In Situ Characterization of Cymbidium Ringspot Tombusvirus Infection-Induced Posttranscriptional Gene Silencing in Nicotiana benthamiana (original) (raw)
Abstract
In plants, posttranscriptional gene silencing (PTGS) is an ancient and effective defense mechanism against viral infection. A number of viruses encode proteins that suppress virus-activated PTGS. The p19 protein of tombusviruses is a potent PTGS suppressor which interferes with the onset of PTGS-generated systemic signaling and is not required for viral replication or for viral movement in Nicotiana benthamiana. This unique feature of p19 suppressor allowed us to analyze the mechanism of PTGS-based host defense and its viral suppression without interfering with other viral functions. In contrast to the necrotic symptoms caused by wild-type tombusvirus, the infection of p19-defective mutant virus results in the development of a typical PTGS-associated recovery phenotype in N. benthamiana. In this report we show the effect of PTGS on the viral infection process for N. benthamiana infected with either wild-type Cymbidium Ringspot Tombusvirus (CymRSV) or a p19-defective mutant (Cym19stop). In situ analyses of different virus-derived products revealed that PTGS is not able to reduce accumulation of virus in primary infected cells regardless of the presence of p19 PTGS suppressor. We also showed that both CymRSV and Cym19stop viruses move systemically in the vasculature, with similar efficiencies. However, in contrast to the uniform accumulation of CymRSV throughout systemically infected leaves, the presence of Cym19stop virus was confined to and around the vascular bundles. These results suggest that the role of p19 is to prevent the onset of mobile signal-induced systemic PTGS ahead of the viral infection front, leading to generalized infection.
Almost all eukaryotes possess a homology-dependent RNA degradation system that has evolved to protect against invasive or mobile foreign genetic elements. This mechanism is referred to as posttranscriptional gene silencing (PTGS) for plants, gene quelling for fungi, and RNA interference for animals (14, 26, 30). The targets could be transgenes, viruses, or transposons or any RNA homologous to the inducer molecules that are degraded by activated PTGS. The mechanism of virus-induced PTGS is not completely understood, but double-stranded (ds) RNA replication intermediates of an RNA virus are strong inducers (28). The accumulation of small 21- to 25-nucleotide-long interfering RNAs (siRNAs) corresponding to both plus and minus strands of the inducer RNA are a characteristic of PTGS (29). In addition to the cell-autonomous defense function in higher plants, PTGS is associated with a mobile signal that instructs target RNA degradation at a distance (16; O. Voinnet and D. C. Baulcombe, Letter, Nature **389:**553, 1997). The mobile signal, which must consist at least in part of homologous RNA, is suggested to spread from cell to cell through plasmodesmata and systemically via the vascular system of plants (15). The findings that plant viruses encode proteins that suppress PTGS provide the most compelling evidence for the function of PTGS as a natural antiviral defense mechanism (12, 13, 25). Several virus-encoded RNA silencing suppressor proteins have been characterized to date (25). These suppressor proteins act at different stages of the silencing pathway, targeting the initiation, the maintenance, or the systemic signaling events of PTGS (4, 12).
The members of the genus of Tombusvirus contain a positive-sense single-stranded RNA genome with five open reading frames (ORFs) (19). ORF5 of tombusviruses encodes a 19-kDa protein (p19) which is one of the best-characterized PTGS suppressor protein and is also a pathogenicity determinant (2, 5, 20). p19 blocks systemic, but not local, virus-induced silencing and binds PTGS-generated ds siRNAs in vitro (21, 22). It may act by sequestering the PTGS specificity determinant ds siRNAs, thereby preventing the induction of systemic PTGS signaling. The majority of viral suppressor proteins identified so far are also involved in additional viral functions (e.g., they are required for cell-to-cell trafficking [27], long-distance movement [7, 11], and replication [11]). Recent data suggest that some of these activities may be connected to PTGS suppressor activity (11). However, the silencing suppressor activity of p19 is uncoupled from viral movement and replication functions in N. benthamiana (17, 18, 22). This feature of p19 provides a unique experimental system for investigating plant responses to a wild-type virus versus a p19 mutant virus that is defective only in PTGS suppressor activity. Infection with p19-defective tombusviruses (e.g., Cym19stop) resulted in the development of a typical PTGS-associated recovery phenotype for N. benthamiana (17, 18, 22). The recovered leaves of Cym19stop-infected plants were resistant to infection by another recombinant virus carrying a sequence element homologous to that of the inducing virus (22).
