AUGUSTUS: ab initio prediction of alternative transcripts (original) (raw)
Abstract
AUGUSTUS is a software tool for gene prediction in eukaryotes based on a Generalized Hidden Markov Model, a probabilistic model of a sequence and its gene structure. Like most existing gene finders, the first version of AUGUSTUS returned one transcript per predicted gene and ignored the phenomenon of alternative splicing. Herein, we present a WWW server for an extended version of AUGUSTUS that is able to predict multiple splice variants. To our knowledge, this is the first ab initio gene finder that can predict multiple transcripts. In addition, we offer a motif searching facility, where user-defined regular expressions can be searched against putative proteins encoded by the predicted genes. The AUGUSTUS web interface and the downloadable open-source stand-alone program are freely available from http://augustus.gobics.de.
INTRODUCTION
Despite considerable efforts in the Bioinformatics community, the performance of existing gene prediction tools is still not satisfactory. In current genome projects, a common approach for gene finding is the following. Several sets of gene predictions are compiled, usually from different gene finders trained specifically for the species at hand. Further, alignments from ESTs and proteins to the genome are constructed. Finally, the predictions and the alignments are combined to find plausible gene structures, either manually or by using meta tools that combine several predictions and alignments [e.g. (1)].
AUGUSTUS is a gene finder based on a Generalized Hidden Markov Model (GHMM) (2,3). The original version of the program was a purely ab initio method, i.e. its prediction was based on information contained in the genomic sequence to be analyzed. An extended version of the program is able to use additional extrinsic information, for example matches to protein databases or alignments of genomic sequences, to improve the prediction accuracy (4). At the recent EGASP workshop in Cambridge, UK, a systematic evaluation of existing gene finders for the human genome has been carried out based on a large set of well-annotated parts of the human genome (5). At this workshop, AUGUSTUS turned out to be the best program in the category of ab initio gene prediction. Its performance could be further improved by using BLAST (6) hits to EST or protein sequences and alignments of syntenic genomic sequences using DIALIGN (7,8); in this category, however, the program was outperformed by N-Scan (9), a new program based on multiple alignments of genomic sequences. Compared to more traditional approaches, gene-finding methods based on genomic sequence alignments have a considerable advantage since they do not depend on EST or protein sequences or statistical models of gene structures (10–13). On the other hand, alignment-based methods work only if genome sequences at an appropriate evolutionary distance are available. Although the performance of ab initio gene-prediction methods is usually improved if information from comparative sequence analysis is added, ab initio gene prediction remains highly important since for many newly sequenced genomes, few EST or related genomic sequences sequences are available and comparison to protein sequences can find only those genes that have close relatives in existing databases.
To make AUGUSTUS available to the research community, we set up a WWW server at Göttingen Bioinformatics Compute Server (GOBICS) (14,15). Like most gene-prediction methods that are currently available, earlier versions of AUGUSTUS predicted exactly one transcript per gene and ignored the fact that one gene often yields more than one distinct mRNA product. It has been estimated that 40–60% of all human genes have alternative splice forms. Of those genes 70–88% of alternative splices change the protein product; the remaining splice variants differ in the untranslated regions only (16). Thus, it is important to have gene-finding tools that are able to deal with this phenomenon. The program SLAM (17), for example, predicts alternative splice variants. This program, however, is based on alignments of genomic sequences, and it requires two syntenic genomic sequences as input data. We recently installed a new version of AUGUSTUS at our server that can predict multiple transcripts for predicted genes. To our knowledge, this is the first ab initio gene finder that can predict multiple transcripts, and our web server is the only gene prediction web server with this option.
With our new alternative-transcripts option, the user can control the number of predicted splice variants per gene. This way, it is possible to influence sensitivity and specificity of the program output. If predicted genes or transcripts are automatically evaluated and post-processed, high prediction sensitivity may be desirable to increase the number of candidate genes that are to be analyzed, even if this increases the number of false-positive predictions. In contrast, if expensive experiments are carried out based on computationally predicted genes, it is preferable to have highly specific tools that minimize the risk of false-positive predictions. Thus, a good gene-finding method should allow the user to chose between high sensitivity and high specificity. At our server, this can be done by specifying the maximum number of predicted splice variants. In addition, we implemented a motif-searching option at our server where predicted genes can be searched for user-specified regular expressions, e.g. PROSITE patterns.
