually depauperate of polymorphism based on this summary statistic relative to the empirical distribution. Nonetheless, they possess additional characteristics, such as low KSmax, S, Tajima's D, Rh, and/or number of protein variants (Table 2), that make them candidates for recent adaptive substitution. To further consider this possibility, we took advantage of the random fragment database to ask whether polymorphism in loci flanking these genes is similarly low, as might be expected if there was a recent selective sweep in the R locus. Silent nucleotide diversity (π_s_) was calculated for two to four flanking fragments located at distances up to 320 kb on each side of these five R genes, and the differences in π_s_ between R genes and their flanking fragments were plotted against the physical distance separating the loci (Figure 4). A similar analysis was performed for the set of 876 random fragments. In these cases, we analyzed π_s_ differences between each fragment and its two nearest neighbors at distances up to 400 kb. For the set of 876 random fragments, π_s_ differences were generally small, and averages did not increase significantly at increasing physical distance (Figure 4). π_s_ differences between the five R genes and their flanking fragments were concentrated below the median of the random fragment distribution: 19 fragments below median versus eight fragments above (χ2 test of independence, P < 0.05; Figure 4).
Figure 4.
Difference in Silent Nucleotide Diversity between Five R Genes and Their Flanking Fragments Located at Increasing Physical Distances.
Fragments that are located to the left and right side of each R gene are indicated with squares and triangles, respectively. Upper and lower 5% tails and median and average values are indicated by polynomial trend lines calculated for 50-kb sliding windows with a 1-kb increment for silent nucleotide differences between the set of 876 random genomic fragments and their nearest neighbors.
None of the five R genes displayed consistently low π_s_ differences below the lower 5% threshold calculated based on observations for the 876 random fragments. Only At5g04720 showed low π_s_ differences, with all fragments on both sides below the median of the empirical distribution, making this locus a candidate for a recent selective sweep. A similar but less clear pattern was observed for At1g33560 and At1g64070, where π_s_ differences with fragments flanking one side of the R gene are in general below the median of the random fragments distribution, whereas π_s_ differences are relatively high on the other side. For two R genes, At1g12290 and At1g17600, we observed large π_s_ differences with fragments on both sides. Such a pattern might be expected at a locus following a selective sweep if the recombination rate is sufficiently high to overcome the linkage disequilibrium (LD) induced by selection at neighboring loci. At face value, however, the data do not indicate a selective sweep for any of these four R genes.
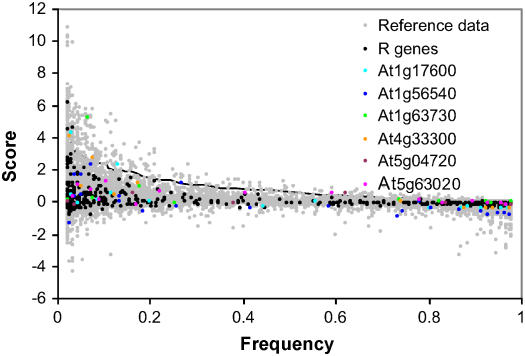
We also looked for evidence of partial selective sweeps of individual alleles using a haplotype sharing approach (Toomajian et al., 2006; see Methods). A haplotype sharing score was assigned to each of 556 R alleles where the minor allele frequency at the corresponding polymorphic site was two or greater (Figure 5). A score was also assigned to each allele that met the same criterion from 1102 random fragments. A sliding window along alleles ordered by frequency was used to produce the 95th percentile of the random fragments distribution. Only six R gene alleles between the frequency of 0.05 and 0.95 are located above this percentile. This number is significantly lower than expected based on the proportion of alleles that come from R genes and the number of alleles above this percentile (χ2 test of independence, P < 0.05). Six R gene alleles are associated with particularly long haplotypes indicative of partial selective sweeps, and each comes from a different gene (At1g17600, At1g56540, At1g63730, At4g33300, At5g04720, and At5g63020). However, only one of these alleles has a haplotype sharing score in the top 2.5% of the distribution (At1g63730); this allele is only found in six accessions, all from the US.
Figure 5.
Haplotype Sharing for 12,403 Alleles in a Set of 1102 Random Fragments and 556 Alleles from the Set of 27 R Genes.
Haplotype sharing for the random fragments and R alleles is depicted by gray and black circles, respectively. The 95th percentile of the distribution is represented by a black line. Alleles from the six R genes that contain an allele in the top 5% of the distribution are color coded.
Finally, for 10 of the R genes we could ask whether there is evidence for the accumulation of adaptive substitutions on the timescale separating A. thaliana with its congener, Arabidopsis lyrata. The Ka:Ks ratio test compares the number of nonsynonymous substitutions (potentially adaptive amino acid replacement changes) with the number of synonymous substitutions (assumed to be evolving neutrally). A conservative test for adaptive evolution is when the Ka:Ks ratio exceeds unity. We obtained corresponding A. lyrata sequence for 10 R genes (marked with an asterisk in Table 2). The maximum Ka:Ks ratios for two of these R genes (At1g27170 and At1g56540) exceeded one for both the solvent-exposed residues in the xxLxLxx motif and the left frame (At1g56540). Monte-Carlo simulations showed, however, that these values were within the confidence intervals for Ka:Ks = 1. The rest had a maximum Ka:Ks ratio significantly lower than one for both the solvent-exposed residues in the xxLxLxx motif and its flanking frames, indicating the action of purifying selection. In summary, we could find no evidence for adaptive protein evolution in the LRR region of any of the 10 genes for which we could carry out this conservative test.
Levels of Recombination
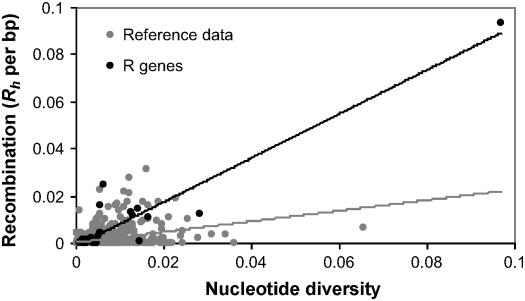
The minimum number of recombination events per base pair (Rh) for the set of 27 R genes is considerably greater (average Rh = 0.0082) than for the set of 876 random fragments (average Rh = 0.0013; Mann-Whitney U-test, P < 0.0001; Table 2). Eight _R_ genes were located in the upper 5% tail of the random fragment distribution (P < 0.0001), far exceeding the 1.35 expected. A greater proportion of _R_ gene loci also have at least one recombination event (70.4% of _R_ genes versus 29.5% of random fragments; Mann-Whitney U-test, P < 0.00001). As expected under standard population genetics theory, _Rh_ and π are correlated (_R2_ = 0.89; Kendall's τ = 0.56), although the strength of the correlation is largely due to _RPP13_ (At5g46530) being an outlier for both _Rh_ and π. With the removal of this outlier, _Rh_ and π were still found to be correlated (_R2_ = 0.45; Kendall's τ = 0.54). Nevertheless, as shown in Figure 6, _R_ genes have higher _Rh_ values for a given π value than the set of 876 random fragments (Mann-Whitney U-test, P < 0.0001). This observation does not change after removal of two outlier loci with π > 0.05 (RPP13 and a locus from the reference data), indicating that the difference is not a simple consequence of a greater detectability of recombination in the more variable R genes.