The work presented here addresses the role of systemic PTGS in the development of a recovery phenotype and the counter-defense strategy of the virus-encoded suppressor protein by applying spatial analyses of virus-derived products to Cymbidium Ringspot Tombusvirus (CymRSV)- and Cym19stop-infected N. benthamiana plants.
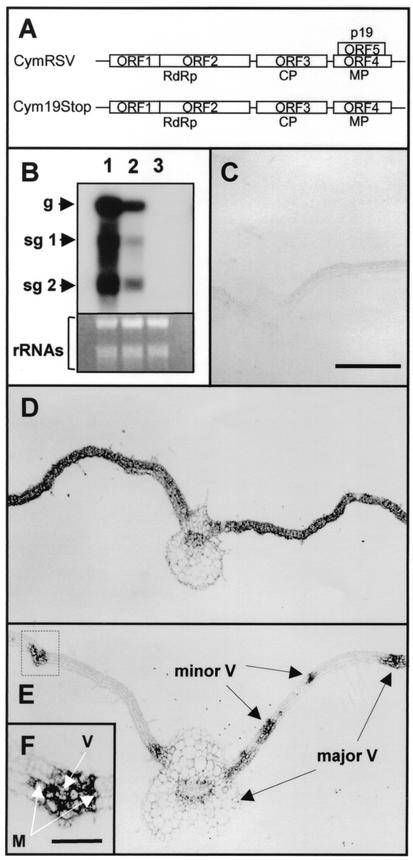
Ten 4-week-old N. benthamiana plants were infected with in vitro RNA transcripts of CymRSV and Cym19stop as described previously (6). Plants were kept at a constant temperature of 24°C in the growth chamber because PTGS acts more effectively at this temperature than at 21°C (23). Infection of test plants with CymRSV or Cym19stop resulted in the development of severe symptoms on the first systemically infected leaves after 6 to 7 days postinoculation (d.p.i.) on both virus-infected plants (data not shown). As described previously, CymRSV infection led to necrosis of the systemically infected leaves by 8 to 10 d.p.i. which eventually culminated in the death of the plant (6). In contrast, Cym19stop-inoculated plants displayed severe symptoms on first systemically infected leaves, but without detectable necrosis. The subsequently developed leaves of Cym19stop-infected plants showed the development of PTGS-associated recovery phenotype (22). In line with previous observations, Northern blot analyses of RNA samples from the first systemically infected leaves at 7 d.p.i. showed a significant reduction in the level of Cym19stop RNA compared to CymRSV RNA accumulation (Fig. 1B) (22).
FIG. 1.
Accumulation of CymRSV and Cym19stop RNA in the first systemically infected leaves at 7 d.p.i. (A) Schematic representation of CymRSV and Cym19stop genomic RNAs. RdRp, RNA-dependent RNA polymerase. (B) Total mRNAs, prepared from systemically infected tissues at 7 d.p.i. or mock-inoculated tissue, were separated under denaturing conditions, blotted, and hybridized with CymRSV-derived probe (upper panel). RNA was extracted from CymRSV (lane 1), Cym19stop (lane 2), and mock-inoculated (lane 3) tissues. Relative gel loadings are shown by ethidium bromide staining of the rRNAs (bottom panel). g, genomic RNA; sg, subgenomic RNA. (C to E) In situ hybridization of 12 μm paraffin-embedded leaf cross sections with digoxigenin-11-UTP-labeled virus-specific RNA probe. Samples were taken from the first systemically infected leaves at 7 d.p.i. (C) Control mock-inoculated tissue. (D) CymRSV-infected tissue. (E) Cym19stop-infected tissue. (F) Magnification (×3) of a vascular bundle and surrounding tissue marked by dotted box in panel E. M, mesophyll cells; V, vascular tissue. Major and minor veins are shown by arrows. Bar in panel C, 400 μm (applies to panels B and E); bar in panel F, 100 μm.
