Gene Expression Profiling of the Cellular Transcriptional Network Regulated by Alpha/Beta Interferon and Its Partial Attenuation by the Hepatitis C Virus Nonstructural 5A Protein (original) (raw)
Abstract
Alpha/beta interferons (IFN-α/β) induce potent antiviral and antiproliferative responses and are used to treat a wide range of human diseases, including chronic hepatitis C virus (HCV) infection. However, for reasons that remain poorly understood, many HCV isolates are resistant to IFN therapy. To better understand the nature of the cellular IFN response, we examined the effects of IFN treatment on global gene expression by using several types of human cells, including HeLa cells, liver cell lines, and primary fetal hepatocytes. In response to IFN, 50 of the approximately 4,600 genes examined were consistently induced in each of these cell types and another 60 were induced in a cell type-specific manner. A search for IFN-stimulated response elements (ISREs) in genomic DNA located upstream of IFN-stimulated genes revealed both previously identified and novel putative ISREs. To determine whether HCV can alter IFN-regulated gene expression, we performed microarray analyses on IFN-treated HeLa cells expressing the HCV nonstructural 5A (NS5A) protein and on IFN-treated Huh7 cells containing an HCV subgenomic replicon. NS5A partially blocked the IFN-mediated induction of 14 IFN-stimulated genes, an effect that may play a role in HCV resistance to IFN. This block may occur through repression of ISRE-mediated transcription, since NS5A also inhibited the IFN-mediated induction of a reporter gene driven from an ISRE-containing promoter. In contrast, the HCV replicon had very little effect on IFN-regulated gene expression. These differences highlight the importance of comparing results from multiple model systems when investigating complex phenomena such as the cellular response to IFN and viral mechanisms of IFN resistance.
Alpha/beta interferons (IFN-α/β) are expressed by many cell types in response to viral or bacterial pathogens. By binding to specific transmembrane receptors, these cytokines trigger a response that culminates in the induction of a large number of genes, many of which encode proteins with antiviral or antiproliferative properties. This increase in gene expression is controlled by multiple transcription factors and regulatory elements, including signal transducer and activators of transcription 1 and 2 (STAT1 and STAT2), which, together with IFN-regulatory factor 9 (IRF9), bind to _cis_-acting IFN-stimulated response elements (ISREs) to induce the transcription of many IFN-stimulated genes (ISGs). Other members of the IFN-regulatory-factor family, such as IRF3 and IRF7, also play a critical role in IFN signaling. These latent cytosolic factors translocate to the nucleus upon phosphorylation by unknown kinases and bind to ISRE-like elements to mediate the expression of specific ISGs. Using gene expression profiling, a number of investigators have begun to explore the complexities of the IFN response (5, 7, 8, 39, 41, 49), and although these studies have revealed an ever-increasing number of IFN-regulated genes, little is known about the tissue-specific nature of the response or the impact of viral infection on ISG expression.
Many viruses have evolved strategies to evade the antiviral effects of the IFN response (18, 22, 26). These strategies differ greatly, but it is common for a virus to encode one or more proteins that interact directly with an IFN-induced gene product or that interfere with some aspect of IFN signaling. Since the IFN response is a central component of innate immunity, a better understanding of the mechanisms used by viruses to counteract this response would be beneficial, particularly in those instances in which IFN is also the primary therapeutic option. A case in point is hepatitis C virus (HCV). HCV has infected over 170 million people worldwide (48) and is a primary risk factor for the development of liver cirrhosis and hepatocellular carcinoma (10). Although a combination of IFN-α and ribavirin is currently the optimal choice for therapy, HCV isolates are often resistant to IFN and this regimen is ineffective in many patients (2, 25).
Much of the search for the molecular basis of HCV resistance to IFN has centered on the viral nonstructural 5A (NS5A) protein. This focus began with evidence that the amino acid sequence of a region of NS5A, termed the IFN sensitivity-determining region (ISDR), correlates with therapeutic outcome (11, 12). Subsequent in vitro studies demonstrated that NS5A from an IFN-resistant isolate inhibits the IFN-induced protein kinase PKR (16), suggesting a mechanism for IFN resistance. However, the link between NS5A and resistance to IFN remains hazy (44). Therapeutic outcome does not always correlate with ISDR sequence (1, 36) or with inhibition of PKR activity (13, 38), and other studies suggest that IFN resistance is related to regions of NS5A outside the ISDR (34, 35) or to the ability of NS5A to interact with one or more of a variety of cellular proteins (6, 24, 33, 45, 47).
Thus, despite all these reports, questions remain about the mechanisms by which NS5A might mediate IFN resistance. In this study, we used global gene expression profiling to evaluate the cellular transcription network regulated by IFN-α/β and to determine whether NS5A has an effect on IFN-regulated gene expression. IFN-induced changes in gene expression were examined in a variety of human cell types, including cultured primary fetal hepatocytes, HeLa cells expressing wild-type or mutant forms of NS5A, and Huh7 cells expressing an HCV subgenomic replicon. These studies revealed IFN-responsive genes that were consistently induced in all cell types examined as well as cell type-specific responses. NS5A partially blocked the IFN-mediated induction of specific ISGs as well as the IFN-mediated induction of an ISRE-driven reporter gene. Through this mechanism, NS5A may partially attenuate the IFN response, which may be an important factor in HCV resistance to IFN.
MATERIALS AND METHODS
Cell lines and IFN treatment.