Figure 6.
Correlation between Minimum Number of Recombination Events per Base Pair and Nucleotide Diversity.
Correlation between minimum number of recombination events per base pair (Rh) and nucleotide diversity (π) is significant for both the set of 27 R genes (black trend line; R2 = 0.89; Kendall's τ = 0.56) and the reference set of 876 random fragments (gray trend line; R2 = 0.1458; Kendall's τ = 0.38).
DISCUSSION
R Genes Have Unusual Polymorphism Patterns
We investigated variation in R genes in relation to a large empirical database of genetic variation in a reference collection of 96 accessions at loci spread across the entire Arabidopsis genome. This comparison was facilitated by the use of the same reference collection to measure R gene variation. Analysis of the reference database showed that the empirical distributions for most summary statistics of polymorphism are broader than those predicted from equilibrium models of neutral variation (Nordborg et al., 2005). With ∼1000 loci in this database, it seems plausible that the tails of the distributions are populated by a certain (unknown) fraction of loci under selection or in close linkage to sites that are under selection. Thus, for example, given the strong skew in the empirical database toward loci with negative Tajima's D values, indicating a relative excess of mutations at low frequency, a locus exhibiting a strongly positive Tajima's D is likely to be the result of selection for intermediate frequency alleles. We would argue on this basis that 5th and 95th percentiles of summary statistic empirical distributions are conservative cutoff values for distinguishing R genes with polymorphism patterns indicative of selection.
In addition to the conservative nature of this criterion for employing the empirical distribution to detect selection, it is also possible for a locus to be under selection but not to be in a tail of the empirical distribution of summary statistics. This can occur when selection is weak, especially if nucleotide changes having an effect on fitness are not known. Barton and Etheridge (2004) used coalescent simulations based on a diffusion approximation to show that the effect of balancing selection is discernibly large only when mutation rates between allelic classes are low and selection is strong. Even when selection is strong, evidence for selection may be hard to obtain, as indicated in Gillespie's (1994) investigation of genetic variation under several models of selection. With respect to R genes, we cannot ignore the possibility that selection for and on resistance alleles is pervasive but that the complexity of the selection only rarely produces patterns of polymorphism that are discernibly similar to patterns expected under simple models of directional selection.
Our strategy was successful in revealing a wide range of evolutionary states among R genes. The diversity in patterns of polymorphism is perhaps best illustrated by the sample genealogies (Figure 1), which range from loci with very little polymorphism and/or only rare alleles to ones with highly diverged alleles at intermediate frequency. Taken as a set, however, the R genes have higher nucleotide diversity, greater numbers of both synonymous and nonsynonymous segregating sites, and more recombination than the background genome. These attributes are consistent with non-neutral evolution of the LRR region.
Nordborg et al. (2005) showed that the empirical allele frequency distribution is skewed toward rare alleles compared with equilibrium neutral expectations, possibly indicating recent population growth (Innan et al., 1997). However, the skew is much greater for nonsynonymous mutations than for synonymous mutations, suggesting negative selective forces acting on amino acid replacement changes. In comparison, the R genes are less skewed toward having rare alleles, both for synonymous and nonsynonymous SNPs, and the average allele frequency tends to be higher than the background genome. In addition, nonsynonymous changes are slightly less skewed toward rare alleles than synonymous changes. Both observations suggest some positive selection acting on R genes.
Our results show that summary statistics for half of the R genes are located in the 5% tails of the empirical distribution. In total, we identified 14 R genes (At1g27170, At1g53350, At1g56540, At1g59620, At1g59780, At1g63750, At1g64070, At3g46530, At3g50950, At4g14370, At4g14610, At5g11250, At5g58120, and At5g63020) where at least one summary statistic was found in the upper 5% tail of the random fragment distribution. We also identified one R gene (At2g16870) with a summary statistic (Tajima's D) in the lower 5% tail of the random fragment distribution. None of the 27 R genes were significant for all seven summary statistics investigated. At3g46530 (RPP13) stood out with five significant summary statistics (KSmax, π, S, Rh, and number of protein variants). This is visible in the PCA (Figure 3), where this R gene is on the extreme right side of the graph. This graph also shows how the other R genes represent a gradient of possible evolutionary states based on their summary statistic values. The same conclusion can be drawn from the comparison of neighbor-joining trees (Figure 1), which display a wide range of tree shapes and depths.
The 14 R genes with at least one summary statistic value in the upper 5% tail of the random fragment distribution are in general highly polymorphic and mostly located in cluster I of the PCA. Exceptions are At1g27170, At1g53350, At3g50950, At5g11250, and At5g63020. Genes in this cluster are characterized by large summary statistic values with high PC1 loadings (KSmax, π, S, Rh, and number of protein variants), which as previously described, are indicators of balancing or frequency-dependent selection. The PCA graph shows that At3g50950 and At5g63020 R genes are grouped in cluster II, which is characterized by positive Tajima's D values and negative loadings of PC2. This group also contains At5g04720, At4g33300, At5g44870, and At4g26090 (RPS2), for which none of the summary statistics were significant. RPS2 was previously identified as being under balancing selection based on analyses of nucleotide polymorphism, but this conclusion was aided by experimental knowledge of the resistance and susceptibility phenotype of the alleles (Caicedo et al., 1999; Mauricio et al., 2003). Cluster III of the PCA graph (Figure 3) consists of genes with low levels of polymorphism and/or low frequency variants. This cluster contains four of the five R genes that were investigated further for possible recent selective sweeps.