To gain better insight into the operation of virus infection-induced PTGS at the tissue level, we investigated CymRSV- and Cym19stop-infected plants by applying in situ hybridization to the first systemic leaves at 7 d.p.i. Samples were taken from 10 infected plants in two separate experiments, and the samples of the independent experiments were handled separately. Two systemically infected leaves per plant were taken, and the leaf samples were collected and processed for paraffin embedding (10). Three paraffin blocks containing about five to six embedded leaves per sample were sectioned and applied to microscopic slides. A digoxigenin-11-UTP-labeled minus-sense probe corresponding to the p19 encoding region was hybridized to tissue sections and detected with alkaline phosphatase-conjugated antidigoxigenin antibody, as described previously (8). The majority of the investigated CymRSV-infected leaves showed uniform high-level accumulation of virus RNA throughout the whole tissue, indicating the establishment of a successful systemic infection (Fig. 1D). In contrast, all of the investigated Cym19stop-infected leaves displayed a marked reduction in the extent of virus infection. Typical representatives of these samples showed accumulation of virus RNA confined to the veins and the neighboring mesophyll cells (Fig. 1E).
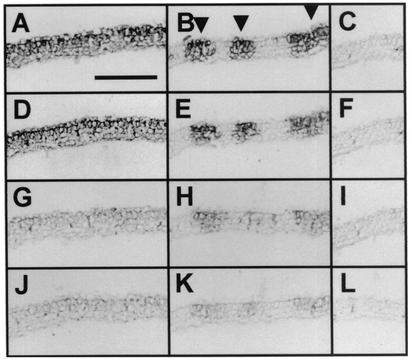
To reveal whether Cym19stop accumulates in the infected cells with an efficiency similar to that of the wild type virus, samples taken from the systemically infected leaves were analyzed by in situ hybridization. Consecutive sections of CymRSV- and Cym19stop-infected leaves were applied onto microscopic slides. The slides were hybridized with the same probe detecting the plus strand of virus RNA. To avoid the misinterpretation of the intensity of hybridization signals, the color reaction was stopped at different time points (20, 40, 90, and 180 min) before reaching the saturation level (Fig. 2). These experiments showed that the CymRSV- and Cym19stop-infected cells showed similar signal intensities at all of the investigated time points, indicating that these cells contain close to equal amounts of virus RNA. The observation that accumulation of virus RNA in cells infected by Cym19stop was similar to that of CymRSV suggests that once the cells become infected, both wild-type and mutant viruses replicate with similar efficiencies at the cellular level. This finding is in line with the previous observation that CymRSV and Cym19stop accumulated at similar levels in transfected protoplasts (21). These results indicate that in primary infected cells, PTGS is not able to restrict the replication of the virus regardless of the presence or absence of p19.
FIG. 2.
Analyses of viral RNA accumulation levels in CymRSV- and Cym19stop-infected tissues. In situ hybridization of 12 μm paraffin-embedded leaf cross sections with digoxigenin-11-UTP-labeled virus-specific RNA probe. Samples were taken from the first systemically infected leaves at 7 d.p.i. (A, D, G, and J) Near-consecutive sections of CymRSV-infected tissue. (B, E, H, and K) Near-consecutive sections of Cym19stop-infected tissue. (C, F, I, and L) Near-consecutive sections of mock-inoculated tissue. The color reaction was stopped at 20 (J, K, and L), 40 (G, H, and I), 90 (D, E, and F) and 180 (A, B, and C) min. Arrowheads in panel B indicate the sites of Cym19stop accumulation. Bar in panel A, 250 μm (applies to panels A to L).
In situ analyses also showed that Cym19stop moved with an efficiency similar to that of CymRSV in the vascular tissue and was able to unload from the veins and move into the mesophyll cells (Fig. 1E and F). Accumulation of Cym19stop RNA in and around veins was detected in parallel with the appearance of the wild-type virus RNA by 4 d.p.i. in systemically infected leaves, indicating an effective long-distance movement of Cym19stop (data not shown). However, the most striking feature of the host defense response in the absence of p19 was the apparent block of the further advance of virus infection in the systemically infected leaves (Fig. 1, compare panels D and E).