MATERIALS AND METHODS
Sampling and posterior probabilities
Let the term parse denote a segmentation of the input DNA sequence s into exons, introns and intergenic regions. Without considering alternative transcripts, each parse would define a distinct gene structure on s. For each species for which AUGUSTUS has been trained, AUGUSTUS has one GHMM. As usual in HMM-based gene prediction, this model defines a probability distribution over all pairs of a DNA sequence s and a parse ϕ. Now, let s be a given fixed input DNA sequence s in which we want to find gene structures. The model implicitly defines a conditional probability distribution over all parses. Let
denote the probability of parse ϕ given input sequence s. We call p(ϕ_∣s_) the posterior probability of ϕ. If the probabilistic modelling is good, the posterior probability is relatively high for true or reasonable parses ϕ. We call the parse with the highest posterior probability the Viterbi parse according to the name of the algorithm that is used to find it (18). For an input sequence s, most HMM-based gene prediction programs output one single parse, namely the gene structure corresponding to the Viterbi parse.
Random sampling has been used in the context of gene prediction, e.g. by Alexanderson et al. (19). To produce alternative splice variants for a sequence s, AUGUSTUS randomly samples n parses ϕ1, ϕ2, … , ϕ_n_ using a sampling algorithm that has been described in (3). In each of these n random experiments, a parse ϕ is picked with probability p(ϕ∣s). Usually, many sampled parses share exons, introns or transcripts. The sampled parses are used to estimate posterior probabilities of exons, introns, transcripts and genes. The posterior probobability of an exon or intron is the probability of this exon or intron to be part of the sampled parse with exactly the same boundaries and on the same strand. The posterior probability of a transcript is the probability with which the sampled parse contains a transcript that is from the start codon to the stop codon completely identical to this transcript. With the posterior probability of a gene we denote the probability that the sampled parse contains some coding region on the strand of the gene within the boundaries of the gene. While the posterior probabilities of exons, introns and transcripts could be computed theoretically using the backward and forward algorithm, the probability of a gene cannot be easily computed. For consistent results, we therefore estimated all posterior probabilities using our sampling approach. In our approach, posterior probabilities of exons, introns, transcripts and genes are estimated by their relative frequencies in the sampling procedure. For example, if an exon occurres 80 times in n = 100 sampled parses, then the posterior probability is estimated as 80%.
Alternative transcripts
Our aim was to construct a predicted set of genes with likely alternative transcripts (Figure 1). The number of alternative transcripts for a gene should be different from gene to gene, depending on how many likely alternatives exist. We first compile a set of transcripts by taking all transcripts from the Viterbi parse (‘Viterbi transcripts’) and from all sampled parses. Then we estimate the posterior probabilities of all transcripts, exons, introns and as described above. We discard transcripts where the coding sequence has a length smaller than a certain minimum length _L_min where the default value for _L_min is 102 bp. Further, we apply the following filtering criteria to the non-Viterbi transcripts to retain only the likely alternatives.
- We throw away transcripts where any exon or intron has a posterior probability below some constant _P_min.
- We throw away transcripts where the geometric mean of the posterior probability of all exons and introns is below a constant Pminav
- If more than _T_max transcripts overlap at the same position, we keep only _T_max of them, giving highest priority to Viterbi-transcripts and further sorting by mean posterior probability of exons and introns.
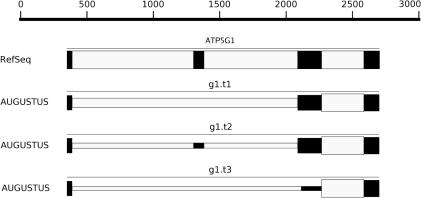
Figure 1.