Evidence for Balancing Selection
Six summary statistics (KSmax, π, S, Tajima's D, number of protein variants, and interallelic Ka:Ks ratios) are potential indicators of balancing selection. Loci harboring balanced polymorphisms tend to differ from neutrally evolving loci in having alleles at more intermediate frequency (more positive Tajima's D), in having a greater number of protein variants segregating, and in having older, more diverged alleles (larger KSmax, π, and S). An additional (and unusual) feature of selection relevant to R genes is adaptive divergence between alleles (large interallelic Ka:Ks ratio). The theory of balancing selection does not require that these summary statistics be large. Indeed, the magnitude of some of the summary statistic values can initially be small under balancing selection and will increase with the age of selected alleles. In addition, many of the statistics are expected to be correlated. On the other hand, there is an obvious multiple testing issue when so many summary statistics and comparisons are being made, and this is an important caveat to the interpretation of the evidence of selection.
We can identify seven R genes (At1g56540, At1g59780, At3g50950, At4g14370, At4g14610, At5g58120, and At5g63020) as candidates for weak or transient balancing selection and can confirm an eighth locus, RPP13 (At3g46530), as a long-lived balanced polymorphism. These genes are located in clusters I and II of the PCA and have one or more summary statistic values located in an upper 5% tail of the empirical distributions. RPP13 (At3g46530) is exceptional in having highly significant summary statistic values (>99th percentile of reference distribution) for five summary statistics (KSmax, π, S, number of protein variants, and interallelic Ka:Ks ratios). We acknowledge that not all of the R genes with a small number of significant summary statistics may have been subject to this form of selection. At the same time, these seven R genes as a group harbor an impressively large average of 17 protein variants in the LRR region and have a positive average Tajima's D = 0.7 (the random fragments average Tajima's D = −0.8). Unless this region is evolving with little or no functional constraint, some form of selection is likely to be acting to draw amino acid replacement mutations into the species and to retain them for some period of time.
We observed interallelic Ka:Ks ratios that were significantly higher than 1.0 (P < 0.005) for four _R_ genes from cluster I (At1g56540, At1g59780, At3g46530, and At5g58120). While interspecies _Ka_:_Ks_ ratios >1.0 imply directional selection, high interallelic Ka:Ks ratios reflect selection that maintains a set of differentiated functional alleles. High interallelic Ka:Ks ratios have been observed for the human and mouse class I major histocompatibility complex loci, including but not limited to the antigen recognition (Hughes and Nei, 1988). In this case, the pattern is presumed to result from overdominant selection. For Arabidopsis, however, with its high degree of selfing, overdominant selection is unlikely. Frequency-dependent selection is a more plausible scenario for maintaining variation in R genes, where alleles are maintained within populations due to either the different and active specificities of the alleles (e.g., L locus of flax; Ellis et al., 1999) or due to a cost of resistance in the absence of the pathogen (Stahl et al., 1999; Tian et al., 2003). These four R genes also have significant values of KSmax, π, S, Rh, or number of protein variants, confirming that these summary statistics are indicators of balancing selection.
We observed a substantial percentage of R genes with values above the upper 5% threshold of the empirical distribution for the summary statistics, Rh, and number of protein variants. Resistance alleles might tend to persist longer in a population than neutral alleles, allowing more time for them to recombine, as has been shown for RPS5 based on the relatively short distance decay of LD (Tian et al., 2002). In this case, the overall level of polymorphism in the short stretch of DNA encompassing the LRR regions would also be expected to increase with the age of the alleles. But relatively high rates of recombination in R genes is not explained by higher levels of polymorphism in this group of genes, as indicated in Figure 6, where it can be seen that at any rate of nucleotide diversity, R genes have higher levels of recombination than random fragments. Therefore, we are left with the intriguing possibility that the high levels of recombination in R gene LRR regions might reflect a selective advantage of recombination for the generation of new R gene specificities, as has been suggested by Hammond-Kosack and Jones (1997).
The maintenance of alternative alleles, especially those encoding proteins that recognize multiple avirulence proteins, has been hypothesized (Dangl and Jones, 2001) to help explain how a surprisingly small number of R genes may mediate recognition of all possible pathogen-encoded ligands. True to this prediction, the genes in our data set segregate for a remarkably large number of protein variants (see column P1 in Table 2). Under a balancing selection hypothesis, one might expect to find alleles encoding distinct recognition specificities being maintained for long periods of time. Yet one of the most striking results from this study is the general lack of highly differentiated R alleles (other than for RPP13). Instead, for most of the R genes, the protein variants are relatively young.
We can gain insight into the extent to which susceptible alleles are maintained by looking at the distribution of alleles rendered nonfunctional due to frameshift or stop codon mutations over the gene tree of the R gene. We observed many such mutations for 17 R genes and could distinguish three different categories. R genes that constituted the first category (Table 2) all had nonfunctional alleles that were concentrated in one or more clades; therefore, we expect that these nonfunctional R gene alleles may be maintained by balancing selection due to a cost of resistance, especially when the clades are highly differentiated (Stahl et al., 1999; Tian et al., 2002, 2003). The second category is comprised of R genes with susceptible alleles scattered across the gene tree. The fact that nonfunctional alleles of At1g59620 were dispersed over the entire star-shaped neighbor-joining tree and that there were a large number of nonsynonymous changes supports previous claims that this R gene is nonfunctional (Meyers et al., 2003). For At1g17600, we also observed nonfunctional alleles scattered over its gene tree. However, this R gene has very low nucleotide diversity and a small number of nonsynonymous changes. Perhaps this gene has experienced a recent selective sweep of a resistance allele, and there is now selection for new susceptibility alleles. The third category of frameshift mutations consisted of eight R genes (Table 2) for which we observed nonfunctional alleles for only one or two accessions; in these cases, the nonfunctional allele is due to a single mutational event. Such a single mutational event was observed for RPS2 (At4g26090); here, the accession Kondara is located in the resistant clade (as determined by a previous study; Mauricio et al., 2003) but contains a frameshift that makes it susceptible (susceptible phenotype was confirmed in greenhouse experiments; Aranzana et al,. 2005). These may be additional examples of selection favoring mutations to susceptibility alleles.