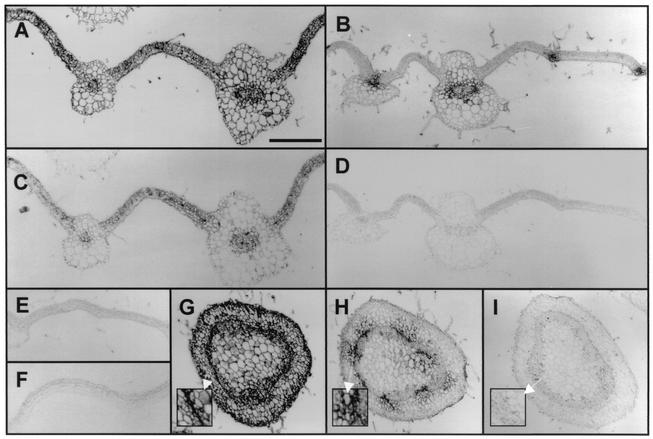
To analyze whether the accumulation of viral proteins other than p19 is affected in Cym19stop-infected plants, the expression of different viral proteins was defined in both CymRSV- and Cym19stop-infected tissues by in situ immunohistochemistry. Virus particles were purified from infected plants (3) and used to raise antibody (Ab) against the CymRSV coat protein (CP). In situ immunohistochemistry of paraffin sections from systemically infected leaves at 7 d.p.i. was conducted as described previously using diluted (1:5,000) anti-CP Ab (8). The detection of CP by immunohistochemistry showed the same accumulation pattern for both CymRSV and Cym19stop (Fig. 3) as was detected by in situ hybridization (Fig. 1). The CP accumulated uniformly in the whole CymRSV-infected leaves, while in Cym19stop-infected leaves the accumulation of CP was confined to and around the vasculature. As a control, consecutive sections of the same tissue samples were also examined for the accumulation of p19 by immunohistochemistry using diluted (1:2,000) anti-p19 Ab (9). As expected, CymRSV-infected tissue contained p19, while no p19 was detected in Cym19stop-infected tissue (Fig. 3, compare C and D). To extend our observations for a different tissue type, cross sections of stems were examined by immunohistochemistry to detect CP accumulation as an indicator of virus infection. Similar to the case with the leaf sections, the stem sections of CymRSV-infected plants displayed a uniform high level of CP accumulation, while Cym19stop infection displayed CP accumulation limited to and around the vasculature (Fig. 3, compare panels G and H). Although the transverse movement of Cym19stop was decreased in the stem, the virus accumulated to high levels in the vascular tissue and the surrounding cells (Fig. 3H). These results collectively indicated that the viral invasion of plant tissues in Cym19stop-infected plants is restricted to mostly in and around the vascular tissues by PTGS, while in cells which already have been infected the cell-autonomous virus replication was not affected.
FIG. 3.
Accumulation of CP in CymRSV- and Cym19stop-infected tissues at 7 d.p.i. Immunohistochemistry applying anti-CP Ab to cross sections of CymRSV-inoculated (A), Cym19stop-inoculated (B), and mock-inoculated (E) systemic leaves, immunohistochemistry applying anti-p19 Ab to consecutive sections of CymRSV-inoculated (C) and Cym19stop-inoculated (D) systemic leaves, (F) immunohistochemistry applying anti-p19 Ab to mock-inoculated leaf (F), and immunohistochemistry applying anti-CP Ab to stem cross sections of CymRSV-inoculated (G), Cym19stop-inoculated (H), and mock-inoculated (I) plants are shown. Bar in panel A, 400 μm (applies to panels A to I). Insets in (G, H, and I) show magnification (×3) of parts of CymRSV- and Cym19stop-infected and mock-inoculated stem sections.