The construction of HeLa S3 cells expressing NS5A under the control of a tetracycline-regulated promoter was described previously (17). The cell lines used were HeLa pTRE-NS5A-1B (expressing wild-type NS5A), HeLa pTRE-NS5A-1B5 (expressing a PKR-binding-deficient mutant), and HeLa pTRE-4 (containing the pTRE vector alone). NS5A expression was induced by the removal of tetracycline from the culture medium. To measure the effects of NS5A on IFN-regulated gene expression, IFN-α/β (Hayashibara Biochemical Laboratories, Inc.; 400 IU/ml) was added to cells 8 h after induction of NS5A and RNA was isolated 16 h later as previously described (19). The optimal duration of IFN treatment was determined by a time course experiment in which HeLa pTRE-4 cells were treated with IFN-α/β (400 IU/ml) or mock treated with medium alone and incubated at 37°C for 2, 4, 6, 8, or 16 h. Total RNA was isolated from cells at each time point and used for microarray analysis. For comparison, total RNA was also isolated from HeLa pTRE-4 cells that were treated for 16 h with Intron-A (recombinant IFN-α2b) (Schering-Plough; 400 IU/ml). Huh7 cells harboring an HCV subgenomic replicon (32) were maintained in medium supplemented with G418 (0.8 mg/ml). For gene expression studies, replicon-containing cells were grown until approximately 70% confluent and then treated with IFN (400 IU/ml) for 16 h.
Fetal hepatocytes were isolated from tissue obtained from aborted second trimester fetuses (92 to 117 days of gestation) provided by the Central Laboratory for Human Embryology at the University of Washington. Excised fetal liver (2 to 4 g) was minced into small pieces in Seglen's collagenase buffer and digested with collagenase A (Roche Diagnostics; 3 mg/ml) at 37°C. The cells were then washed twice and suspended in Dulbecco's minimal Eagle's medium. Typical cell yields were 1 × 107 cells, which were 95% viable as determined by trypan blue dye exclusion. Hepatocytes were then plated on collagen-coated dishes in medium containing 5% fetal bovine serum. After a 4-h attachment period, the medium was changed to serum-free William's medium supplemented with epidermal growth factor (Becton Dickinson; 20 ng/ml), 10 mM nicotinamide, 0.2 mM ascorbic acid-2-phosphate, 20 mM HEPES, 17 mM NaHCO3, pyruvate (550 mg/liter), 14 mM glucose, 2 mM glutamine, 10 M dexamethasone, ITS+ premix (Becton Dickinson), and antibiotics. Cultures were maintained at 37°C at 6% CO2.
Adult hepatocytes were obtained from surgical resections (negative for hepatitis B and C virus). Tissue wedges (approximately 50 g) were trimmed and perfused with collagenase (1 mg/ml). The yield was approximately 1.7 × 107 cells, which were 85 to 90% viable as measured by trypan blue dye exclusion. The primary hepatocytes were infected (at 72 h after plating) with a virus stock obtained from the PA317 amphotrophic packaging cell line stably transfected with the plasmid pLXSN16 E6/E7, which contains the E6-E7 genes of human papillomavirus 16. Briefly, cells in 60-mm-diameter plates were infected with 250 μl of virus stock in 2.5 ml of medium containing Polybrene (4 μl/ml) and epidermal growth factor (20 μg/ml) for 24 h. The resulting cell line was designated HH2.
Microarray and Northern blot analysis.
Microarrays containing 4,608 human cDNAs were constructed by the University of Washington Center for Expression Arrays. The I.M.A.G.E. identification numbers and gene identities can be found at http://ra.microslu.washington.edu/. Plates 7 through 17 plus a control were used. First-strand cDNA synthesis, microarray hybridizations, and Northern blot analyses were performed as described previously (19). For each condition, duplicate slides were hybridized with the same probes, but with the fluorescent labels reversed, to control for dye-specific effects. In addition, each analysis was repeated independently for a total of four slides per condition (eight measurements per gene). Intensity values in Cy3 and Cy5 channels were extracted from each image, and the Cy3/Cy5 ratio was determined using Spot-on Image software. Data for all replicates were combined and normalized with custom software (Spot-on Unite). Briefly, Spot-on Unite normalizes the data, rejects outliers, and calculates the mean and standard deviation for each replicate measurement (19).
Data processing and analysis.
Using an Avalanche II scanner (Molecular Dynamics), microarrays were scanned at two wavelengths, and the resulting images were quantified using Spot-On Image, a spot-finding program (19). Raw data and sample information were then entered into Expression Array Manager, a custom-designed gene expression database that automatically uploads data and two-color images into the Rosetta Resolver system (Rosetta Biosoftware), a gene expression data analysis package that integrates a variety of sophisticated analysis tools, including hierarchical clustering analysis. The output from this type of analysis permits the simultaneous evaluation of multiple experiments and the visualization of coordinate patterns of gene regulation. In accordance with proposed standards (4), all data described in this report, including sample information, intensity measurements, gene lists, error analysis, microarray content, and slide hybridization conditions, are available in the public domain through Expression Array Manager at http://www.expression.washington.edu/public.
Luciferase reporter assays.
HeLa pTRE-4 cells were seeded at a density of 3 × 105 cells per 35-mm-diameter tissue culture dish and transiently cotransfected with 1.0 μg of either pISRE-Luc or pCMV-Luc (Stratagene) and with 0.2 or 1.0 μg of pTRE-NS5A-1B or pTRE-β-galactosidase (as a control). Expression of the firefly luciferase gene in pISRE-Luc is controlled by a basic promoter element (TATA box) plus 5 direct repeats of an ISRE (TAGTTTCACTTTCCC). Transfections were performed using SuperFect (Qiagen) according to the manufacturer's instructions. Cells were then treated with human IFN-α/β (400 IU/ml), incubated for 24 h, and lysed. An aliquot of each lysate was mixed with Luciferase Assay Reagent (Promega), and luciferase activity was measured in a Beckman-Coulter LS6500 scintillation system in the single-photon mode. The expression of NS5A and β-galactosidase (B-Gal) proteins was confirmed by immunoblotting using antibodies specific for NS5A (ID Labs) or β-Gal (Santa Cruz Biotechnology). Experiments were performed in triplicate.
RESULTS
IFN induces conserved and cell type-specific changes in gene expression.
