Systematic Identification of Novel Protein Domain Families Associated with Nuclear Functions (original) (raw)
Abstract
A systematic computational analysis of protein sequences containing known nuclear domains led to the identification of 28 novel domain families. This represents a 26% increase in the starting set of 107 known nuclear domain families used for the analysis. Most of the novel domains are present in all major eukaryotic lineages, but 3 are species specific. For about 500 of the 1200 proteins that contain these new domains, nuclear localization could be inferred, and for 700, additional features could be predicted. For example, we identified a new domain, likely to have a role downstream of the unfolded protein response; a nematode-specific signalling domain; and a widespread domain, likely to be a noncatalytic homolog of ubiquitin-conjugating enzymes.
Large proteins are often composed of domains. These are polypeptide regions that adopt compact three-dimensional (3D) structures and are often found in diverse molecular contexts (Janin and Chothia 1985). The independent evolutionary histories of domains found within the same protein lead to an assumption that the domain is the fundamental unit of protein structure and function (Doolittle 1995). Domains are most readily observable in known 3D structures, but because of the relative paucity of available structural data, the majority of protein domain families have been identified first by sequence analysis. Many domains are ‘genetically mobile’, meaning that they can be found associated with different domain combinations in different proteins. The term ‘module’ is sometimes used to distinguish between mobile domains and those that are invariably found in identical molecular contexts.
Sequence characterization of domain families represents a first step toward the determination of their 3D structures and molecular functions. Domain identification from sequence is usually performed on a case-by-case basis, by applying a variety of automatic methods supplemented with careful manual analysis. The number of protein domain families characterized from sequence has been increasing steadily over the years and has led to the development of Web-based resources such as SMART and Pfam (Schultz et al. 1998, Bateman et al. 2000) for effective and reliable domain identification.
We have systematically searched for new domain families, using proteins annotated by the SMART (Simple Modular Architecture Research Tool) database of domains as our starting point. We have targeted our strategy to all proteins that contain at least one of 107 types of predominantly nuclear domains in the SMART collection. Crucial to our technique is the accurate knowledge of known domain boundaries provided by databases such as SMART and Pfam (Schultz et al. 1998, Bateman et al. 2000). Using sequence regions not covered by previously characterized domains, we have searched for homologs in nonredundant sequence databases and used previously computed domain architectures to determine which of the initial search regions could correspond to new domain families. A manual analysis of the various candidate families led to the final characterization of novel domain types and their sequence borders.
RESULTS
Classification of the Novel Domains
The protocol described earlier revealed a variety of novel domains that could be classified into four broad categories:
Fifteen novel domain families with representatives in diverse molecular contexts in different species (Table 1, Part A). Of these, three have recently been described on separate occasions (Table1, Part A, Callebaut et al. 2001; Clissold and Ponting 2001;Doerks et al. 2001).
Three domain families were found to be specific to single or closely related species (Table 1, Part B).
Seven further domain families are likely to be divergent members of previously recognized families, with weak (but not statistically significant) similarity to previously described domains. (One of these, the BED domain, has been recently published independently (Aravind 2000)) (Table 1c).
Three additional families were recognized as representing family-specific N or C-terminal extensions of previously known domains (Table 1, Part D). These regions always co-occur with a particular neighboring domain, although their domain context within the protein as a whole may vary. Because of their size, they are likely to have well-defined structures, but might only exist in the context of the domain that they extend. In three of these cases, the extension is only found in closely related species. We do not consider these sequence families to be modules, and we do not discuss them further.
Table 1.
Table of Novel Domains
| Domain | Description | Length (AS) | Sec.struct.pred. | Pred.function | No. of proteins | Associated domains | Species | Acc. no. of a representative sequence (domain borders) |
|---|---|---|---|---|---|---|---|---|
| Part A.—Domains Present in Different Species | ||||||||