A number of the well-studied R genes in A. thaliana (RPM1, RPS2, RPS5, RPP1, RPP13, and RPP8), all of which were identified from the positional cloning of segregating resistance and susceptibility alleles in the ecotype collection, have indications of being under some form of balancing selection (Botella et al., 1998; McDowell et al., 1998; Caicedo et al., 1999; Stahl et al., 1999; Tian et al., 2002; Rose et al., 2004). Two of them, RPS2 and RPP13, were included in this study. RPS2 was not significant for any of the summary statistics in this study. Since previous studies by Caicedo et al. (1999) and Mauricio et al. (2003) made use of phenotypic information and sequence data for the entire gene, this suggests that if we had this additional information for all 27 R genes, we might be able to identify additional genes to be under some sort of balancing selection, especially the ones located in cluster II of the PCA graph (Figure 3). Nevertheless, it becomes clear from our study that a large subset of R genes are not segregating for old alleles, suggesting that previous studies unknowingly focused only on a biased subset of genes. However, since our approach required sequence from a vast majority of the accessions to include a locus, this excludes possible cases of gene presence/absence polymorphisms, which may be more likely to be under strong balancing selection. This was recently shown to be the case in a study by Shen et al. (2006) of seven previously uncharacterized R gene loci (and two previously characterized R gene loci RPM1 and RPS5) with intermediate frequency presence/absence polymorphisms. Inclusion of R genes that segregate for presence or absence would simply strengthen our main finding that balancing selection (or weak balancing selection) is a more likely outcome than selective sweeps.
Worldwide Selective Sweeps Are Uncommon in R Gene Evolution
We did not observe evidence for a recent selective sweep for any of the 27 R genes based on significantly low values for their nucleotide diversity and/or Tajima's D relative to the empirical distribution. However, there are five R genes (At1g12290, At1g17600, At1g33560, At1g64070, and At5g04720) that might have undergone one or more selective sweeps in the recent past; these loci have relatively young allele ages and a low nucleotide diversity, of the same order of magnitude as the insect defense gene, TGG1, which is believed to have experienced a recent selective sweep (Stranger and Mitchell-Olds, 2005). Although their nucleotide diversity was lower than other apparently neutrally evolving A. thaliana functional genes (e.g., Kuittinen and Aguadé, 2000; Aguadé, 2001; Hauser et al., 2001), nucleotide diversity of these five R genes was not significantly low when compared with the random fragment distribution. For all five R genes, silent nucleotide diversity (π_s_) values fall within the 15th and 49th percentile of the empirical distribution. Furthermore, for none of the 27 R genes did π_s_ = 0, whereas this was the case for 13.4% of the 876 random fragments. Nordborg et al. (2005) have suggested that this skew toward lower polymorphism values may be the result of background selection acting to remove deleterious alleles. In fact, despite having low nucleotide diversity for synonymous changes, all five R genes code for a relatively large number of protein variants, ranging between 4 and 15.
We also investigated whether any of these genes stood out in terms of extended LD when compared with the 876 random fragments. One signature of a recent selective sweep should be that LD between the site under selection and neighboring fragments extends beyond the 25 to 50 kb over which LD generally decays (Maynard-Smith and Haigh, 1974; Aguadé et al., 1989; Nordborg et al., 2005). A proxy for extended LD is an extended region of low π_s_ differences between the R gene and its neighboring fragments. π_s_ differences between the five R genes and fragments up to 320 kb away were in general low (below median of the empirical distribution) but were not located in the lower 5% tail of the empirical distribution. Thus, although this analysis does not provide strong evidence for selective sweeps, it does not rule them out. It is also possible that the physical distances between the R genes and the nearest neighbor in this analysis are too far to reveal the effects of genetic hitchhiking.
We also used a haplotype sharing approach (Toomajian et al., 2006) to investigate the possibility of partial selective sweeps in 556 R gene alleles. This analysis is designed to detect an allele that has rapidly changed its allele frequency. We first analyzed haplotype length of the R gene alleles as well as alleles from a set of 1102 random genomic fragments and used this distribution to obtain the 95th percentile for different allele frequency bins. R genes with haplotypes longer than the 95th percentile are candidates for a regional selective sweep. Comparison between haplotype length in the set of 27 R genes and the set of 1102 random fragments shows that significantly fewer of the R gene alleles were found in the upper 5% of the distribution than expected by chance, and none were found in the upper 1% of the distribution. Either R genes have stable allele frequencies because pathogen isolates are relatively ubiquitous in Arabidopsis populations, or resistance alleles have been present in the species long enough to have diversified onto different genetic backgrounds.
This observation may in part be the result of smaller physical distances between R genes and their flanking fragments compared with the distances between random fragments alone. However, it may also indicate that adaptive substitution of novel R gene alleles is not a common scenario. This is an important finding, as it provides support for the trench warfare model of R gene evolution, where frequency-dependent selection rather than adaptive substitution dominates the selective landscape. The analysis also revealed several possible instances of a partial selective sweep, three of which have additional interesting features (modulo the caveat raised in the preceding paragraph). A group of >60% of accessions, which contains all North American accessions, shared an allele of At5g04720 that is associated with a long haplotype (P < 0.05). With the exception of two accessions that share a single synonymous mutation, alleles of this locus differ by only nonsynonymous substitutions. This R gene, therefore, is a candidate for having undergone a quick turnover of alleles such as expected under an arms race model of selective sweeps. Arms race dynamics have also been suggested for RPS4. Since this R gene shares characteristics with At5g04720 (RPS4 has only a single amino acid polymorphism and no synonymous differences in its LRR region; Bergelson et al., 2001), current evidence still supports this conclusion. Two additional R genes, At1g56540 and At5g63020, are in the top 5% of the haplotype sharing distribution for intermediate frequency alleles (30 and 59% of accessions, respectively). Summary statistics for both loci show indications for balancing selection (Table 2). The fact that we observe evidence for a partial selective sweep and for balancing selection confirms earlier observations for RPM1, where there is evidence for historical allele frequency fluctuations (Stahl et al., 1999). It is possible that these two R genes have experienced a recent rise in the frequency of a resistance allele resulting in an extended haplotype sharing surrounding the site(s) under selection.
No Difference between Single-Copy and Multiple-Copy R Genes
We did not observe different patterns of variation in the multiple-copy R genes compared with the single-copy R genes (Mann-Whitney U-test, P = 0.26, 0.36, 0.38, 0.33, 0.94, 0.07, and 0.81 for KSmax, π, S, Tajima's D, Rh, Fst, and number of protein variants, respectively) perhaps because we also found no evidence for exchange between clustered loci. Indeed, it is likely that the group of eight multiple-copy R genes included in this study is biased toward cluster members that are highly differentiated and do not interchange genetic material with other cluster members. Future research needs to focus on less differentiated clusters of R genes to draw conclusions about multiple-copy R gene evolution by exchange of genetic material between cluster members.