Since an important goal of this study was to determine whether NS5A has an effect on IFN-regulated gene expression, we began our analyses by profiling the changes in gene expression that occur in response to IFN treatment. These experiments were performed using a variety of cell types, including HeLa cells, in which our NS5A-expressing cell lines were developed; Huh7 cells, which are used for the HCV replicon system; HH2 cells, an immortalized hepatocyte line; and cultured primary human fetal hepatocytes. An initial time course of IFN treatment using HeLa cells revealed that the greatest number of gene expression changes occurred 16 h after IFN treatment (data available at http://www.expression.washington.edu/public). Although some of these changes may be due to secondary effects of IFN or to factors such as differential RNA stability, this time point was judged to provide the best snapshot of IFN-mediated changes in gene expression and was used for all subsequent experiments in which we examined the ability of NS5A to alter these gene expression patterns.
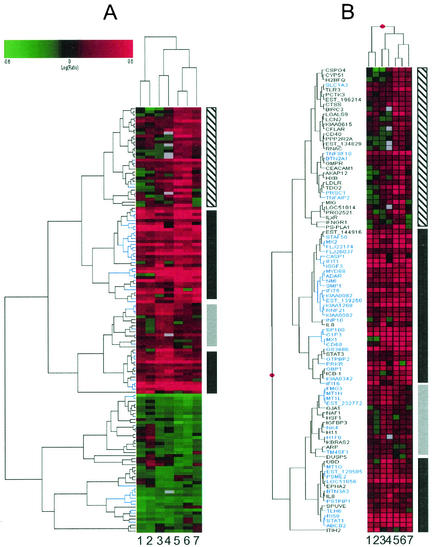
Agglomerative hierarchical clustering analysis of the union set of all differentially regulated genes revealed differences in the number and identity of differentially regulated genes in each of the four cell types as well as differences in the magnitude of gene expression changes (Fig. 1; additional data available at http://www.expression.washington.edu/public). However, some changes were common to all cell types and the gene expression profiles for cells of liver origin were more similar to one another (Fig. 1A, lanes 3 to 7) than they were to those of HeLa cells, which formed their own distinct cluster (Fig. 1A, lanes 1 and 2). In general, primary human fetal hepatocytes were the most responsive to IFN treatment and HeLa cells were the least responsive. A view of the ISGs identified by this analysis is shown in Fig. 1B, where genes that were significantly induced (P ≤ 0.005) in all cell types examined are highlighted in blue. These genes likely represent a core response to IFN that is conserved across cell types, and subsequent bioinformatics analyses were focused on this set of genes. A complete listing of differentially regulated genes and all primary data used to generate these clusters (including severalfold change in expression and error analysis) are available at http://www.expression.washington.edu/public.
FIG. 1.
IFN induces conserved and cell type-specific changes in gene expression. Lanes 1 and 2, HeLa cells; lanes 3 and 4; Huh7 cells; lanes 5 and 6, primary fetal hepatocytes; lane 7, HH2 cells. (A) Hierarchical clustering of the union set of genes that were differentially regulated in at least one cell type (P ≤ 0.005 for each pair of duplicate experiments). The scale at top indicates the magnitude of induction or repression (log10). Red bars represent genes that were induced by IFN, green bars represent genes that were repressed by IFN, black bars represent genes that were not differentially expressed, and gray bars represent genes that were below the intensity threshold in that experiment. IFN-induced changes in gene expression that were conserved across all cell types are represented by the vertical black bars to the right, primary fetal hepatocyte-specific changes in gene expression are indicated by the vertical hatched bar, and Huh7 cell-specific changes are indicated by the vertical gray bar. A magnified view of genes that were down-regulated by IFN is available at http://www.expression.washington.edu/public. (B) Magnified view of ISGs detected by microarray analysis. The symbols for genes that were induced by IFN in all cell types are highlighted in blue.
Genome analysis reveals novel ISREs.
Some of the ISGs identified by our microarray analyses are known to contain _cis_-acting DNA elements that participate in IFN-regulated transcription. However, for many of the genes we identified, little or no information is available regarding the sequence elements that regulate their responsiveness to IFN. As a means both to validate our microarray results and to look for alternative mechanisms of regulation, we conducted a search for ISRE-like sequences in genomic regions upstream of the 50 genes that were induced by IFN in all cell types. We reasoned that since these genes were the most consistently induced by IFN, they were the most likely to have recognizable IFN-responsive elements. To account for the possibility of alternative transcription initiation sites, we examined 1,000-bp regions upstream of both the predicted ATG start codon and the start of the 5′ untranslated region. Because of ambiguity about what constitutes a bona fide ISRE, we used a single consensus sequence that worked well in identifying known ISREs. Using this approach, we identified all previously described ISREs that occur upstream of known ISGs as well as new ISRE-like sequences in alternative locations upstream of known ISRE-containing genes such as PKR, GBP1, and MX1 (Table 1). Previously unidentified ISRE-like sequences were also found in the promoter regions of CASP1, GS3686, STAT1, IRF9, KIAA1268, and others. Since these genes were induced by IFN, it is likely that many of these sequences function as IFN-responsive elements, although this has not been systematically verified. It is also possible that these sequences do not mediate direct responsiveness to IFN but rather serve as binding sites for secondary factors, such as IRF1 (46). In all, 20 of the 50 IFN-regulated genes examined contained ISRE-like sequences in their promoter regions and 8 genes had more than one element. In contrast, no consensus IFN-responsive elements were found in genes that were down-regulated by IFN or in random 1,000-bp fragments, suggesting that ISGs are enriched for ISRE-like sequences. Only 10% of the more than 60 genes that were regulated by IFN in at least one cell type, but which failed to meet statistical criteria for differential expression in all cell types, had recognizable ISRE-like sequences.
TABLE 1.