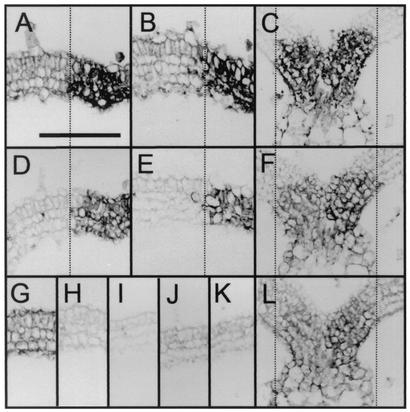
The viral movement protein (MP) and p19 are translated from the same subgenomic RNA containing two nested ORFs (Fig. 1A) (19). To exclude the possibility that the introduced mutations in Cym19stop interfere with the expression of MP, leading to restricted cell-to-cell movement, the accumulation of MP was investigated by use of immunohistochemistry. Since the role of MP is to facilitate the cell-to-cell movement of viruses, we used samples of infected plants at 5 d.p.i. to reveal the accumulation of MP at the infection front. Consecutive sections were hybridized with either anti-CP Ab to identify the virus infection front or anti-MP Ab (diluted 1:200) to investigate MP accumulation. To analyze the level of MP accumulation, the color reaction was stopped before reaching the saturation point. The MP accumulation patterns showed no significant differences between CymRSV- and Cym19stop-infected tissues (Fig. 4, compare panels D and E), demonstrating that MP is present in Cym19stop-infected tissue.
FIG. 4.
Accumulation of MP and p19 in systemically infected leaves at 5 d.p.i. Immunohistochemistry of consecutive cross sections using anti-CP Ab (A) and anti-MP Ab (D) with CymRSV-infected leaves and immunohistochemistry of consecutive cross sections using anti-CP Ab (B) and anti-MP Ab (E) with Cym19stop-infected leaves are shown. The alignment is shown relative to the edge of the virus infection front (dotted lines). Immunohistochemistry of control mock-inoculated sections applying anti-CP Ab (G) and anti-MP Ab (H) is shown. (C, F, and L) Colocalization of p19 with virus RNA and CP. Consecutive sections of CymRSV-infected systemic leaves analyzed by in situ hybridization to detect the accumulation of virus RNA (C) and immunohistochemistry to detect p19 (F) and CP (L) are shown. The alignment is shown relative to the edge of the virus infection front (dotted lines). Bar in panel (A, 200 μm (applies to panels A to L).
The 2b systemic suppressor protein of Cucumber mosaic virus has been proposed to autonomously enter and probably translocate through the phloem (7). To test whether p19 also has the capacity to move autonomously ahead of the infection front, we have analyzed the spatial accumulation pattern of p19 relative to the expression profile of viral RNA and CP. Consecutive sections of the first systemic leaves of CymRSV-infected plants at 5 d.p.i. were used to analyze p19 accumulation relative to the infection front. In situ hybridization and immunohistochemical staining showed that viral RNA, p19, and CP colocalize in the infected tissue (Fig. 4, compare panels C, F, and L); therefore, we conclude that p19 acts in cells where viral replication has been established by abolishing the production or emission of the mobile PTGS signal. These findings are in line with those of our previous studies, which demonstrated that p19 is able to inhibit the transgene-induced systemic PTGS and binds ds siRNAs in vitro (21).
In summary, this work revealed that in the absence of a potent systemic silencing suppressor, the virus infection-induced systemic PTGS confines accumulation of virus to the veins and the surrounding tissues. Based on the gained data, we suggest that in the absence of systemic PTGS suppressor protein (p19), the systemic signal moves faster than the virus in the infected plant, thereby establishing PTGS in cells ahead of the infection front. When virus enters such cells it will be immediately destroyed by PTGS-mediated RNA degradation. As a result of this mechanism, the spread of the virus is inhibited in the infected leaves; however, once its replication has been established in a cell, the mutant virus accumulates to wild-type levels.
It has been suggested that the ability of a virus to move in the infected tissue depends on its ability to block the PTGS-generated systemic signaling (1, 24). Consistent with this hypothesis, we suggest that CymRSV-encoded PTGS suppressor protein acts by blocking the spread of systemic PTGS-associated mobile signals from cells accommodating active virus replication.
Alternatively, the observed phenomenon might be explained by dual functions of p19. In addition to the PTGS suppression function, p19 may have an additional biochemical function contributing to virus penetration in the infected tissue. However, our recent observation (23) does not support this alternative explanation. It has been demonstrated that at a low temperature (15°C) whereby virus and transgene-induced PTGS are inhibited, both CymRSV and Cym19stop viruses invade the whole leaves of virus-infected plants. These results strongly suggest that Cym19stop possesses all the factors necessary for effective long-distance and cell-to-cell movement and p19 acts solely as a PTGS suppressor protein.
Acknowledgments
This research was supported by a grant from the Hungarian Scientific Research Fund (OTKA) (T038313).