R Gene Specificity Resides Not Only in the xxLxLxx Motif
LRR domains are found in diverse proteins and are involved in protein–protein interaction, peptide-ligand binding, and protein–carbohydrate interaction (Jones and Jones, 1997; Kajava, 1998). Pathogen recognition is assumed to mainly take place at the R gene LRR region (Staskawicz et al., 1995). However, to date there has only been one observed physical interaction between a pathogen avirulence protein and the LRR region of a resistance protein (i.e., Pi-ta resistance protein from rice [Oryza sativa]; Jia et al., 2000). The functional importance of LRRs as the domains of specificity is further supported by results from domain swaps and mutational analyses of R genes (Ellis et al., 1999; Hwang et al., 2000; Luck et al., 2000). More specifically, R gene specificity appears to reside in the solvent-exposed residues in the xxLxLxx motif that is located within the LRR region. This has been shown based on a significant clustering of nonsynonymous substitutions in this motif of the Xa21 gene in rice (Wang et al., 1998) and significant interallelic Ka:Ks ratios observed for this motif for the A. thaliana resistance genes RPP1 (Botella et al., 1998), RPP5 (Noël et al., 1999), RPP8 (McDowell et al., 1998), and RPP13 (Rose et al., 2004), the flax (Linum usitatissimum) L gene (Ellis et al., 1999), rice Xa21 gene (Wang et al., 1998), and the RGC2 gene in lettuce (Lactuca sativa) (Meyers et al., 1998). In addition, there is functional evidence of nonsynonymous replacements in the xxLxLxx motif that are sufficient to disrupt RPS5 function (Warren et al., 1998). Although most studies have demonstrated that R gene specificity resides largely in the LRR region, there are indications at least for the L class of flax rust resistance genes, that the TIR region may also be involved in determining specificity (Ellis et al., 1999; Luck et al., 2000). Furthermore, the N-terminal domain of the tomato (Solanum lycopersicum) Mi protein could be involved in negatively regulating its own LRR to prevent mistimed activation of disease resistance and death (Hwang et al., 2000). We therefore acknowledge that our survey of polymorphism patterns in the LRR region of 27 R genes probably does not capture all variation involved in R gene specificity.
Five independent observations in our data are consistent with some determination of R gene specificity residing outside the xxLxLxx motif. First, we found significant interallelic Ka:Ks ratios for the solvent-exposed residues in the xxLxLxx motif for only three of the 27 R genes. Second, we observed significant interallelic Ka:Ks ratios in the flanking domains of the xxLxLxx motif for four R genes (At1g56540, At1g59780, At3g46530, and At5g58120). For one of these R genes (At3g46530 [_RPP13_]), Ka values for the xxLxLxx motif equaled those for the domain to the right of the xxLxLxx motif. Third, the number of nonsynonymous substitutions in the xxLxLxx motif was not significantly greater than in the flanking domains for any of the 27 R genes. Fourth, for three R genes (At1g33560, At3g50950, and At5g38850), we observed only one protein variant for the xxLxLxx motif but more protein variants for the entire LRR region. Fifth, for four R genes (At1g59780, At3g46530, At5g11250, and At5g44870), we observed amino acid indels outside the xxLxLxx motif. All these observations suggest that selection may well be acting on sites outside the xxLxLxx motif.
The function of amino acids outside the xxLxLxx motif is not well understood. Replacements of some of these amino acids could lead to a disruption of the general structure of a ligand binding surface as has been speculated for Pro residues, which cause kinks in the peptide backbone and may function in positioning the conserved core motifs (Jones and Jones, 1997). This is supported by Mondragón-Palomino et al. (2002)'s genome-wide analysis of positive selection in members of the NBS-LRR gene family of A. thaliana. This study revealed positive selection not only in the LRR region, particularly in the xxLxLxx motif, but also outside the LRR region. One such functionally important region is the TIR region, as has been observed by Ellis et al. (1999) for the flax L gene. It would be worthwhile in the future to investigate selection pressures on different LRR modules. For example, since LRR regions are large relative to predicted avirulence gene products, different binding specificities could be generated by interactions with different LRRs within a protein, and a single protein could interact with a variety of ligands. Such dual specificity has been observed for RPM1 (Bisgrove et al., 1994) and the tomato Mi gene (Rossi et al., 1998), where regions that bind ligands will be subject to stronger adaptive selection than regions that play a structural role. If this is general, we would expect to find regions within the LRR region with a significant excess of amino acid substitutions, as has been observed for the Xa21 gene in rice (Wang et al., 1998).
Evolution of ligand binding in the set of R genes included in this study may take place mostly by recombination and gene conversion between alleles that alter the combination and/or orientation of the arrays of solvent-exposed residues in the LRR region. Such a dynamic has been observed by Richter et al. (1995), where intragenic cross-over events at the Rp1 complex locus in maize resulted in four combinations that represented novel alleles that were resistant to at least one rust biotype against which neither parent showed resistance. The presence of large numbers of recombinant alleles is perhaps the strongest pattern we observed in our data set. However, we cannot exclude other factors influencing the evolution of ligand binding, including indels in the LRR domain, hypervariability in the xxLxLxx motif, and changes in the secondary structure of the LRR domain resulting from amino acid substitutions outside of the xxLxLxx motif.
Summary
The wide variety in gene trees and summary statistics observed for a set of 27 NBS-LRR R genes depicts a continuum of possible states in the evolutionary process, ranging from (regional) selective sweeps to long-lived balancing selection with many stages in between, including loss of function and maintenance of nonfunctional alleles. Several loci could be identified as candidates for recent selective sweeps, but this scenario is not common, and the evidence is not unequivocal. If anything, the set of R genes show an underrepresentation of signals for regional and worldwide selective sweeps when compared with genome-wide patterns of polymorphism. Therefore, the data do not support the coevolutionary arms-race hypothesis as a general evolutionary model for this group of genes. We found strong signatures of balancing selection acting to maintain various alleles at intermediate frequencies over prolonged periods of time for only one R gene, RPP13, a locus previously shown to have strong signatures of balancing selection (Rose et al., 2004). Weaker indications of balancing selection were evident in seven genes. Nevertheless, almost all of the genes in our survey segregate for unusually large numbers of amino acid replacement variants that we hypothesize reflects continuous selection pressure for plants to produce new R alleles. However, these alleles appear not to be maintained by selection for long periods of time. The R genes, as a group, are also unusually recombinogenic, which we speculate might be a further adaptation to produce novel resistance alleles.