Bioinformatic identification of candidate IFN-regulatory elementsa
| Gene | Position (nt) | Strand | Locus | Sequence |
|---|---|---|---|---|
| ABCB2 | ATG (−765) | Plus | 6p21 | GAAAGCGAAAGC |
| ABCB2 | ATG (−24) | Minus | GAAATCGAAAGC | |
| ABCB2 | ATG (−18) | Minus | GAAAGCGAAATC | |
| ADAR | ATG (−197) | Plus | 1q21.1 | GAAACGAAAGC |
| ADAR | ATG (−192) | Plus | GAAAGCGAAATT | |
| CASP1 | ATG (−438) | Minus | 11q23 | GAAATAGAAAC |
| CASP1 | ATG (−42) | Minus | GAAACTGAAAGT | |
| G1P3 | ATG (−627) | Minus | 1p35 | GAAAAAGAAAC |
| G1P3 | Exon 1 (−150) | Plus | GAAAATGAAACT | |
| G1P3 | Exon 1 (−109) | Plus | GAAAATGAAACT | |
| G1P3 | Exon 1 (−87) | Plus | GAAATAGAAAC | |
| GBP1 | Exon 1 (−128) | Minus | 1p22 | GAAACTGAAAGT |
| GS3686 | Exon 1 (−878) | Minus | 1p31 | GAAATGAAAGC |
| IFI16 | Exon 1 (−41) | Plus | 1q22 | GAAACGAAAGC |
| IFIT1 | ATG (−470) | Plus | 10q25 | GAAACTGAAAAT |
| IFIT1 | ATG (−208) | Minus | GAAAGTGAAACT | |
| INP10 | ATG (−287) | Plus | 4q21 | GAAAGTGAAACC |
| ISGF3 | ATG (−172) | Plus | 14q11 | GAAAGGAAAC |
| KIAA1268 | ATG (−79) | Plus | 3q21 | GAAACGAAAGC |
| MX1 | ATG (−218) | Minus | 21q22 | GAAAGAGAAAC |
| MX1 | Exon 1 (−101) | Minus | GAAACGAAAC | |
| MX1 | Exon 1 (−56) | Minus | GAAATGAAAC | |
| MYD88 | ATG (−98) | Plus | 3p22 | GAAAGCGAAAGC |
| NMI | Exon 1 (−78) | Plus | 2p24 | GAAAGTGAAATT |
| PRKR | Exon 1 (−7728) | Plus | 2p21 | GAAAACGAAACT |
| PRKR | Exon 1 (−3617) | Plus | GAAAGGAAAC | |
| PRKR | Exon 1 (−374) | Minus | GAAAGGAAAC | |
| PSME2 | ATG (−57) | Plus | 14q11 | GAAAGCGAAAGC |
| RI58 | ATG (−278) | Minus | 10q23 | GAAACCGAAACT |
| RI58 | ATG (−252) | Minus | GAAATCGAAACT | |
| RI58 | ATG (−237) | Minus | GAAACTGAAACT | |
| RNF21 | Exon 1 (−107) | Minus | 11p15 | GAAAGTGAAATT |
| STAT1 | Exon 1 (−492) | Minus | 2q32 | GAAAGCGAAACT |
| TNFSF10 | Exon 1 (−96) | Minus | 8p21 | GAAATGAAAGC |
We also noticed that a number of ISGs were located in close proximity to one another at several locations throughout the genome, suggesting the possibility of a coordinate mechanism of gene regulation based on local alterations in chromatin structure (42). Examples of such genes include IFIT1 and RI58 on chromosome 10q23.31, BTN2A1 and BTN3A3 on chromosome 6p21.1, MT1G, MT1H, and MT1L on chromosome 6q13, and STAF50/TRIM22 and RNF21/TRIM34 on chromosome 11p15. Given this observation, we devised a strategy to map our consensus ISRE sequence to the entire noncoding region of the human genome. This allowed us to look for additional clusters of ISRE-like sequences as well as to visualize in detail the positions of these sequences relative to genes in those same regions (methods and detailed analysis are to be presented elsewhere) (27). An example of the output obtained from this analysis for chromosome 10q23.31 as viewed using the University of California Santa Cruz genome browser (http://genome.ucsc.edu) (27) is available at http://www.expression.washington.edu/public. In addition to IFIT1 and RI58, both of which have ISREs, there were other genes in this region that contained ISRE-like sequences. For instance, IFIT4/ISG60, which is located directly upstream of the IFIT1 gene and which is a member of the same gene family (9), has two putative ISREs. Indeed, an alternative form of the IFIT4 gene has been described, which is consistent with the multiple EST species that correspond to this region. Thus, a genome-wide scan for ISRE-like sequences has the potential to reveal the positions of all putative ISREs (even those located in introns), locate hotspots of IFN-regulated genes that exhibit coordinate patterns of regulation, and provide information regarding the potential IFN responsiveness of genes not present on the microarray. We caution, however, that this type of bioinformatic approach is not meant to be a definitive analytical tool but rather is a high-throughput method to identify potential regulatory elements.
NS5A alters IFN-regulated changes in gene expression.