We thank Rick Nelson, Loránt Lakatos Attila Molnár, Dániel Silhavy, and György Szittya for helpful comments.
REFERENCES
- 1.Baulcombe, D. 2002. Viral suppression of systemic silencing. Trends Microbiol. 10**:**306-308. [DOI] [PubMed] [Google Scholar]
- 2.Burgyan, J., C. Hornyik, G. Szittya, D. Silhavy, and G. Bisztray. 2000. The ORF1 products of tombusviruses play a crucial role in lethal necrosis of virus-infected plants. J. Virol. 74**:**10873-10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgyan, J., and M. Russo. 1998. Tombusvirus isolation and RNA extraction. Plant Virol. Protocols 81**:**225-230. [DOI] [PubMed] [Google Scholar]
- 4.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281**:**1-5. [DOI] [PubMed] [Google Scholar]
- 5.Chu, M., B. Desvoyes, M. Turina, R. Noad, and H. B. Scholthof. 2000. Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology 266**:**79-87. [DOI] [PubMed] [Google Scholar]
- 6.Dalmay, T., L. Rubino, J. Burgyan, A. Kollar, and M. Russo. 1993. Functional analysis of cymbidium ringspot virus genome. Virology 194**:**697-704. [DOI] [PubMed] [Google Scholar]
- 7.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21**:**398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havelda, Z., and A. J. Maule. 2000. Complex spatial responses to cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell 12**:**1975-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havelda, Z., G. Szittya, and J. Burgyan. 1998. Characterization of the molecular mechanism of defective interfering RNA-mediated symptom attenuation in tombusvirus-infected plants. J. Virol. 72**:**6251-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, D. P. 1991. In situ hybridisation in plants, p. 163-174. In S. J. Gurr, M. J. McPherson, and D. J. Bowles (ed.), Molecular plant pathology: a practical approach. Oxford University Press, Oxford, England.
- 11.Kasschau, K. D., and J. C. Carrington. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285**:**71-81. [DOI] [PubMed] [Google Scholar]
- 12.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12**:**150-154. [DOI] [PubMed] [Google Scholar]
- 13.Marathe, R., R. Anandalakshmi, T. H. Smith, G. J. Pruss, and V. B. Vance. 2000. RNA viruses as inducers, suppressors and targets of post-transcriptional gene silencing. Plant Mol. Biol. 43**:**295-306. [DOI] [PubMed] [Google Scholar]
- 14.Matzke, M., A. J. Matzke, and J. M. Kooter. 2001. RNA: guiding gene silencing. Science 293**:**1080-1083. [DOI] [PubMed] [Google Scholar]
- 15.Mlotshwa, S., O. Voinnet, M. F. Mette, M. Matzke, H. Vaucheret, S. W. Ding, G. Pruss, and V. B. Vance. 2002. RNA silencing and the mobile silencing signal. Plant Cell 14(Suppl.)**:**S289-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16**:**4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu, W., J. W. Park, and H. B. Scholthof. 2002. Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol. Plant Microb. Interact. 15**:**269-280. [DOI] [PubMed] [Google Scholar]
- 18.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant Microb. Interact. 15**:**193-202. [DOI] [PubMed] [Google Scholar]
- 19.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of tombusviridae. Adv. Virus Res. 44**:**381-428. [DOI] [PubMed] [Google Scholar]
- 20.Scholthof, H. B., K. B. Scholthof, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7**:**1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21**:**3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14**:**359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyan. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22**:**633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants—defense and counterdefense. Science 292**:**2277-2280. [DOI] [PubMed] [Google Scholar]
- 25.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17**:**449-459. [DOI] [PubMed] [Google Scholar]
- 26.Voinnet, O. 2002. RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5**:**444-451. [DOI] [PubMed] [Google Scholar]
- 27.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103**:**157-167. [DOI] [PubMed] [Google Scholar]
- 28.Waterhouse, P. M., M. W. Graham, and M. B. Wang. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95**:**13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse, P. M., M. B. Wang, and E. J. Finnegan. 2001. Role of short RNAs in gene silencing. Trends Plant Sci. 6**:**297-301. [DOI] [PubMed] [Google Scholar]
- 30.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411**:**834-842. [DOI] [PubMed] [Google Scholar]