| JmjC* | Jumonji related family | 100 | β | Metallo-enzymes | 140 | BRIGHT, jmjN PHD, FBOX, LRR, C2, TPR PLAc, CXXC†, ZnF_C2H2 | Eu, y, a, c, d, h | O14607§ (1042–1205) |
| CSZ | Domain in chromatin remodeling S1 domain containing and Zinc finger proteins | 750 | α/β | DNA-binding, Chromatin modulation | 35 | S1, SH2, C2HC, HhH | Eu y, a, c, d, h | P34703 (389–1120) |
| RPR | Proteins involved in regulation of nuclear pre-mRNA | 120 | α | Protein-interaction | 40 | RRM, PWWP, SURP†, G-Patch | y, a, c, d, h | Q9SJQ7 (88–225) |
| DDT* | Different transcription and chromosome remodeling factors | 60 | α | DNA-binding | 30 | AT_Hook, PHD, HOX, BROMO, MBD | y, a, c, d, h | Q9UIG2§ (102–161) |
| TLDc | TBC, LysM and other proteins | 220 | α/β+β | Enzyme | 30 | TBC, LysM, R3H, FBOX | y, a, c, d, h | Q9VNA1§ (1163–1325) |
| PUG | Protein knases, UBA or UBX domain containing proteins and glycanases | 60 | α/β | RNA-binding | 25 | C2H2, UBA, TGc, UBX, S_TKc, STYKc | y, a, c, d, h | Q9MAT3 (323–386) |
| HSA | Helicases and SANT domains | 70 | α | DNA-binding | 20 | SANT, BROMO DEXDc, HELlc | y, a, c, d, h | P25439§ (501–573) |
| PSP | Proline-rich, in spliceosome associated proteins | 60 | α | RNA- or snRNP-binding | 15 | SAP, C2HC | y, a, c, d, h | O16997 (200–357) |
| FYRN | Trithorax and X-chromosome inactivating proteins | 40 | α/β | Unknown | 25 | PHD, SET, PWWP | a, c, d, h | Q24742§ (1869–1914) |
| FYRC | Trithorax and X-chromosome inactivating proteins | 90 | α/β | Unknown | 25 | PHD, SET, PWWP | a, c, d, h | Q24742§ (3495–3583) |
| RUN* | TBC, PH, FYVE and other proteins | 65 | α | GTPase signalling | 40 | DENN†, TBC, PLAT, PH, C1, FYVE, GST, SH3 | c, d, h | BAB14033 (115–178) |
| TCH | Transcription factors and CHROMO domain helicases | 50 | α/β | Unknown | 20 | CHROMO, PHD, TFSM2, DEXDc, HELIc, SANT, BROMO | c, d, h | O15025§ (882–931) |
| DZF | DSRM or ZnF_C2H2 domain containing proteins | 250 | α/β | Unknown | 40 | C2H2, DSRM | c, d, h | O88531 (762–1016) |
| NEUZ | Domain in neuralized-like proteins | 120 | β | Unknown | 10 | SOCS, RING, SPRY, SH2 | c, d, h | Q19299 (199–321) |
| ZnF_TTF | Domain in transposases and transcription factors | 100 | α + β | Metal-binding | 20 | KRAB, BTB | a, d, h | Q9ZWT4 (100–199) |
| Part B.—Domains Species–Specific | ||||||||
| FBD | Domain in FBOX and other domain containing plant proteins | 80 | α/β | Unknown | 160 | FBOX, LRRcap, BRCT, AAA | a | Q9LXJ7§ (304–382) |
| ZnF_PMZ | Plant mutator transposase zinc finger domain | 27 | α/β | Metal-binding | 125 | AT_Hook, ZnF_C2HC, PHD | a | Q9SH73 (3212–3239) |
| SPK | SET and PHD domain containing proteins and protein kinases | 120 | α/β | Protein-interaction | 40 | SET, ICE_p10†, ICE_p20†, ZnF_C2HC, PHD, STYKc | c | Q9XU06 (139–250) |
| Part C.—Domains, Newly Recognized Divergent Subfamilies | ||||||||
| ZnF_BED* | BED zinc finger, Related to C2H2/C2H2 zinc fingers (based on pattern similarity) | 60 | β | Metal binding | 50 | AT_Hook, PTPc_DSPc | y, a, c, d, h | Q9LWM2 (169–224) |
| CPDc | Catalytic domain of ctd-like phosphatases, related to phosphatase superfamily (based on pattern similarity) | 120 | α/β | Phosphatase | 70 | BRCT, DSRM, UBQ | y, a, c, d, h | Q9PTJ8 (93–236) |
| RWD | RING finger and WD repeat containing proteins and DEXDc helicases, related to the UBCc domain (revealed by hmm searches) | 110 | α/β | Protein-interaction | 60 | S_TKc, RING, WD, UPF29†, DEXDc, HELIc | y, a, c, d, h | Q9QZ05§ (25–137) |
| BTP | Bromodomain transcription factors and PHD domain containing Proteins, related to archaeal histone-like transcription factors, defined by PFAM (revealed by PSI-Blast results with less significance (E = 0.041)) | 90 | α | DNA-binding | 25 | AT_Hook, BROMO, PHD | y, a, c, d, h | Q9S7R9 (41–131) |
| MADF | Zinc finger, PHD domain and WD repeats containing proteins, related to SANT domain (after the second iteration Q9SR68 bridges to SANT domains (E = 0.002)) | 90 | α | DNA- or Potein-binding | 60 | C2H2, PHD, WD | Virus, a, c, d | Q9V5Y9 (22–110) |
| Znf_DBF | Zinc finger in DBF-like proteins, related to C2H2 zinc fingers (revealed by pattern similarity and hmm searches, E value = 1.4) | 50 | α | Metal-binding | 10 | BRCT, AT_Hook | y, d, h | O93843 (590–638) |
| CHK | C4-zinc finger and HLH domain containing kinase subfamily of choline kinases (after the second iteration P35790 bridges to choline kinases, defined by PFAM (E = 0.003)) | 200 | α/β | Enzyme | 70 | ZnF_C4, HLH, i.c† | Eu, c, d | Q9VBT6 (129–321) |
| Part D.—Family Specific Extensions of Known Domains | ||||||||
| AWS | Associated with SET domain, subdomain of PRESET‡ (hmm searches, E value = 0.52) | 50 | α/β | Histone modification | 25 | SET, PWWP, AT_Hook, WW, PHD, POSTSET, BAH | y, a, c, d, h | P46995 (63–119) |
| POX | Domain associated with HOX-domains | 50 | α | Unknown | 20 | HOX | a | Q38897 (199–337) |
| PRE_C2HC | Associated with zinc fingers | 70 | α/β | Unknown | 15 | ZnF_C2HC | d | O44939§ (546–616) |
Alignments of the novel domains, the proteins they are found in, and their phyletic distribution are publicly available in the SMART database (http://smart.embl-heidelberg.de/).