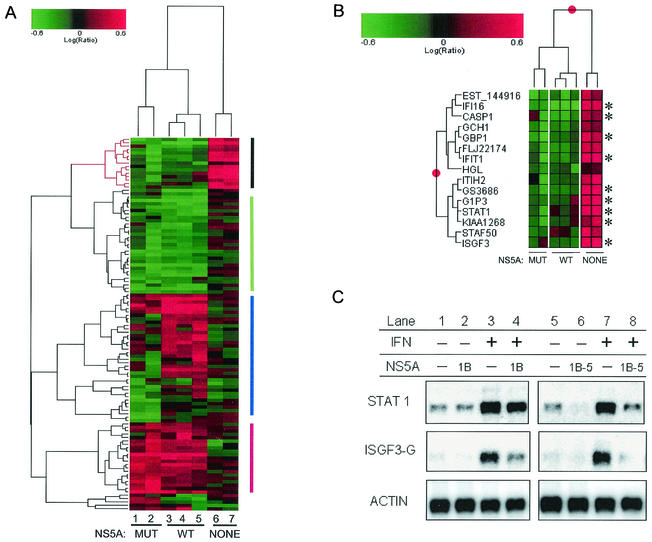
To determine whether NS5A has an effect on IFN-regulated gene expression, we performed microarray analyses on IFN-treated HeLa cells expressing either wild-type NS5A (NS5A-1B) or a mutant form of NS5A (NS5A-1B5) that contains multiple mutations within the ISDR and that does not bind to or inhibit PKR (15). Using hierarchical clustering analysis, we identified 14 genes that were induced more than twofold by IFN in the absence of NS5A expression (Fig. 2A, black bars). In contrast, when NS5A expression was induced, the IFN-mediated induction of all 14 of these genes was at least partially blocked (Fig. 2B). Both forms of NS5A can impose this block, although the induction of some genes, such as STAT1 and ISGF3 (IRF9), was more strongly repressed by NS5A-1B5. The results for STAT1 and ISGF3 (IRF9), both of which cooperate to bind the ISRE, were verified by Northern blot analysis (Fig. 2C). Since NS5A-1B and NS5A-1B5 had similar effects in these experiments, it is likely that regions of NS5A outside of the ISDR and PKR-binding domain are responsible for the effects of NS5A on IFN signaling.
FIG. 2.
NS5A alters IFN-regulated changes in gene expression. (A) Clustering of differentially expressed genes detected in IFN-treated cells in the presence or absence of wild-type or mutant forms of NS5A. Genes induced by IFN and down-regulated by NS5A are indicated by the vertical black bar to the right, genes down-regulated by both forms of NS5A are indicated by the vertical green bar, genes induced by both forms of NS5A are indicated by the vertical red bar, and genes that were regulated differently by NS5A-1B and NS5A-1B5 are indicated by the vertical blue bar. Genes were selected based on a ≥2-fold change and a P value of ≤0.005 in at least two experiments. Lanes 1 and 2, NS5A-1B5 (mutant); lanes 3 to 5, NS5A-1B (wild type); lanes 6 and 7, no NS5A. Each lane represents a unique experiment (and separate RNA isolation) using the cell line indicated. (B) Magnified view of ISGs whose expression was inhibited by NS5A. *, genes with known or putative ISREs (Table 1). Lane designations are the same as described for panel A. (C) Northern blot analysis of STAT1 and ISGF3G (IRF9) mRNAs during IFN treatment of HeLa cells expressing wild-type or mutant NS5A.
A further analysis of the effects of NS5A on IFN-treated cells revealed that in addition to attenuating the induction of ISGs, NS5A had a broad impact on cellular gene expression. Hierarchical clustering revealed two groups of differentially expressed genes that exhibited similar patterns of regulation in cells expressing either NS5A-1B or NS5A-1B5 (Fig. 2A, red and green bars). Conversely, other clusters contained genes that were differentially expressed in the presence of one form of NS5A but not the other (Fig. 2A, blue bar), perhaps due to differences in the PKR-binding properties of the two proteins. Gene expression profiling of NS5A-expressing cells in the absence of IFN treatment indicated that at least 200 genes were differentially regulated in response to NS5A expression (additional data are available at http://www.expression.washington.edu/public). Although a detailed analysis of these genes is beyond the scope of the work presented here, it was apparent that several signaling pathways were affected by NS5A expression. For instance, NS5A appeared to have a significant effect on calcium signaling, since the expression of multiple genes that encode for calcium-binding proteins, or which function in a calcium-dependent manner, was altered in response to NS5A expression. We also observed that NS5A expression resulted in the activation of a number of genes that cluster in a region of chromosome 19 (additional data available at http://www.expression.washington.edu/public), again suggesting the possibility of coordinate regulation related to chromatin structure.
NS5A inhibits the transcriptional activation of ISRE-containing promoters.
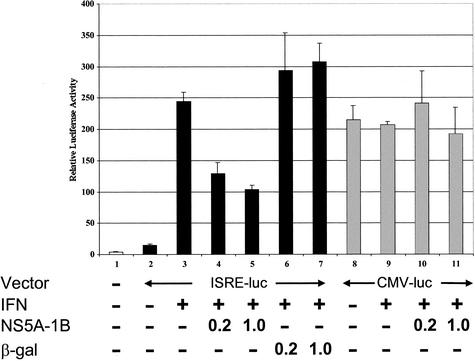
Two of the ISGs (ISGF3 and STAT1) that were negatively affected by NS5A are transcription factors that participate in ISRE-mediated gene expression. In addition, an examination of the promoters of the 14 ISGs whose expression was attenuated by NS5A revealed that 9 of these genes contained either known or predicted ISREs in their promoter regions (Fig. 2B and Table 1). Therefore, to more directly study the effect of NS5A on ISRE-containing promoters, we used a luciferase reporter assay to examine the ability of NS5A to inhibit transcription from an ISRE-driven promoter. IFN treatment induced the transcriptional activity of an ISRE-containing reporter gene, as measured by luciferase activity, by more than 10-fold (Fig. 3, lanes 2 and 3). In contrast, in the presence of NS5A, the ability of IFN to induce ISRE-dependent transcription was partially blocked (Fig. 3, lanes 4 to 7). This block was specifically mediated by NS5A, since a control construct encoding β-Gal failed to reduce the luciferase activity driven by the ISRE-containing promoter (Fig. 3, lanes 6 and 7). Importantly, the NS5A-mediated reduction of luciferase activity was specific to the ISRE-containing promoter, since NS5A had no effect on luciferase activity driven by a CMV promoter (Fig. 3, lanes 10 and 11). Therefore, NS5A does not appear to have a global effect on luciferase RNA translation but more likely functions to block ISRE-mediated transcription. The ability of NS5A to down-regulate ISRE-mediated transcription is consistent with our microarray results and provides independent evidence that NS5A can attenuate IFN-regulated gene expression.