METHODS
DNA Samples and Sequencing
DNA was extracted from fresh leaves of 96 Arabidopsis thaliana accessions (see Supplemental Table 1 online) and one Arabidopsis lyrata accession. The A. thaliana accessions were chosen to include a worldwide sample, where 50 were sampled from local populations in a hierarchical fashion. The same set of 96 accessions has been extensively characterized for polymorphism in 876 genomic fragments that average 583 bp in length and represent 0.48 Mbp of the genome (Nordborg et al., 2005). R gene sequences for the 96 A. thaliana accessions were obtained by directly sequencing PCR amplification products. PCR was done using a hotstart Taq polymerase (TaKaRa) with the following conditions: 95°C for 2.30 min, 35 cycles of 95°C for 30 s, 56°C for 40 s, and 72°C for 1.30 min, followed by 72°C for 7.00 min. The 876 genomic fragments were selected so that they would equally cover the Arabidopsis genome with an average spacing of 100 kb. Since the 27 R genes were not selected in this way, their distances to neighboring fragments are in general shorter than the distances between each of the 876 fragments and their nearest neighbors. R gene sequences for the (heterozygous) A. lyrata accession involved first cloning one of the two possible alleles using an M13mp18 perfectly blunt cloning kit (Novagen). Orthologous fragments were identified based on synteny of multiple (overlapping) PCR fragments (for At1g27170, At1g53350, and At1g63730) and on the high similarity of the PCR product (for the remaining R gene loci). Since A. thaliana and A. lyrata are closely related (diverged 5.1 to 5.4 million years ago; Koch et al., 2000), they have equivalent linkage group organization and are separated by only a limited number of chromosomal rearrangements (∼5; Kuittinen et al., 2004). It is therefore assumed that highly similar PCR products are orthologous. Sequencing (in both directions) was performed using dye terminator cycle-sequencing chemistry (Quick Start Kit; Beckman Coulter) and a CEQ 8000XL sequencer (Beckman Coulter). Up to five (overlapping) fragments were sequenced per locus to cover the LRR region of each of these 27 R genes.
Primer Design
Primers were designed based on Pfam analysis of the LRR regions. For instances where Pfam predicted less than half the actual number of LRR modules as described by Meyers et al. (2003), we designed extra primers for an additional part of the LRR region. We designed and tested primers for all 40 single-copy NBS-LRR R genes that have been described for the Col-0 genome (Arabidopsis Genome Initiative, 2000; Richly et al., 2002; Meyers et al., 2003) and only those multiple-copy NBS-LRR R genes that are located on chromosome 1 of Col-0. Primers were tested on a subset of eight A. thaliana accessions. In cases where <100% of the set of eight accessions showed the desired amplification product, new primers were designed and tested. This process was repeated multiple times until we obtained a selection of primer pairs that could be used to resequence the LRR regions of 27 R genes (19 single-copy and 8 multiple-copy genes) for most, if not all, of the 96 A. thaliana accessions. Up to five primer pairs were designed to cover the LRR region of each of these 27 R genes (see Supplemental Tables 2 to 4 online). For the remaining single-copy NBS-LRR genes, we encountered problems with amplification (presence/absence polymorphisms) and sequencing (overlapping sequences), even after multiple rounds of primer redesign. Since BLAST analysis of the primers showed that none of the primers had a match elsewhere in the genome, we assume that overlapping sequences are the result of a duplication of the R gene, indicating that some of the single-copy R genes are segregating as copy number polymorphisms of closely related genes. We similarly interpret the occurrence of accessions for which particular LRR regions would not amplify as an indication of presence/absence polymorphism, as has been observed for RPM1, RPS5, and seven other R genes (Stahl et al., 1999; Tian et al., 2002; Shen et al., 2006). For most of the multiple-copy R genes, high identities between gene family members resulted in overlapping sequences.
Structural Features of LRR Regions
We resequenced the LRR region of 12 CNL-type R genes and 15 TNL-type R genes covering most of the subgroups as described by Meyers et al. (2003) (Table 2). We obtained data for 19 single-copy R genes and eight loci that are located in R gene clusters. We were able to resequence the LRR region of all three members of cluster 10 (Meyers et al., 2003) as well as single members of four other clusters on chromosome 1. We additionally resequenced one multiple-copy R gene on chromosome 5 that was initially assigned as being a single-copy R gene due to its copy number in Col-0. Since primers were initially designed based on Pfam analysis of the LRR regions, which later was proven to inadequately predict correct LRR region bounds (Meyers et al., 2003), some of the sequenced LRR regions lack up to three LRR modules at the beginning and/or end. On average, we obtained a sequencing product for 85% of the accessions for each R gene locus.
The recognition specificity of six of the 27 R genes is known or can be predicted based on similarity to known proteins. At1g12290 is a paralog of RPS5, which recognizes Pseudomonas syringae (Simonich and Innes, 1995). At1g33560 is also known as ADR1, which recognizes microbial pathogens (Grant et al., 2003). At3g46530 is also known as RPP13, which recognizes Peronospora parasitica (Bittner-Eddy et al., 2000). At4g26090 is also known as RPS2, which recognizes P. syringae (Kunkel et al., 1993). At4g33300 and at5g04720 are also known as ADR1-L1 and ADR1-L2, respectively; both recognize microbial pathogens (Grant et al., 2003). Sequence lengths ranged between 547 and 1861 bp. Positions of introns were obtained by comparing DNA sequences with amino acid sequence information for corresponding LRR regions (http://niblrrs.ucdavis.edu). Most members of the TNL group and members of the CNL-A subgroup possess an intron in the LRR region (Meyers et al., 2003). In total, we examined introns in the sequenced LRR region of seven R genes (At1g17600, At1g63730, At1g63740, At4g14370, At4g33300, At5g11250, and At5g17680); these introns ranged between 28 and 123 bp. An additional eight R genes possess an intron in their LRR region, but relevant LRR modules were not sequenced because the original prediction methods cut them off.
Data Analysis
We compared the set of 27 R genes to a set of 876 random fragments of ∼500 bp, generated by Nordborg et al. (2005), spanning the Arabidopsis genome, for a number of population genetic summary statistics. The following summary statistics were calculated using C++ programs written by E.G.B. for the set of 27 R genes and the set of 876 random fragments: nucleotide diversity π (Nei and Li, 1979), number of segregating sites standardized by sequence length S, Tajima's D (Tajima, 1989), FST (Nei, 1973), and minimum number of recombination events, Rh (Myers and Griffiths, 2003). A subset of 236 random coding fragments for which at least 400 bp consisted of exon sequence was used for comparisons involving coding regions. Summary statistics involving coding regions included numbers of synonymous (Ks) and nonsynymous (Ka) substitutions, calculated according to Nei and Gojobori (1986), maximum number of synonymous substitutions between accession pairs (KSmax), and the number of protein variants based on nonsynonymous substitutions. Each of the above mentioned summary statistics for the R genes was compared with the distributions of the two sets of random fragments using the Mann-Whitney U-test in R 1.8.1. Grouping of R genes based on combinations of their summary statistics was visualized by PCA based on the correlation matrix of the variables in SAS 8.02.