Of the total 28 regions discovered, 8 were found by simple single-passBLAST searches. For the remaining 20,PSI-BLAST searches were necessary to provide statistically significant links between proteins with different domain architectures. This is broadly consistent with the reported three-fold sensitivity of PSI-BLAST overBLAST (Park et al. 1998).
Conserved protein domains are most useful when they can be used to make predictions of likely function. For the domains presented here, this was possible to varying degrees. We provide three examples to illustrate the more important categories described earlier, and show the types of (necessarily conjectural) functional information that can be inferred from the present identifications.
A Widespread Module in Diverse Species: A Novel Domain in Peptide N-glycanases and Other Putative Nuclear Proteins
The majority of our novel domains are found in diverse species and in different protein contexts without significant sequence similarity to other domains. A particularly interesting example is described here.
A hypothetical Arabidopsis protein (SpTREMBL accession:Q9MAT3) is predicted to contain two N-terminal zinc finger motifs (ZnF_C2H2), followed by a UBA domain (Hofmann and Bucher 1996). A predicted coiled-coil region links this to a C-terminal half that contains no currently described domains.PSI-BLAST searches initiated with this C-terminal region show significant sequence similarity (E-value <10–5) to UBX domain-containing proteins and metazoan homologs of peptide:N-glycanases (PNGases).
Searching of preliminary protein predictions from the Plasmodium falciparum genome, with the conserved region identified in an_Arabidopsis_ sequence (SpTREMBL accession no. Q9FKI1), revealed an additional association with a UBCc domain-containing protein (E-value 3 × 10–4).
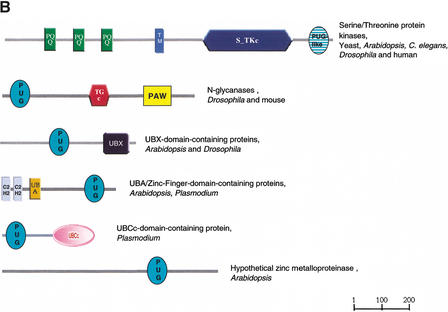
We refer to these homologous regions as PUG domains, after thePeptide:N-Glycanases and other putative nuclearUBA or UBX domain-containing proteins. PNGases are believed to have a role in the unfolded protein response (UPR) (Suzuki et al. 2000). The UPR results in increased levels of transcription of endoplasmic reticulum (ER)-resident protein-coding genes, following accumulation of unfolded proteins in the ER. The PUG domain is found to co-occur in proteins with three domains that are central to ubiquitin-mediated proteolysis: UBA, in_Arabidopsis_, UBCc in Plasmodium, and UBX in mammals and Arabidopsis. This indicates that PUG domain-containing proteins might link the UPR to ubiquitin-mediated protein degradation. Other links between the UPR and UBQ-mediated proteolysis have been shown previously (Travers et al. 2000).
The candidate orthologs of PNGases in Saccharaomyces cerevisiae, Saccharomyces pombe, and Arabidopsis_do not appear to encode this domain, indicating its presence in these proteins is a metazoan innovation. Of these putative PNGases, only the_S. cerevisiae protein has been directly characterized; it appears to be localized to the nucleus, with a lower level occurring in the cytosol (Suzuki et al. 2000). As the apparent orthologs in metazoan genomes appear to be present singly, rather than as multiple paralogs (which might imply functional variation), it seems likely that the proteins encoded by them will have a similar cellular localization.
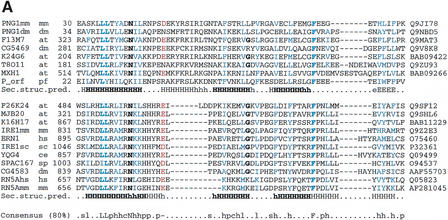
Additional HMMer2 searches, using an HMM derived from these PUG domain sequences, showed marginal similarity to IRE1p-like kinases (SpTREMBL accession: Q9SHL6) (_E_-value: 0.21) within a region known to be homologous to the C-terminal tail of 2‘-5′ oligo (A)-dependent ribonuclease (Zhou et al. 1993) (see Fig.1). Although of only marginal significance, the similarity also extends to cellular function because IRE1p-like kinases are known to initiate the UPR (Shamu and Walter 1996). The C-terminal tail of IRE1p is required for induction of the UPR (Shamu and Walter 1996), and has been shown to possess site-specific endoribonuclease activity (Sidrauski and Walter 1997). This activity is consistent with the C-terminal location for RNase activity found in its homolog, 2‘-5′ oligo (A)-dependent ribonuclease (Bork and Sander 1993). Consequently, we tentatively suggest the presence of divergent PUG domains in the C termini of IRE1p-like kinases.
Figure 1.