FIG. 3.
NS5A inhibits ISRE-mediated gene transcription. HeLa cells were transfected with plasmid constructs as indicated. Lane 1, pTRE; lanes 2 and 3, pISRE-Luc; lane 4; pISRE-Luc + NS5A (0.2 μg of DNA); lane 5, pISRE-Luc + NS5A (1.0 μg of DNA); lane 6, pISRE-Luc + β-Gal (0.2 μg of DNA); lane 7, pISRE-Luc + β-Gal (1.0 μg of DNA); lanes 8 and 9, pCMV-Luc; lane 10, pCMV-Luc + NS5A (0.2 μg of DNA); lane 11, pCMV-Luc + NS5A (1.0 μg of DNA). Following transfection, cells were mock treated or treated with IFN for 16 h as indicated. Cell lysates were then prepared, and luciferase activity was measured. Luciferase activity is expressed as relative light units (RLU) on the y axis. All experiments were performed in triplicate. Mean values and error bars are shown.
An HCV subgenomic replicon has little effect on ISG expression.
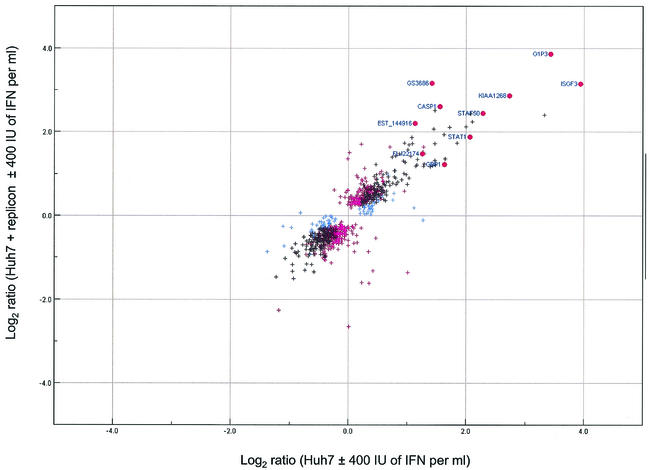
Given that NS5A was capable of down-regulating a subset of ISGs, we were interested in determining whether the same phenomenon occurred in Huh7 cells expressing an HCV subgenomic replicon. We began by comparing the gene expression profile of Huh7 cells expressing the replicon with that of Huh7 cells alone. Somewhat surprisingly, relatively few differences in gene expression were observed (additional data available at http://www.expression.washington.edu/public). We also noted that despite a high level of double-stranded RNA (dsRNA) production in cells expressing the replicon, there was an absence of ISG induction. The absence of ISG induction was not due to a lack of IFN responsiveness, since IFN treatment of Huh7 cells resulted in the induction of many ISGs (Fig. 1, lanes 3 and 4). However, there is a block in dsRNA signaling in cells expressing the replicon (37) which may inhibit the autocrine production of IFN, thereby blocking the induction of ISGs. We then examined gene expression profiles in IFN-treated cells to evaluate the effect of the replicon on the IFN-mediated induction of ISG expression (Fig. 4). For most ISGs, including the majority of those which were down-regulated by NS5A, the level of IFN-mediated induction was the same in replicon-containing cells as in cells lacking the replicon. However, the replicon did attenuate the IFN-mediated induction of ISGF3-γ and GBP1 to a level similar to that observed in NS5A-expressing cells (Fig. 4). Thus, compared to NS5A alone, the replicon had very little effect on ISG expression. The reasons for this difference are unknown but may be related to the IFN-sensitive phenotype of the replicon (14, 23) or the presence of other HCV proteins that might alter the IFN-attenuating properties of NS5A. It is also possible that the selection of replicons for replication efficiency affects nonreplicative functions of NS5A, including its effect on ISG expression.
FIG. 4.
IFN-regulated changes in gene expression in Huh7 cells in the presence and absence of an HCV subgenomic replicon are similar. A scatter plot of log2 expression ratios for differentially regulated genes (P ≤ 0.01) in Huh7 cells (x axis) and Huh7 cells harboring the replicon (y axis) in the presence or absence of IFN is shown. Blue crosses represent genes that were differentially expressed in IFN-treated Huh7 cells, magenta crosses represent genes that were differentially expressed in IFN-treated Huh7-replicon cells, and black crosses represent genes that were differentially expressed in both experiments. The ISGs that were down-regulated by NS5A in HeLa cells are highlighted by red circles. (Note that 4 of the 14 ISGs repressed by NS5A in HeLa cells were not induced by IFN in Huh7 cells).
DISCUSSION
Using global gene expression profiling, we identified a set of 50 genes that were induced by IFN in all cell types examined and another 60 genes that were induced in at least one cell type. In addition, we identified 31 genes that were repressed by IFN treatment. Different cell types differed in their response to IFN treatment, and of the cells tested, primary fetal hepatocytes exhibited the largest number of changes in gene expression in response to IFN (96 differentially regulated genes). We stress, however, that our analyses were not designed to identify all IFN-regulated genes, and despite the large number of differentially regulated genes identified in this study, these genes likely represent only a subset of the transcriptome that is responsive to IFN treatment. Analyses (using more comprehensive arrays) of other cell lines (G. K. Geiss, unpublished observations) (7, 8) or of cells overexpressing IFN-regulatory factors (21) have identified additional genes that are responsive to IFN treatment. Other variables, such as the amount or duration of IFN treatment, also influence the number and identity of IFN-regulated genes.