The human gene ATP5G1 and the AUGUSTUS ab initio prediction for this region. The first transcript (g1.t1) is also the one predicted by standard AUGUSTUS using the Viterbi algorithm only. It misses the second exon of the gene. The second transcript (g1.t2) contains that exon and is correct. The height of a box (black: exon, light gray: intron) reflects the posterior probability of that exon or intron: The higher the posterior probability, the higher the box.
The third criterion ensures that only _T_max tracks are needed when the predictions are displayed in a genome browser (in Figure 2, _T_max = 3). Finally, predicted transcripts are clustered to genes in such a way that any two overlapping transcripts on the same strand are in the same gene. Our web server has four different options for the number of reported alternative transcripts, single transcript and few, medium number or many transcripts. For these options, the following parameter settings are used:
- single transcript: only Viterbi-transcripts are reported.
- few transcripts: _P_min = 20%, Pminav=50% _T_max = 2
- medium number of transcripts: _P_min = 8% Pminav=40%, _T_max = 3
- many transcripts: _P_min = 8%, Pminav=30%, _T_max = 20
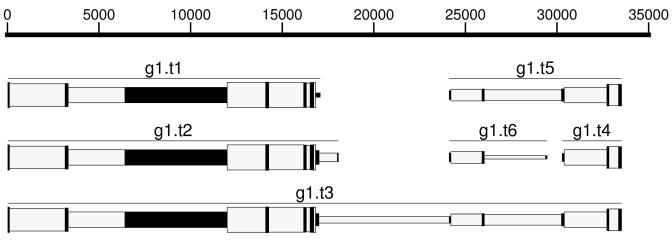
Figure 2.
Region of a human gene on the forward strand for which AUGUSTUS predicted six transcripts (gene SON, chromosome 21, 33 837 000–33 872 000, ncbi build 35). The long intron of transcript g1.t3 containing position 20 000 has low posterior probability. Thus, the model is unsure whether this is actually one gene, two or three genes. In fact, for this gene there exists EST evidence both for the short transcript g1.t1 and for longer transcripts with exons mostly agreeing with those predicted above.
WEB SERVER DESCRIPTION
Input
At the AUGUSTUS web server, the user can upload their sequences in FASTA format or paste them into a web form. The maximal total length of the sequences submitted to the server is 3 million base pairs. AUGUSTUS has species-specific parameter sets that can be chosen at the web site. For the following species, pre-calculated sets of parameters are available: Homo sapiens, Drosophila melanogaster, Arabidopsis thaliana, Brugia malayi, Coprinus cinereus, Tribolium castaneum, Schistosoma mansoni, Tetrahymena thermophila and Galdieria sulphuraria.
Output
AUGUSTUS outputs its results both in graphical and text format. The text output consists of exon, intron, transcript and gene boundaries in the common General Feature Format (GFF) as well as predicted amino acid sequences and predicted coding sequences in FASTA format. The results page of our web server shows for each sequence a thumbnail and links to images in pdf and postscript format similar to the example shown in Figures 1 and 2. The graphical output is generated with the program gff2ps (20) from the text output.
SEARCHING FOR REGULAR EXPRESSIONS
To analyze putative protein products of predicted genes, our server offers a pattern-searching option. Here, the user can enter arbitrary patterns as regular expressions; these patterns are then searched against all predicted splice variants from all genes predicted in the input sequence. This can be helpful if the user is looking for members of a particular protein family with conserved positions that are already described by a pattern, for example as an entry in the PROSITE database. In particular, in connection with the above described option for alternative splice forms, our pattern-searching option can be applied to filter out those splice variants that are consistent with the described pattern.