(A) Multiple sequence alignment of PUG domains of N-glycanases (PNG1mm, PNG1dm), UBX domain-containing proteins (F13M7, CG5469), HOX domain containing proteins (F3M18, MLN1), UBA/Zinc-finger-domain-containing proteins (K24G6, T8011), and hypothetical zinc metalloproteinase (MXH1) and multiple sequence alignment of PUG-like domains in serine/threonine protein kinases / RNAses (F26K24, MJB20, K16H17, IRE1mm, ERN1, IRE1sc, YQG4, SPAC167, CG4583, RN5Ahs, RN5Amm). First column, protein names; second column, species names (at, Arabidopsis thaliana; ce,Caenorhabditis elegans; dm, Drosophila melanogaster; hs, Homo sapiens; mm, Mus musculus; pf,Plasmodium falciparum; sc, _Saccharomyces cerevisiae;sp, Schizosaccharomyces pombe); third column, start of the domain in the respective sequences; rightmost column, database accession numbers. Conserved positively charged residues are shown in pink; conserved hydrophobic residues are shown in blue; other conserved residues are shown in bold. The predicted secondary structure taken from the consensus of the alignments (B/H, strand/helix predicted with expected average accuracy >82%; b/h, strand/helix predicted with expected average accuracy <82%) (Rost et al. 1994) is shown_below, respectively (consistent secondary structure in bold letters). The consensus sequence (conserved in 80% of the sequences) for both alignments is shown below; s, l, p, h, c, -, L, N, and F indicate small, aliphatic, hydrophobic, polar, charged, negatively charged residues, conserved Leucines, Asparagine, and Phenylalanin. (B) Domain architecture of proteins containing the PUG domain (green) and the PUG-like domain (green dark horizontal pattern). Only proteins with distinct modular organizations are shown. The domain names are those of the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de) (Schultz et al. 1998, 2000). C2H2, zinc finger C2H2 DNA-binding domain; PAW, domain in PNGases and other worm proteins; PQQ, β-propeller repeat; S_TKc, serine/threonine protein kinase catalytic domain; TGc, transglutaminase/protease-like homologs catalytic domain; UBA, biquitin-associated domain; UBCc, catalytic domain of ubiquitin-conjugating enzymes; UBX, domain present in ubiquitin regulatory proteins; TM, transmembrane region.
Further analysis of the metazoan PNGase sequences revealed a conserved region that is also present in multiple copies in hypothetical_Caenorhabditis elegans_ proteins (e.g., four copies in C17B7.5). This domain was not found in the initial rounds of searching because it does not occur with any of our starting set of nuclear domains. We have included this domain in the SMART collection and have named it PAW (domain present in PNGases and other worm proteins).
Novel Modules Found in Narrow Phyletic Ranges: A Nematode-Specific Putative Signaling Domain in C. elegans
Lineage-specific expansions of protein domain families (i.e., a large increase in the number of a particular domain in one genome compared with other genomes) are a widespread phenomenon (e.g., International Human Genome Sequencing Consortium 2001). In extreme cases, it may not be possible to establish links between a domain that is widespread in one organism and known domains seen in other species. Such cases may represent genuine ‘invention’ of new domains, or, perhaps more likely, instances where the tempo of molecular evolution has risen to the extent that sequence similarity with known domains is no longer detectable. Alternative scenarios of massive loss from other lineages are less parsimonious. Three (i.e., ∼11%) of our new domains appear to occur in very restricted phylogenetic lineages; these exclude species-specific N- or C-terminal extensions of known domains (see Table 1, Part B).
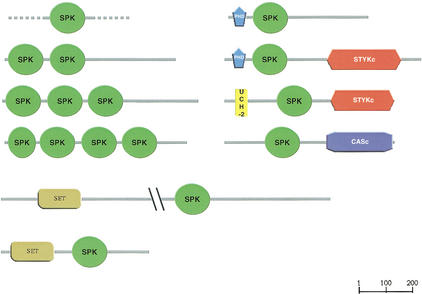
PSI-BLAST searching with the region C-terminal to a SET domain (Cui et al. 1998) of the hypothetical protein Y43F11A.5 (SpTREMBL accession: Q9U2G8) detected a novel domain found in many different predicted proteins from C. elegans but thus far in no other species. The domain is ∼120 residues in length, and found associated with the catalytic domain of caspases (CASc), protein kinases of undetermined specificity (STYKc), and the SET methyltransferase domain. Multiple tandem copies of the domain may be present in the same sequence (Fig. 2). We named this domain SPK [associated with SET,PHD (Aasland et al. 1995), protein Kinase]. The alignment is provided on the Web (seehttp://www.embl-heidelberg.de/∼doerks/alignment_fig3.html/)
Figure 2.
Domain architecture of proteins containing the SPK domain. Only proteins with distinct modular organizations are shown. The domain names are those of the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de) (Schultz et al. 1998, 2000). CASc, catalytic domain of caspases; PHD, PHD C4HC3 zinc finger; SET, (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain; STYKc, catalytic domain of protein kinases. The UCH-2 (ubiquitin carboxy-terminal hydrolase family 2) domain is defined by Pfam (Bateman et al. 2000)
Further analysis of nucleic acid sequence databases revealed SPK domains in the Caenorhabditis briggsae sequence, in regions for which no proteins have been predicted (e.g., NCBI GI:11095060, data not shown). No other species were found to contain the domain. It is possible that the domain exists in nematode lineages other than Caenorhabditis, but is simply not found due to insufficient sequence coverage of these species.
