Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome (original) (raw)
Abstract
In Drosophila, Piwi (P-element-induced wimpy testis), which encodes a protein of the Argonaute family, is essential for germ stem cell self-renewal. Piwi has recently been shown to be a nuclear protein involved in gene silencing of retrotransposons and controlling their mobilization in the male germline. However, little is known about the molecular mechanisms of Piwi-dependent gene silencing. Here we show that endogenous Piwi immunopurified from ovary specifically associates with small RNAs of 25–29 nucleotides in length. Piwi-associated small RNAs were identified by cloning and sequencing as repeat-associated small interfering RNAs (rasiRNAs) derived from repetitive regions, such as retrotransposon and heterochromatic regions, in the Drosophila genome. Northern blot analyses revealed that in vivo Piwi does not associate with microRNAs (miRNAs) and that guide siRNA was not loaded onto Piwi when siRNA duplex was added to ovary lysate. In vitro, recombinant Piwi exhibits target RNA cleavage activity. These data together imply that Piwi functions in nuclear RNA silencing as Slicer by associating specifically with rasiRNAs originating from repetitive targets.
Keywords: Piwi, retrotransposon, rasiRNA, Slicer, Drosophila
RNA interference (RNAi) is a gene silencing pathway triggered by double-stranded RNA (dsRNA) (Tomari and Zamore 2005). dsRNA is processed by Dicer to short interfering RNA (siRNA) (21–23 nucleotides [nt] in length), unwound, and loaded onto the RNA-induced silencing complex (RISC) as a single-stranded guide siRNA (Hammond 2005). RISC then binds the target RNA and cleaves it at sites completely complementary to the siRNA (Hutvagner 2005). microRNAs (miRNAs) are a large group of small noncoding RNAs (21–22 nt in length) functioning in gene silencing by repressing translation of the target mRNA (Ambros 2004; Du and Zamore 2005). The expression of miRNA is developmentally regulated, implying an important role for miRNAs in regulating gene silencing through development (Ambros 2004; Du and Zamore 2005). Members of the Argonaute family, defined by the presence of the PAZ and PIWI domains, play essential roles in gene silencing triggered by small RNAs such as siRNA and miRNA (Carmell et al. 2002).
In Drosophila, there are five genes encoding members of the Argonaute family, AGO1, AGO2, AGO3, Aubergine, and Piwi (P-element-induced wimpy testis). Among these, AGO1 and AGO2, members of the Argonaute sub-family, are expressed ubiquitously through development, whereas the other three, members of the Piwi sub-family, are expressed only in germline cells (Williams and Rubin 2002). AGO1 and AGO2 in Drosophila function in gene silencing through specific binding with miRNA and siRNA, respectively (Okamura et al. 2004). miRNA and siRNA are derived from different sources of precursors; miRNA is produced from a hairpin-shaped precursor (pre-miRNA) by Dicer1/Loquacious (R3D1) (Forstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005), whereas siRNA is produced from long dsRNA by Dicer2/R2D2 (Liu et al. 2003). siRNA-loaded AGO2 functions in RNAi as a factor directly responsible for cleaving a target completely complementary to siRNA (Miyoshi et al. 2005). AGO2 has recently been shown to also be involved in RISC formation in RNAi by slicing the passenger strand of the siRNA duplex (Matranga et al. 2005; Miyoshi et al. 2005; Leuschner et al. 2006). miRNA-associated AGO1 is thought to repress translation of mRNA targets without cleaving them; however, AGO1 also possesses target RNA cleavage (Slicer) activity, as does AGO2, and is capable of cleaving target RNAs when their sequences show complete complementarity to miRNA (Okamura et al. 2004; Miyoshi et al. 2005).