Following the PROSITE syntax, the conserved positions of a protein family may be specified as in the following example describing the hydrophobin family: <{C}(17,85)-C-{C}(5,10)-C-C-{C}(11,44)-C-{C}(8,23)-C-{C}(5,9)-C-C-{C}(6,18)-C-{C}(2,13)>. This pattern consists of eight conserved cystein residues with certain restrictions on the space between two subsequent cysteins. If a genome is to be searched for members of a protein family characterized by a regular expression, it is advisable to let AUGUSTUS predict a large number of splice variants to increase the prediction sensitivity, possibly at the expense of its specificity. With this strategy, the number of false positive splice variants may be increased, but a large number of predicted transcripts also increases the probability of finding all instances of the specified motif in the genome sequence under study.
RESULTS
We tested our method on a large set of test data from the recent EGASP workshop that was organized in the context of the ENCODE project (5). This dataset comprises a total of 296 genes with an average of 2.2 transcripts per gene. Table 1 shows the relation between the posterior probability of exons as defined above and the specificity at the exon level. As one may expect, the posterior probability is a good indicator of exon specificity. Predicted exons with low posterior probability are much less likely to be correct than predicted exons with high posterior probability. Therefore, the posterior probability gives a good criterion for prioritising putative exons, e.g. for experimental verification.
Table 1.
Percentage of correctly predicted human exons and introns in coding regions grouped by their posterior probability
| Posterior probability P | Exon specificity |
|---|---|
| 0 ≤ P ≤ 50% | 46/242 ≈ 19.0% |
| 50% < P ≤ 70% | 132/356 ≈ 37.1% |
| 70% < P ≤ 80% | 84/175 ≈ 48.0% |
| 80% < P ≤ 90% | 171/275 ≈ 62.2% |
| 90% < P ≤ 95% | 140/195 ≈ 71.8% |
| 95% < P ≤ 99% | 338/422 ≈ 80.1% |
| 99% < P ≤ 100% | 545/612 ≈ 89.1% |
| Total | 1456/2277 ≈ 63.9% |
Table 2 summarizes the prediction accuracy of AUGUSTUS on the EGASP test data. We used AUGUSTUS with the original single-transcript option and with the new options for ‘few transcripts’, ‘medium number of transcripts’ and ‘many transcripts’. Sensitivity and specificity of these program versions are given at four different levels, namely at the base, exon, transcript and gene level. At the gene level, a gene is considered a true positive if one predicted transcript coincides with an existing transcript of this gene. As expected, the program sensitivity increases at all four levels if the number of transcripts per gene is increased. Correspondingly, the specificity at the base, exon and transcript level decreases if the number of predicted transcript increases. In contrast, at the gene level the specificity of AUGUSTUS increases if more transcripts per gene are predicted. The reason for this seemingly paradoxical result is as follows: with an increased number of predicted transcripts, it is more likely that one of the real transcripts in a gene is matched by a predicted transcript. The number of predicted genes, on the other hand, does not increase significantly if more transcripts are predicted because most of the additionally predicted transcripts belong to one of the previously predicted genes. Thus, the ratio of true positive genes to predicted genes increases if the number of predicted transcripts is increased.
Table 2.
Accuracy values of variants of AUGUSTUS on the ENCODE test set with 296 genes and an average of 2.2 transcripts per gene
| Single transcript | Few transcripts | Medium number of transcripts | Many transcripts | |
|---|---|---|---|---|
| Gene sensitivity | 0.233 | 0.273 | 0.294 | 0.345 |
| Gene specificity | 0.170 | 0.200 | 0.211 | 0.239 |
| Transcript sensitivity | 0.106 | 0.125 | 0.137 | 0.165 |
| Transcript specificity | 0.170 | 0.144 | 0.112 | 0.053 |
| Exon sensitivity | 0.527 | 0.540 | 0.557 | 0.600 |
| Exon specificity | 0.639 | 0.615 | 0.571 | 0.490 |
| Base sensitivity | 0.775 | 0.779 | 0.790 | 0.814 |
| Base specificity | 0.764 | 0.757 | 0.738 | 0.705 |
| Average transcripts/gene | 1.0 | 1.4 | 2.0 | 5.1 |
Letting AUGUSTUS predict many transcripts is most useful when the focus is on finding at least one correct splice form for a gene: as shown in Table 2, the gene-level sensitivity increases from 23.3 to 34.5% if the option ‘many transcripts’ is used (on average, 5.1 transcripts per gene are predicted with this option). This is substantially more than the gene-level sensitivity of standard gene finders; e.g. for the commonly used programs GENSCAN (21), GENEID (22) and GENEZILLA (23), the gene-level sensitivity values on the EGASP dataset are 15.5, 10.5 and 19.6%, respectively.