The association of SPK with SET, PHD, catalytic protein kinases or caspase domains (see Fig. 2) hints at an important role in metabolic, developmental, or evolutionary processes that are unique to_Caenorhabditis_. However, none of the putative proteins in which the domain has been found have been characterized by any experimental technique other than RNAi screening. All homologs tested by RNAi are wild type according to wormbase (http://www.wormbase.org/). This technique would not be expected to reveal more subtle phenotypes associated with later developmental stages.
Modules in New Contexts: A Noncatalytic Subfamily of Ubiquitin-Conjugating Enzyme Homologs
The protocol presented here detects regions of homology between sequences where no domains have previously been assigned. Some of our newly identified regions appear to be distantly related to known domains, but correspond to new molecular contexts. Such cases indicate potential changes of domain function or add new insights to the function of the proteins in which the domain has been newly identified. An increasing number of known domains are being realized as members of wider superfamilies because of the availability of 3D structures. For example, the UBX domain has recently been reclassified as a subfamily of the ubiquitin fold superfamily (Buchberger et al. 2001). In addition to protein structure determination, carefully applied sensitive sequence searching methods can also provide such insights. This is exemplified by the following example detected in this study.
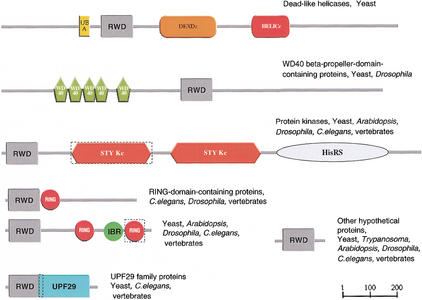
The mouse GCN2 eIF2α kinase and histidyl-tRNA synthetase (SpTREMBL accession: Q9QZ05) is an essential component of translation control (Jentsch et al. 1991; Sattleger et al. 1998). A PSI-BLASTsearch initiated with the region N-terminal to an inactive protein kinase domain (see Fig. 3) in the GCN2 protein revealed significant similarity to presumed orthologs in other eukaryotic species from yeast to vertebrates. FurtherPSI-BLAST iterations and additional HMM searches reveal significant similarity to WD-repeat-containing proteins; yeast DEAD (DEXD)-like helicases; UPF0029, an uncharacterized protein family from the Pfam database (accession no. PF01205); a range of hypothetical proteins; and many RING finger-containing proteins. We called the newly defined region RWD after the better characterized RING finger and WD-domain-containing-proteins andDEAD-like helicases. PSI-BLAST searches initiated with different seeds also revealed homology with ubiquitin-conjugating enzymes (UBCc) domain, (e.g. SpTREMBL acc: Q94721hits Q9SDY5 on iteration 3, _E_value = 9 × 10–4), although the catalytic cysteine critical for ubiquitin-conjugating activity is not conserved in most members of the novel subfamily (seehttp://www.embl-heidelberg.de/∼doerks/aligment_fig4.html/)). This observation is particularly interesting in light of previous experimental studies on A07 (SpTREMBL accession: Q9QZR0), a protein that includes both an RWD and a RING finger domain, that have shown that a region between 85 and 363 amino acids in A07 (including the RING finger) binds ubiquitin-conjugating enzyme E2 and acts as a substrate for E2-dependent ubiquitination (Lorick et al. 1999).
Figure 3.
Domain architecture of proteins containing the RWD domain. Only proteins with distinct modular organizations are shown. The domain names are according to the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de) (Schultz et al. 1998,2000). DEAD (DEXDc)-like helicases superfamily (N-terminal domain); HELICc, helicase superfamily (C-terminal domain); RING, RING finger domain; STYKc, protein kinases (unclassified specificity); UBA, ubiquitin-associated domain; WD40, WD40 repeats. The RING finger domain in the dashed box is not recognized by SMART or Pfam. The STYKc domain in the dashed box is degenerated (partial and noncatalytic). The IBR (In between Ring fingers) domain and UPF29 (uncharacterized protein family) are defined by Pfam (Bateman et al. 2000). The HisRS (histidyl-tRNA synthetase) domain is defined by literature (Sattleger et al. 1998).
Predictions of Function
On the basis of reports in the literature and/or co-occurrence with previously identified domains, some functional features can be predicted for 78.6% of our newly identified set of 28 domain families. This represents an increase in the state of functional prediction for ∼700 proteins (i.e., the total number of distinct proteins that are covered by novel domains with a putative function; see Table 1, Parts A–D). The predicted functions represent a variety of different cellular processes and molecular functions such as DNA/RNA- or metal-binding protein–protein interactions.
Five further cases of function prediction are outlined as follows.
Chromatin-Binding Domains
The CSZ domain-containing protein SPT6 and orthologs regulate transcription through establishment or maintenance of chromatin structure (Chiang et al. 1996; Winston 2001). A histone-binding capability for SPT6 has been experimentally confirmed (Bortvin et al. 1996). Here the CSZ domain is associated with an S1– and two SH2 domains, which are unlikely responsible for histone or chromatin binding. By this process of elimination, we predict a histone- or chromatin-binding function for the novel CSZ domain. The presence of HhH motifs in some copies of the CSZ domain raises the alternative or complementary possibility of a DNA and/or RNA binding function.