Aubergine and Piwi, two of the Piwi subfamily members in flies, were shown to play important roles in germline cell formation. Aubergine is required for pole cell formation (Harris and Macdonald 2001) and for activating RNAi during Drosophila oocyte maturation (Kenner-dell et al. 2002). It has also been shown that Aubergine is involved in silencing retrotransposons in the female germline (Vagin et al. 2004) and Stellate genes by the homologous Suppressors of Stellate [_Su(Ste)_] repeats that produce small RNAs in testis (Aravin et al. 2004). Piwi is essential in germline stem cell (GSC) self-renewal (Cox et al. 1998, 2000; Szakmary et al. 2005). It has been proposed that niche Piwi maintains GSCs by repressing bagof-marbles expression in GSCs, and the germline Piwi functions by promoting germ cell division (Szakmary et al. 2005). One of the piwi mutations, piwi1, causes male sterility due to severe defects in spermatogenesis. piwi1 ovarioles are mostly devoid of germline cells (Lin and Spradling 1997). Piwi has also recently been shown to be involved in silencing LTR retrotransposons such as copia and mdg1 in testis (Kalmykova et al. 2005). Transposition burst of mdg1 was also observed in piwi mutants. However, little is known at the molecular levels about how Piwi is involved in gene silencing and in GSC self-renewal.
In this study, we investigated the molecular function of Piwi in gene silencing. We found that the Piwi complex immunopurified from ovary contained a class of small RNAs, distinct in their size from miRNAs and siRNAs. Through cloning and sequencing the Piwi-associated small RNAs, we identified them as repeat-associated siRNAs (rasiRNAs) derived from various kinds of repetitive elements in the genome such as the retrotransposon and heterochromatin regions. rasiRNAs are a subset of small RNAs and were originally found in a small RNA profiling study in fly testis and embryo (Aravin et al. 2003). Since rasiRNAs are cognate to transposable elements and other repetitive elements in the fly genome, it is suggested that they participate in defining chromatin structures and regulating transposon activities. We noticed that the Piwi-associated rasiRNAs are often found in both sense and antisense orientations and that the 5′ ends of the rasiRNAs are predominantly uri-dine, suggesting that they are processed by an RNase III domain enzyme. Specificity of Piwi association with rasiRNAs was assessed by Northern blot analyses, which showed that Piwi does not associate with miRNAs in vivo and that guide siRNA was not loaded onto Piwi when siRNA duplex was added to ovary lysate. We found that Piwi does exhibit Slicer activity, as do AGO1 and AGO2; however, Piwi localizes in the nucleus while AGO1 and AGO2 are found in the cytoplasm. Taken together, the results imply that Piwi functions in nuclear RNA silencing as Slicer by associating specifically with rasiRNAs that originate from the repetitive targets.
Results
Piwi expression in ovary, testis, and embryo
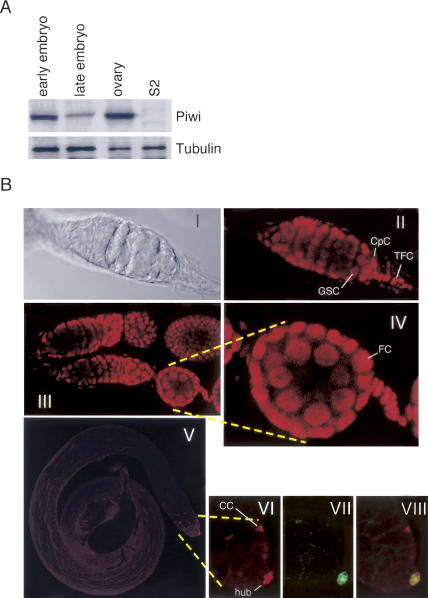