Acknowledgments
The option of predicting multiple transcripts per gene was developed in part when training AUGUSTUS for Nicotiana tabacum in the context of the Tobacco Genome Initiative http://tgi.ncsu.edu, (24) in Philip Morris USA. This work was supported by BMBF grant 01AK803G (Medigrid) to B.M. and by a fellowship within the Postdoc-Program of the German Academic Exchange Service (DAAD) to M.S. Funding to pay the Open Access publication charges for this article was provided by Philip Morris USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Allen J.E., Pertea M., Salzberg S.L. Computational gene prediction using multiple sources of evidence. Genome Res. 2004;14:142–148. doi: 10.1101/gr.1562804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanke M., Waack S. Gene prediction with a hidden markov model and a new intron submodel. Bioinformatics (ECCB 2003 special issue) 2003;19:ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 3.Stanke M. Universität Göttingen; 2004. Gene Prediction with a Hidden Markov Model. PhD Thesis. [Google Scholar]
- 4.Stanke M., Schöffmann O., Morgenstern B., Waack S. Gene prediction in eukaryotes with a Generalized Hidden Markov Model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guigó R., Reese M.G. EGASP: collaboration through competition to find human genes. Nature Meth. 2005;2:575–577. doi: 10.1038/nmeth0805-575. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S.F., Gish W., Miller W., Myers E.M., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Brudno M., Chapman M., Göttgens B., Batzoglou S., Morgenstern B. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics. 2003;4:66. doi: 10.1186/1471-2105-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross S.S., Brent M.R. Using Multiple Alignments to Improve Gene Prediction. Proceedings RECOMB'05; 2005. pp. 374–388. [DOI] [PubMed] [Google Scholar]
- 10.Batzoglou S., Pachter L., Mesirov J.P., Berger B., Lander E.S. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bafna V., Huson D.H. The conserved exon method for gene finding. Bioinformatics. 2000;16:190–202. [PubMed] [Google Scholar]
- 12.Korf I., Flicek P., Duan D., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001:S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 13.Rinner O., Morgenstern B. AGenDA: gene prediction by comparative sequence analysis. In Silico Biol. 2002:195–205. [PubMed] [Google Scholar]
- 14.Stanke M., Steinkamp R., Waack S., Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanke M., Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 17.Cawley S.L., Pachter L. HMM sampling and applications to gene finding and alternative splicing. Bioinformatics. 2003;19:ii36–ii41. doi: 10.1093/bioinformatics/btg1057. [DOI] [PubMed] [Google Scholar]
- 18.Viterbi A. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inform. Theory. 1967;13:260–269. [Google Scholar]
- 19.Alexandersson M., Cawley S., Pachter L. SLAM—cross-species gene finding and alignment with a generalized pair Hidden Markov Model. Genome Res. 2003;13:496–502. doi: 10.1101/gr.424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abril J.F., Guigó R. gff2ps: visualizing genomic annotations. Bioinformatics. 2000;16:743–744. doi: 10.1093/bioinformatics/16.8.743. [DOI] [PubMed] [Google Scholar]
- 21.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 22.Guigó R., Knudsen S., Drake N., Smith T. Prediction of gene structure. J. Mol. Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- 23.Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 24.Gadani F., Hayes A., Opperman C.H., Lommel S.A., Sosinski B.R., Burke M., Hi L., Brierly R., Salstead A., Heer J. 5èmes Journées Scientifiques du Tabac de Bergerac. 2003. Large scale genome sequencing and analysis of Nicotiana tabacum: the tobacco genome initiative; pp. 117–130. [Google Scholar]