We identified a novel domain as a tandem repeat in several hypothetical human proteins, as a single copy associated with PHD and TFS2M in the_Drosophila_ gene CG6525, and as the Drosophila brahma and kismet genes. The kismet protein in Drosophila and its orthologs have been shown to be chromatin-remodeling factors, required for segmentation and segmentation identity. Our domain includes a recently reported conserved region (BRK) in brahma and kismet that is thought to bind chromatin (Daubresse et al 1999). We thus propose a chromatin-binding function for the newly identified domain.
Protein Interaction Domains
Recent studies reveal the interaction of the RPR domain in protein pcf11 with the C-terminal domain of the largest subunit of RNA polymerase II (Yuryev et al. 1996). Consequently, a similar function, or, less specifically, a protein-interaction function, is predicted for the RPR domain.
PSP domains appear to be protein-binding domains. The PSP domain-containing protein Cus1p is a component of a spliceosomal complex, associated with U2 snRNA (Gozani et al. 1996). Cus1p interacts directly with the snRNP Hsh155p by a region that overlaps with the PSP domain (Pauling et al. 2000).
The nuclear factor 90 (NF90) is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. The novel DZF domain in NF90 overlaps with a region known as NF45 homology domain, which is assumed to be responsible for conformation establishing of NF90 in the complex, where it may bind NF45 or other proteins (Parker et al. 2001). Thus, it is assumed that the DZF domain is a protein–protein interaction domain.
Several other functional predictions for novel domains are proposed in Table 1. Even where no functional role is postulated, delineation of conserved domain boundaries provides a starting point from which to undertake further experiments aimed at elucidating molecular function and cellular roles.
Predicted Localization of the Novel Domains
Context can also be used to predict whether a novel domain is associated with a certain cellular localization. For example, some of our novel domains are only found with representatives from our initial set of predominantly nuclear domains (i.e., those used to seed the searching procedure). This logic indicates a putative nuclear function and role for 10 of the domain families presented here, representing ∼500 proteins. Others among the novel domain families are likely to have roles in both nucleus and cytoplasm.
Novel Domains Related to Human Diseases
Four (14%) of the newly discovered domain families and one of the family-specific domain extensions occur in proteins whose deficiencies are implicated in severe human diseases. The respective genes or chromosomal regions are known to be responsible for cancer, neurodegenerative processes, or chromosomal aberrations (Table2).
Table 2.
Table of Novel Domains or Family-Specific Extensions Which are Putatively Correlated with Phenotypic Dysfunctions
| Domain name | Protein acc. No.a | Disease | OMIM Acc.Nov.b |
|---|---|---|---|
| AWS | O96028 | Wolf-Hirschhorn syndrome (Stec et al. 1998) | 602952 |
| RWD | CAB88085 | Monosomy 21 (Orti et al. 2000) | --- |
| DNP | O70656 | Malignent astrocytoma (Nakamura et al. 1998) | --- |
| FYRN/FYRC | Q03164 | Acute leukemia (Djabali et al. 1992) | 159555 |
Although the extent to which the domains themselves are responsible for the phenotypic affects observed with these diseases is not known, the new domains are likely to assist in ascertaining the normal functions of these genes, and by implication, a better understanding of their dysfunction.
DISCUSSION
Some well-characterized signaling domains, such as SH2 or PH, are present in a huge number of proteins and occur in combination with a large number of other domains. The fact that they are so widespread no doubt facilitated their early detection and characterization. Perhaps unsurprisingly, the domains found in the present analysis have more limited distributions than examples such as those. Even so, each new domain is found, on average, in 4.0 different architectures in ∼30 proteins. More widespread domains have been detected by our approach [e.g., the BRK domain occurs in more than seven different settings and a total of 30 proteins (Table 1, Part A)].
Only three (11%) of the newly discovered domains are species specific; of these, two are limited to plants and one is nematode specific (Table1, Part B). This could simply reflect the fact that even when species-specific pathways exist, proteins involved in them are likely to be recruited from preexisting components. Alternatively, species-specific domains may more likely be found only with other species-specific domains, rather than with domains found in a large phyletic range, and so would be underrepresented in the results of the search methods applied here.
In general, we cannot answer the question of whether the domains presented here have distant homologs that are not detectable using present methods (in common with any other new domain discovery report). The general evolutionary principle of reuse of preexisting components indicates that this is likely. However, we believe that, even if this is the case, the domains presented here, by dint of considerable sequence variation, are likely to have acquired new biological functions that are worthy of independent investigation.
In conclusion, we have identified a total of 28 novel domain families, 4 of which have been independently reported in the recent literature. Some of the domains are likely to be found in proteins localized to the nucleus. The predicted functions range from enzymatic activities to nucleotide binding. The systematic search for novel domains led to a 26% increase over the known nuclear domains that have been discovered in the last 15 yr, when the C2H2 zinc finger was first described (Miller et al. 1985).