Bioinformatic analysis of ISG promoter regions revealed that 26 of the 110 ISGs identified had putative ISREs and that 20 of those (Table 1) were among the 50 ISGs induced in all cell types. We not only identified known ISREs in the promoter regions of genes such as PKR, MX1, and IFIT1 but also identified novel candidate ISRE-like sequences in the promoter regions of several other genes with known roles in innate immunity, including TLR3, TNFRSF10, and CASP1. A similar analysis of 20 ISGs by Su et al. (43), using a slightly different ISRE consensus sequence that allowed a single mismatch, identified a number of ISREs not found using our search criteria. It is therefore likely that additional ISRE-containing genes remain to be identified. We again note that although this approach provides a rapid means for the identification of candidate regulatory elements, biochemical validation of the function of any specific element is required to confirm its biological activity.
A genome-wide search for common DNA elements in the promoter regions of non-ISRE-containing ISGs, and refinement of the technique to include other relevant information (such as the locations of TATA boxes relative to ISRE sites), is currently under way. ISGs that do not contain an ISRE-like sequence may include genes that contain IFN-γ-activated sites as well as genes that are not directly induced by IFN or which are induced by some other factor that is a component of a signaling pathway that overlaps with IFN signaling, such as NF-κB or TRAIL (29). In addition, we noted that some ISGs cluster with one another at various regions throughout the genome. This observation raises the possibility that alterations in chromatin structure play a role in the coordinate regulation of multiple IFN-regulated genes. This hypothesis should be testable by chromatin immunoprecipitation analysis in combination with microarrays, a procedure for studying genome-wide protein-DNA interactions (30, 40).
We used HeLa cell lines (expressing either wild-type NS5A or a mutant form of NS5A that lacks the ability to bind to or inhibit PKR) to examine the effect on ISG expression of the presence of NS5A. In the absence of NS5A, IFN treatment induced the expression of 14 ISGs by more than twofold. The expression of either form of NS5A inhibited the IFN-mediated induction of all 14 of these ISGs. This finding suggests that NS5A mediates HCV resistance to IFN by attenuating specific aspects of IFN signaling. In addition, since both forms of NS5A had similar effects on ISG expression, this property of NS5A appears to be independent of ISDR sequence or its ability to inhibit PKR. This might partly explain why ISDR sequence does not always correlate with HCV resistance to IFN. Interestingly, in a similar analysis using Huh7 cells, Girard et al. demonstrated that NS5A reduced the IFN-mediated induction of 9 of 50 ISGs (20). However, none of the ISGs that were down-regulated by NS5A in Huh7 cells were among the 14 ISGs identified in our analyses. This difference most likely reflects the cell type-specific nature of the IFN response but might also be due to other factors, including differences in the parameters used to identify and select differentially expressed genes. Still, both studies point to similar roles for NS5A in mediating IFN resistance.
NS5A also had a profound effect on overall cellular gene expression in the absence of IFN treatment. Over 200 genes were differentially regulated in at least four of nine experiments. It is likely that the large number of genes altered by NS5A expression have a profound impact on multiple cellular pathways, which might partially explain the diverse set of biological functions that have been assigned to NS5A in various systems. By combining gene expression analysis with NS5A functional studies, such as analysis of cell growth characteristics or tumorigenic potential (17), it might be possible to identify sets of genes that contribute to these phenotypes.
We also compared the expression profiles of genes induced by NS5A in HeLa cells with those of Huh7 cells expressing an HCV subgenomic replicon. The effect of the replicon on the IFN-mediated induction of ISG expression in Huh7 cells was much less pronounced than that observed for NS5A alone. However, the replicon did down-regulate two ISGs, including ISGF3_-_γ, a subunit of the ISRE-binding complex, and GBP1, a guanylated binding protein of unknown function. Again, the differences observed in these systems might be due to a number of factors, including the level of NS5A expression, cell-specific factors, or the presence of other HCV proteins. These factors may also contribute to the disparate results that have contributed to the controversy surrounding the role of NS5A in mediating IFN resistance (44). Moreover, the high error rate of the HCV RNA-dependent RNA polymerase introduces adaptive mutations in the replicon, which can result in distinct properties with respect to replication efficiency (3, 28, 31) and dsRNA signaling (37). Not surprisingly, some of these mutations have been mapped to NS5A. The results of this study, in conjunction with previous work, point to the need for a way of comparing results from the diverse surrogate systems used to investigate the biology of HCV. Clustering of gene expression profiles from these different systems, ideally to a standard reference sample, may eventually allow us to predict which system, if any, is most similar to a bona fide HCV infection either in tissue culture or in patients.
Acknowledgments
We thank Jeff Furlong for development of Expression Array Manager, Michael Torgov for his contributions to microarray analysis of IFN-treated cells, and Haruhisa Nakao for the NS5A-expressing cell lines. We also acknowledge the University of Washington Center for Expression Arrays for microarrays and software.