If amino acid differences have been fueled by natural selection, then the location of amino acid replacements can provide insight into regions of functional importance. We therefore decided to test for significant clustering of amino acid replacements in the solvent-exposed residues in the xxLxLxx motif according to Wang et al. (1998). Each LRR module consists of three domains, where the middle domain is an xxLxLxx motif (L = Leu or other aliphatic amino acid; x = any amino acid) that is predicted to form a solvent-exposed β-sheet. The hypervariable framed residues in this motif are, according to molecular modeling, exposed to solvent and are expected to interact with avirulence ligands (Ellis et al., 2000). Amino acid replacements in the xxLxLxx motif and in the rest of the LRR region were evaluated for deviations from a random distribution of amino acid replacements over all three domains within each LRR module by Fisher's exact test (as implemented in R 1.8.1). Subsequently, all accession pairs were ranked for their interallelic Ka:Ks ratios at the xxLxLxx motif and the two flanking domains. Confidence intervals for the estimates of Ka, Ks, and Ka:Ks ratio were obtained through Monte-Carlo simulation using the program K-estimator (Comeron, 1999). P values were subsequently compared with a Bonferroni-corrected threshold value based on all interallelic comparisons. Estimates and confidence intervals of Ka, Ks, and Ka:Ks for all comparisons with A. lyrata were calculated by the same procedures.
A recent selective sweep can result in a reduced level of variation at neighboring loci as a result of genetic hitchhiking (Maynard-Smith and Haigh, 1974; Aguadé et al., 1989). R genes with low nucleotide diversity were investigated for indications of a recent selective sweep using two approaches. The first approach investigated differences in silent nucleotide diversity between an R gene and its nearest neighbors contained in the empirical database (http://walnut.usc.edu/2010/) for distances up to 320 kb from the target locus. An empirical distribution of differences was produced in an identical manner for the 876 random fragments for distances up to 400 kb from each target.
The second approach to identify selective sweep candidates involved a genome-wide scan of haplotype sharing according to Toomajian et al. (2006). Data from 1102 fragments that come from an ongoing survey of genomic DNA sequence polymorphism (i.e., the 876 random fragments together with an additional 226 random fragments that were collected in the same manner; Nordborg et al., 2005) were used in this analysis. The density of these sequenced regions along each chromosome allows the identification of unusually long identical haplotypes among subsets of these individuals. The rationale behind this approach is that chromosomes that are identical by descent (IBD) at a polymorphic site must also share a short region surrounding that site. The length of this IBD region is influenced primarily by the age of the shared allele at the polymorphic site and the recombination rate in the region (Nordborg and Tavaré, 2002; Innan and Nordborg, 2003). Subsequently, haplotype sharing around each R gene was compared with what is observed in the rest of the genome. It should be noted that this analysis will miss instances where a sweep has gone to complete or near fixation; it can only detect a recent partial sweep where the swept allele is still at intermediate or low frequency. See Toomajian et al. (2006) for a more detailed description of the haplotype sharing statistic.
Accession Numbers
Sequence data for the A. thaliana NBS-LRR R genes described in this article can be found in the GenBank nucleotide sequence database under the following accession numbers: At1g12290, DQ526453 to DQ526531; At1g17600, DQ526532 to DQ526601; At1g27170, DQ526602 to DQ526670; At1g33560, DQ526671 to DQ526754; At1g53350, DQ526755 to DQ526845; At1g56540, DQ526846 to DQ526927; At1g59620, DQ526928 to DQ526993; At1g59780, DQ526994 to DQ527077; At1g63730, DQ527078 to DQ527168; At1g63740, DQ527169 to DQ527254; At1g63750, DQ527255 to DQ527345; At1g64070, DQ527346 to DQ527423; At1g65850, DQ527424 to DQ527503; At2g16870, DQ527504 to DQ527587; At3g46530, DQ527588 to DQ527674; At3g50950, DQ527675 to DQ527747; At4g14370, DQ527748 to DQ527831; At4g14610, DQ527832 to DQ527919; At4g26090, DQ527920 to DQ528011; At4g33300, DQ528012 to DQ528092; At5g04720, DQ528093 to DQ528179; At5g11250, DQ528180 to DQ528248; At5g17680, DQ528249 to DQ528304; At5g38850, DQ528305 to DQ528393; At5g44870, DQ528394 to DQ528471; At5g58120, DQ528472 to DQ528554; At5g63020, DQ528555 to DQ528643. The GenBank nucleotide sequence database accession numbers for the A. lyrata NBS-LRR R genes described in this article are as follows: At1g27170, DQ528644; At1g33560, DQ528645; At1g53350, DQ528646; At1g56540, DQ528647; At1g64070, DQ528648; At1g63730, DQ528649; At1g63740, DQ528650; At2g16870, DQ528651; At4g26090, DQ528652; At5g44870, DQ528653.
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Table 1. Accession Names and Locations.
- Supplemental Table 2. Primer Information for the Amplification of A. thaliana LRR Region Fragments.
- Supplemental Table 3. Primer Information for the Amplification of A. lyrata LRR Region Fragments.
- Supplemental Table 4. Regions and Number of Accessions Sequenced.
Supplementary Material
[Supplemental Data]
Acknowledgments
We thank Tina Hu, Jue Wang, and Keyan Zhao for their help with primer design and discussions about the data, Arnar Palsson for statistical help, Jean Gladstone for laboratory assistance, and Magnus Nordborg and Richard Hudson for discussions about data analysis. We also thank Brandon Gaut and three anonymous reviewers for valuable comments on the manuscript. This work was supported by a grant from the Dropkin Foundation to E.G.B., a National Institutes of Health postdoctoral fellowship to C.T., and National Science Foundation Grant DEB-0115062 and National Institutes of Health Grant GM57994 to J.B. and M.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joy Bergelson (jbergels@midway.uchicago.edu).
[W]
Online version contains Web-only data.