The novel domains were all detectable using standard search methods (i.e., PSI-BLAST), within default E-value thresholds. The novelty of our approach has been to search using all candidate sequences that could contain a new domain of interest. In contrast, it would appear from our results that only using well-characterized sequences to search prevents the detection of some domains.
Although the majority of domains reported here are present in a wide variety of species, indicating that they have crucial biological roles, they are, on average, present in fewer proteins than previously reported domains. Taken together with the increasing volumes of data being produced by genome projects, targeted approaches to domain detection, such as those presented here, must have a role in enumerating the evolutionarily conserved components required for life.
METHODS
Definition of Nuclear Domains
A subset of SMART database families represents domains often found in nuclear proteins, as defined by annotation in sequence databases (Schultz et al. 2000). The computer program,Meta-A(nnotator) (Eisenhaber and Bork 1998), which assigns protein localizations based on Swiss-Protannotations, was used to predict the most likely localization for a domain family. A domain family was included in this analysis if more than 80% of Swiss-Prot entries of proteins containing the domain were annotated by Meta-A as nuclear. By this method, 86 domains were assigned a nuclear location. Eleven suspected false positives were removed following literature searches, and an additional 32 signaling domains with partial nuclear localization were added when literature searches could confirm this assignment.
Thus, a set of 107 predominantly nuclear domain families was derived (see http://www.embl-heidelberg.de/∼doerks/nuclear_subset.html/). Many domains, such as those with RNA-binding functions, are found in proteins that translocate between the cytoplasm and the nucleus or are found in both cytoplasmic and nuclear proteins. Consequently, some of these ‘nuclear’ domain families may contain cytoplasmic protein representatives. However, according to our protocol, based onSwiss-Prot annotations, the majority of proteins containing these domains will possess a significant population in the nucleus.
Automatic Screening for New Domains
All proteins containing one or more domains represented in the nuclear subset were extracted from public sequence databases, and their complete domain structure characterized using SMART. Regions not annotated using known SMART domain models were extracted, along with their domain context (i.e., position in the protein relative to other domains). Interdomain sequences shorter than 30 amino acids were regarded as less likely to represent novel globular domains (although such short domains do exist) and discarded. Noncontiguous regions of the same sequence were analyzed independently of each other. All of these sequence regions were then clustered into groups using thegrouper program of the SEALS package with a default single linkage clustering threshold of 50 bits (Walker and Koonin 1997). The longest member of each of these groups was filtered for coiled-coil and low complexity sequences (Lupas et al. 1991; Wootton and Federhen 1996) and then used to search a nonredundant sequence database, using the iterative search algorithmPSI-BLAST (Altschul et al. 1997), with an _E_-value inclusion threshold of E<0.001. Eight search rounds were performed, unless the database searching procedure converged in a prior iteration (see Altschul et al. 1997 for details of thePSI-BLAST procedure). The domain organizations of all homologs identified by PSI-BLAST searches were retrieved from the precalculated SMART database. The homologous regions identified in the searches were considered as the candidate domain family. Candidate regions that were found in different domain contexts (see following) in different proteins indicated a possible novel module family. These families were analyzed further using the methods described as follows.
Manual Confirmation and Refinement of Predicted Domains
To be considered as a module (i.e., a genetically mobile domain), homologous sequences were required to be present in at least two diverse molecular contexts (‘domain architectures’). Domain architectures (i.e., the linear arrangement of domains within a protein) were predicted using the SMART and Pfam databases. When a sequence contained no predicted domain other than that of the candidate family, this, too, was regarded as a distinct architecture. When a sequence invariably occurred either N- or C-terminal to a single known domain, it was regarded as an extension of the known domain.
Inaccurate prediction of gene structure (i.e., artificial fusion of adjacent genes) might lead to new domain architectures being counted spuriously. Domain architectures were inspected manually for such apparently erroneous fusions; for example, protein sequences containing both nuclear and extracellular domains were excluded. Similarly, a sequence was discarded if it had no homologs of similar domain architecture, but instead was similar to several pairs of nonhomologous proteins and each pair corresponded to the presumed erroneously fused gene.
At this stage, multiple alignments were generated (Thompson et al 1994) for all candidate domains. In conjunction with known locations of domains and other sequence features (e.g., N and C termini, transmembrane regions), these were used to define the borders of the putative new domains. In 10 cases, HMM-based searches of databases using HMMer2 (Eddy 1998) were needed to detect additional family members. The results were checked manually for consistency, with respect to amino acid conservation and phyletic distribution, to exclude false positives, which would be expected from our 10 HMM searches, given the _E_-value threshold of 0.1. Newly detected sequences were incorporated into the alignment, and the search procedure iterated. When these further analyses led to the identification of distant, but significant, similarity to annotated Pfam or SMART domains, the candidate domain was not pursued further. In cases in which we were unable to connect a family to a known domain with significant sequence similarity, but in which hits with marginal similarity were present, we recorded the family as representing possible divergent members of previously known protein domain families.