This investigation was supported by Public Health Service grants AI47304 and AI48214 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Aizaki, H., S. Saito, T. Ogino, N. Miyajima, T. Harada, Y. Matsuura, T. Miyamura, and M. Kohase. 2000. Suppression of interferon-induced antiviral activity in cells expressing hepatitis C virus proteins. J. Interferon Cytokine Res. 20**:**1111-1120. [DOI] [PubMed] [Google Scholar]
- 2.Battaglia, A. M., and K. O. Hagmeyer. 2000. Combination therapy with interferon and ribavirin in the treatment of chronic hepatitis C infection. Ann. Pharmacother. 34**:**487-494. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290**:**1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. P. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME) toward standards for microarray data. Nat. Genet. 29**:**365-371. [DOI] [PubMed] [Google Scholar]
- 5.Certa, U., M. Seiler, E. Padovan, and G. C. Spagnoli. 2001. High density oligonucleotide array analysis of interferon-α2a sensitivity and transcriptional response in melanoma cells. Br. J. Cancer 85**:**107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, K. M., J. Lee, J. E. Kim, O. K. Song, S. Cho, J. Lim, M. Seedorf, B. Hahm, and S. K. Jang. 2000. Nonstructural protein 5A of hepatitis C virus inhibits the function of karyopherin β3. J. Virol. 74**:**5233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Der, S. D., A. Zhou, B. R. G. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95**:**15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69**:**912-920. [PubMed] [Google Scholar]
- 9.de Veer, M. J., H. Sim, J. C. Whisstock, R. J. Devenish, and S. J. Ralph. 1998. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics 54**:**267-277. [DOI] [PubMed] [Google Scholar]
- 10.Di Bisceglie, A. M. 1998. Hepatitis C. Lancet 351**:**351-355. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Maruno, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334**:**77-81. [DOI] [PubMed] [Google Scholar]
- 12.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murankami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. J. Clin. Investig. 96**:**224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francois, C., G. Duverlie, D. Rebouillat, H. Khorsi, S. Castelain, H. E. Blum, A. Gatignol, C. Wychowski, D. Moradpour, and E. F. Meurs. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74**:**5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-α inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82**:**723-733. [DOI] [PubMed] [Google Scholar]
- 15.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S.-L. Tan, M. Dossett, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18**:**5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale, M., Jr., M. J. Korth, N. M. Tang, S.-L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230**:**217-227. [DOI] [PubMed] [Google Scholar]
- 17.Gale, M., Jr., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 73**:**6506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Sastre, A. 2002. Mechanisms of inhibition of the host interferon α/β-mediated antiviral responses by viruses. Microbes Infect. 4**:**647-655. [DOI] [PubMed] [Google Scholar]
- 19.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75**:**4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard, S., P. Shalhoub, P. Lescure, A. Sabile, D. E. Misek, S. Hanash, C. Bréchot, and L. Beretta. 2002. An altered cellular response to interferon and up-regulation of interleukin-8 induced by the hepatitis C viral protein NS5A uncovered by microarray analysis. Virology 295**:**272-283. [DOI] [PubMed] [Google Scholar]
- 21.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76**:**5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15**:**259-267. [DOI] [PubMed] [Google Scholar]
- 23.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75**:**8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, Y., H. Nakao, S.-L. Tan, S. J. Polyak, P. Neddermann, S. Vijaysri, B. L. Jacobs, and M. G. Katze. 2002. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J. Virol. 76**:**9207-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoofnagle, J. H. 1999. Management of hepatitis C: current and future perspectives. J. Hepatol. 31**:**264-268. [DOI] [PubMed] [Google Scholar]
- 26.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2**:**675-687. [DOI] [PubMed] [Google Scholar]
- 27.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12**:**996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreiger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75**:**4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar-Sinha, C., S. Varambally, A. Sreekumar, and A. M. Chinnaiyan. 2002. Molecular cross-talk between the TRAIL and interferon signaling pathways. J. Biol. Chem. 277**:**575-585. [DOI] [PubMed] [Google Scholar]
- 30.Liu, X. S., D. L. Brutlag, and J. S. Liu. 2002. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20**:**835-839. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75**:**1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Körner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285**:**110-113. [DOI] [PubMed] [Google Scholar]
- 33.Majumder, M., A. K. Ghosh, R. Steele, R. Ray, and R. B. Ray. 2001. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75**:**1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, M. D., H. R. Rosen, G. I. Marousek, and S. Chou. 2002. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig. Dis. Sci. 47**:**1195-1205. [DOI] [PubMed] [Google Scholar]
- 35.Noguchi, T., S. Satoh, T. Noshi, E. Hatada, R. Fukuda, A. Kawai, S. Ikeda, M. Hijikata, and K. Shimotohno. 2002. Effects of mutation in hepatitis C virus nonstructural protein 5A on interferon resistance mediated by inhibition of PKR kinase activity in mammalian cells. Microbiol. Immunol. 45**:**829-840. [DOI] [PubMed] [Google Scholar]
- 36.Paterson, M., C. D. Laxton, H. C. Thomas, A. M. Ackrill, and G. R. Foster. 1999. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology 117**:**1187-1197. [DOI] [PubMed] [Google Scholar]
- 37.Pflugheber, J., B. Fredericksen, R. J. Sumpter, C. Wang, F. Ware, D. L. Sodora, and M. J. Gale. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. USA 99**:**4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podevin, P., A. Sabile, R. Gajardo, N. Delhem, A. Abadie, P. Y. Lozach, L. Beretta, and C. Brechot. 2001. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology 33**:**1503-1511. [DOI] [PubMed] [Google Scholar]
- 39.Radaeva, S., B. Jaruga, F. Hong, W. H. Kim, S. Fan, H. Cai, S. Strom, Y. Liu, O. El-Assal, and B. Gao. 2002. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology 122**:**1020-1034. [DOI] [PubMed] [Google Scholar]
- 40.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290**:**2306-2309. [DOI] [PubMed] [Google Scholar]
- 41.Satoh, J., and Y. Kuroda. 2001. Differing effects of IFNβ vs IFNγ in MS: gene expression in cultured astrocytes. Neurology 57**:**681-685. [DOI] [PubMed] [Google Scholar]
- 42.Smale, S. T., and A. G. Fisher. 2002. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 20**:**427-462. [DOI] [PubMed] [Google Scholar]
- 43.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99**:**15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, S.-L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284**:**1-12. [DOI] [PubMed] [Google Scholar]
- 45.Tan, S.-L., H. Nakao, Y. He, V. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 96**:**5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, N., T. Kawakami, and T. Taniguichi. 1993. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol. 13**:**4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. M. Wen, A. E. Gorbalenya, S. B. Hwang, and M. C. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263**:**30-41. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization. 2002. Global surveillance and control of hepatitis C. J. Viral Hepatitis 6**:**35-47. [PubMed] [Google Scholar]
- 49.Zimmer, R., and P. Thomas. 2002. Expression profiling and interferon-beta regulation of liver metastases in colorectal cancer cells. Clin. Exp. Metastasis 19**:**541-550. [DOI] [PubMed] [Google Scholar]