References
- Aranzana, M.J., et al. (2005). Genome-wide association mapping in Arabidopsis thaliana identifies genes responsible for variation in flowering time and pathogen resistance. PLoS Genet 1 e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., Miyashita, N., and Langley, C.F. (1989). Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M. (2001). Nucleotide sequence variation at two genes of the phenylpropanoid pathway, the FAH1 and F3H genes, in Arabidopsis thaliana. Mol. Biol. Evol. 18 1–9. [DOI] [PubMed] [Google Scholar]
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Jean Finnegan, E., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey, J.M., Zhang, G., Zhang, K., Jin, L., and Shriver, M.D. (2002). Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey, J.M., Eberle, M.A., Rieder, M.J., Carlson, C.S., Shriver, M.D., Nickerson, D.A., and Kruglyak, L. (2004). Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Barton, N.H., and Etheridge, A.M. (2004). The effect of selection on genealogies. Genetics 166 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J., Kreitman, M., Stahl, E.A., and Tian, D. (2001). Evolutionary dynamics of plant R-genes. Science 292 2281–2285. [DOI] [PubMed] [Google Scholar]
- Bisgrove, S.R., Simonich, M.T., Smith, N.M., Sattler, A., and Innes, R. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., Crute, I.R., Holub, E.B., and Beynon, J.L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21 177–188. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis Rpp1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A.L., Schaal, B.A., and Kunkel, B.N. (1999). Diversity and molecular evolution of the Rps2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J.M. (1999). K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15 763–764. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). The generation of plant disease resistance gene specificities. Trends Plant Sci. 5 373–379. [DOI] [PubMed] [Google Scholar]
- Falush, D., Stephens, M., and Pritchard, J.K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1956). The complementary genic system in flax and flax rust. Adv. Genet. 8 29–54. [Google Scholar]
- Gillespie, J.H. (1994). Alternatives to the neutral theory, in: Non-neutral evolution. Theories and molecular data. B. Golding (ed.). Chapman and Hall, New York. Pp. 1–17.
- Grant, J.J., Debrabata, A.C., and Loake, G.J. (2003). Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant Microbe Interact. 16 669–680. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 575–607. [DOI] [PubMed] [Google Scholar]
- Hauser, M.-T., Harr, B., and Schlotterer, C. (2001). Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: molecular analysis of the candidate gene Glabrous1. Mol. Biol. Evol. 18 1754–1763. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L., and Nei, M. (1988). Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335 167–170. [DOI] [PubMed] [Google Scholar]
- Hwang, C.-F., Bhakta, A.V., Truesdell, G.M., Pudlo, W.M., and Moroz Williamson, V. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., Terauchi, R., and Miyashita, N.T. (1997). Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., and Nordborg, M. (2003). The extent of linkage disequilibrium and haplotype sharing around a polymorphic site. Genetics 165 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Jones, J.D.G. (1997). The role of leucine-rich repeat proteins in plant defenses. Adv. Bot. Res. 24 89–167. [Google Scholar]
- Kajava, A.V. (1998). Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277 519–527. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of Chalcone Synthase and Alcohol Dehydrogenase loci in Arabidopsis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kreitman, M. (2000). Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 1 539–559. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., and Aguadé, M. (2000). Nucleotide variation at the CHALCONE ISOMERASE locus in Arabidopsis thaliana. Genetics 155 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuittinen, H., de Haan, A.A., Vogl, C., Oikarinen, S., Leppala, J., Koch, M., Mitchell-Olds, T., Langley, C.H., and Savolainen, O. (2004). Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular Evolutionary Genetics Analysis software for microcomputers. Bioinformatics 17 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., Bent, A.F., Dahlbeck, D., Innes, R.W., and Staskawicz, B.J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J.E., Lawrence, G.J., Dodds, P.N., Shepherd, K.W., and Ellis, J.G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates _Rps2_-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mauricio, R., Stahl, E.A., Korves, T., Tian, D., Kreitman, M., and Bergelson, J. (2003). Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics 163 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith, J., and Haigh, J. (1974). The hitch-hiking effect of a favorable gene. Genet. Res. 23 23–35. [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G.M., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the Rpp8 locus of Arabidopsis. Plant Cell 10 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino, M., Meyers, B.C., Michelmore, R.W., and Gaut, B.S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, S.R., and Griffiths, R.C. (2003). Bounds on the minimum number of recombination events in a sample history. Genetics 163 375–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and Li, W.-H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3 418–426. [DOI] [PubMed] [Google Scholar]
- Noël, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., and Tavaré, S. (2002). Linkage disequilibrium: what history has to tell us. Trends Genet. 18 83–90. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E., Kurth, J., and Leister, D. (2002). Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 19 76–84. [DOI] [PubMed] [Google Scholar]
- Richter, T.E., Pryor, T.J., Bennetzen, J.L., and Hulbert, S.H. (1995). New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics 141 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, L.E., Bittner-Eddy, P.D., Langley, C.H., Holub, E.B., Michelmore, R.W., and Beynon, J.L. (2004). The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, M., Goggin, S.B., Kaloshian, I., Ullman, D.E., and Williamson, V.M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K.J., Ramos-Onsins, S., Ringys-Beckstein, H., Weisshaar, B., and Mitchell-Olds, T. (2005). A multiple sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Araki, H., Chen, L., Chen, J.-Q., and Tian, D. (2006). Unique evolutionary mechanism in _R_-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonich, M.T., and Innes, R.W. (1995). A disease resistance gene in Arabidopsis with specificity for the avrPph3 gene of Pseudomonas syringae pv. Phaseolicola. Mol. Plant Microbe Interact. 8 637–640. [DOI] [PubMed] [Google Scholar]
- Stahl, E.A., Dwyer, G., Mauricio, R., Kreitman, M., and Bergelson, J. (1999). Dynamics of disease resistance polymorphism at the RPM1 locus of Arabidopsis. Nature 400 667–671. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D. (1995). Molecular genetics of plant disease resistance. Science 268 661–667. [DOI] [PubMed] [Google Scholar]
- Stranger, B.E., and Mitchell-Olds, T. (2005). Nucleotide variation at the myrosinase-encoding locus, TGG1, and quantitative myrosinase enzyme activity variation in Arabidopsis thaliana. Mol. Ecol. 14 295–309. [DOI] [PubMed] [Google Scholar]
- Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., Araki, H., Stahl, E., Bergelson, J., and Kreitman, M. (2002). Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., Traw, M.B., Chen, J.Q., Kreitman, M., and Bergelson, J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423 74–77. [DOI] [PubMed] [Google Scholar]
- Toomajian, C., Hu, T.T., Aranzana, M.J., Lister, C., Tang, C., Zheng, H., Zhao, K., Calabrese, P., Dean, C., and Nordborg, M. (2006). A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4 e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.-L., Ruan, D.-L., Song, W.-Y., Sideris, S., Chen, L.L., Pi, L.-Y., Zhang, S., Zhang, Z., Fauquet, C., Gaut, B.S., Whalen, M.C., and Ronald, P.C. (1998). Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downey mildew resistance genes. Plant Cell 10 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]