Acknowledgments
We thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of Plasmedium falciparum (3D7) public prior to publication of the completed sequence. The Sanger Centre (UK) provided sequence for chromosomes 1, 3–9, and 13, with financial support from the Wellcome Trust. A consortium composed of The Institute for Genome Research, along with the Naval Medical Research Center (USA), sequenced chromosomes 2, 10, 11, and 14, with support from NIAID/NIH, the Burroughs Wellcome Fund, and the Department of Defense. The Stanford Genome Technology Center (USA) sequenced chromosome 12, with support from the Burroughs Wellcome Fund. The Plasmodium Genome Database is a collaborative effort of investigators at the University of Pennsylvania (USA) and Monash University (Melbourne, Australia), supported by the Burroughs Wellcome Fund
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL doerks@embl-heidelberg.de; FAX 49 622 1517.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.203201.
REFERENCES
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci. 2000;25:421–423. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- Balciunas D, Ronne H. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem Sci. 2000;25:274–276. doi: 10.1016/s0968-0004(00)01593-0. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C. A hybrid protein kinase-RNase in an interferon-induced pathway? FEBS Lett. 1993;334:149–152. doi: 10.1016/0014-5793(93)81701-z. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Howard MJ, Proctor M, Bycroft M. The UBX domain: A widespread ubiquitin-like module. J Mol Biol. 2001;307:17–24. doi: 10.1006/jmbi.2000.4462. [DOI] [PubMed] [Google Scholar]
- Callebaut I, de Gunzburg J, Goud B, Mornon J. RUN domains: A new family of domains involved in Ras-like GTPase signaling. Trends Biochem Sci. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- Chiang PW, Wang S, Smithivas P, Song WJ, Ramamoorthy S, Hillman J, Puett S, Van Keuren ML, Crombez E, Kumar A, et al. Identification and analysis of the human and murine putative chromatin structure regulator SUPT6H and Supt6h. Genomics. 1996;34:328–333. doi: 10.1006/geno.1996.0294. [DOI] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP. JmjC: Cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA, Tamkun JW. The Drosophilakismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126:1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias Nat. Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Bork P. DDT, a novel domain in different transcription and chromosome remodeling factors. Trends Biochem Sci. 2001;26:145–146. doi: 10.1016/s0968-0004(00)01769-2. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Eisenhaber F, Bork P. Wanted: Subcellular localization of proteins based on sequence. Trends Cell Biol. 1998;8:169–170. doi: 10.1016/s0962-8924(98)01226-4. [DOI] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The UBA domain: A sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Janin J, Chothia C. Domains in proteins: Definitions, location, and structural principles. Methods Enzymol. 1985;115:420–430. doi: 10.1016/0076-6879(85)15030-5. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Seufert W, Hauser H-P. Genetic analysis of the ubiquitin system. Biochim Biophys Acta. 1991;1089:127–139. doi: 10.1016/0167-4781(91)90001-3. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissmann AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Yoshida M, Tsuiki H, Ito K, Ueno M, Nakao M, Oka K, Tada M, Kochi M, Kuratsu J, et al. Identification of a human homolog of the Drosophilaneuralized gene within the 10q25.1 malignant astrocytoma deletion region. Oncogene. 1998;16:1009–1019. doi: 10.1038/sj.onc.1201618. [DOI] [PubMed] [Google Scholar]
- Orti R, Rachidi M, Vialard F, Toyama K, Lopes C, Taudien S, Rosenthal A, Yaspo M-L, Sinet PM, Delabar JM. Characterization of a novel gene, C21orf6, mapping to a critical region of chromosome 21q22.1 involved in the monosomy 21 phenotype and of its murine ortholog, orf5. Genomics. 2000;64:203–210. doi: 10.1006/geno.1999.6109. [DOI] [PubMed] [Google Scholar]
- Park J, Karplus K, Barrett C, Hughey R, Haussler D, Hubbard T, Chothia C. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J Mol Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- Parker LM, Fierro-Monti I, Mathews MB. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded rna-activated protein kinase. J Biol Chem. 2001;276:32522–32530. doi: 10.1074/jbc.M104408200. [DOI] [PubMed] [Google Scholar]
- Pauling MH, McPheeters DS, Ares M., Jr Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol Cell Biol. 2000;20:2176–2185. doi: 10.1128/mcb.20.6.2176-2185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, Rallapalli R, Yano T, Alder H, Croce CM, Huebner K, et al. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophilatrithorax. Oncogene. 1997;15:549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C, Schneider R. PHD—An automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Sattleger E, Hinnebusch AG, Barthelmess IB. cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2α kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J Biol Chem. 1998;273:20404–20416. doi: 10.1074/jbc.273.32.20404. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signalling domains. Proc Natl Acad Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley R, Doerks T, Ponting C, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJB, de Boer PAJ, van Haeringen A, Moorman AFM, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophiladysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Park H, Hollingsworth NM, Sternglanz R, Lennarz WJ. PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. J Cell Biol. 2000;149:1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Walker DR, Koonin EV. SEALS: A system for easy analysis of lots of sequences. Ismb. 1997;5:333–339. [PubMed] [Google Scholar]
- Winston F. Control of eukaryotic transcription elongation. Genome Biol. 2001;2:1006.1–1006.3. doi: 10.1186/gb-2001-2-2-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Hassel BA, Silverman RH. Expression cloning of 2–5A-dependent RNAase: A uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]