To investigate the molecular function of Piwi in gene silencing, we produced monoclonal antibodies against the protein. As an antigen, we used the N-terminal region of Piwi that shows much less similarity to the other members of the fly Argonaute family. Western blotting using the Piwi-specific antibody we raised revealed that Piwi is strongly expressed in the ovary and in the early embryo, but later in development the protein levels in the embryo decrease (Fig. 1A). Piwi was not detected in Schneider 2 (S2) cell lysate (Fig. 1A). The specificity of the anti-Piwi antibody is shown in Supplementary Fig. S1. To examine the expression pattern of Piwi in fly testis, ovary, and embryo, immunofluorescent staining was carried out (Fig. 1B). In ovarioles, Piwi was found to be clearly accumulated in nuclei of both GSCs and somatic cells such as terminal filament cells (TFCs), cap cells, and follicle cells (Fig. 1B, panels II,III, IV). It has previously been shown that piwi mRNA is detected in TFCs (Cox et al. 1998), and that myc-tagged Piwi protein is found in both somatic and germline cells in adult ovary (Cox et al. 2000). Relatively low expression was observed in cystoblasts, as in the case of myc-Piwi (Cox et al. 2000; Szakmary et al. 2005). In embryos, Piwi was found accumulated in nuclei of pole cells (Supplementary Fig. S1). In testis, Piwi appeared to be expressed in the hub, a tiny cluster of post-mitotic somatic cells localized at the apical tip of the testis (Fig. 1B, panels V,VI; panel VII shows the location of the hub stained with anti-Drosophila E[_D_E]-cadherin) and in cyst progenitor cells. The hub functions in the maintenance of GSC identity and influences its behavior (Ohlstein et al. 2004), while Piwi is required for the asymmetric division of GSCs to produce and maintain a daughter GSC but is not essential for further differentiation of committed daughter cells (Cox et al. 1998). We found that Piwi seems not to be expressed in male germline stem cells (GSCs) (Fig. 1B, panels V,VI). This observation agrees well with the Western data showing that the relative amount of Piwi in testis is quite low (data not shown).
Figure 1.
(A) Piwi expression is detected in Drosophila ovaries and embryos, but not in S2 cells. Western blotting was performed on lysates of fly embryo (at early and late stages), ovary, and S2 cells using an antibody against Piwi. Anti-tubulin was utilized as a loading control. (B) Piwi expression pattern in ovary and testis. All the images shown in this figure represent one confocal section. (Panels II,III,IV) The images of ovariole staining show that Piwi (red) is found clearly to be accumulated in nuclei of both GSCs and somatic cells such as TFCs, cap cells (CpC), and follicle cells (FC). Panel I shows the DIC image of panel II. (Panel V–VIII) In testis, Piwi (red) is expressed in the hub (shown in green; stained with anti-_D_E-cadherin) at the apical tip of the testis and in the cyst progenitor cells (CC). Male GSCs are not stained with anti-Piwi. Panel VI shows the enlarged image of a part of panel V. Panel VIII shows a merged image of panels VI and VII.
Piwi in ovary associates with small RNAs distinct from miRNAs
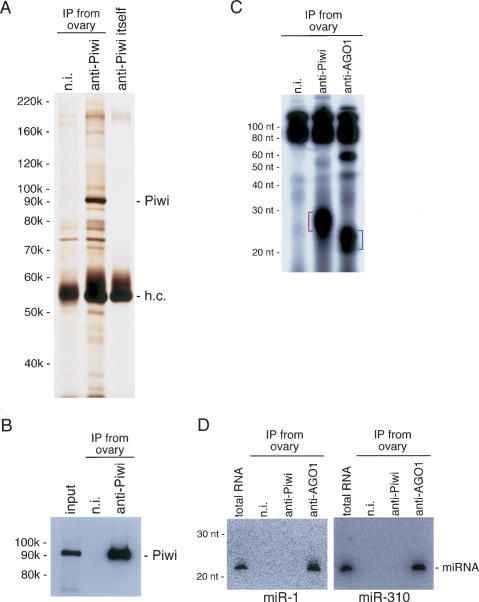
We next performed immunoprecipitation from ovary ly-sate using the anti-Piwi antibody. Silver staining of the immunoprecipitate failed to reveal any prominent bands copurifying with Piwi under physiological conditions (Fig. 2A), although some weak bands were perceived. Western blotting confirmed that the discrete band of ∼92 kDa was Piwi itself (Fig. 2B). We then investigated if immunoprecipitated Piwi associates with any small RNAs. Total RNAs were isolated from the immunopurified Piwi complex, labeled with 32P-pCp, and separated on a denaturing acrylamide gel. As a control, the AGO1 complex—an essential factor in miRNA-mediated gene silencing—was also immunopurified from the ovary, and miRNAs associated with AGO1 were simultaneously visualized. The Piwi-associated small RNAs migrated slightly slower than miRNAs (Fig. 2C). Compared with the size markers, Piwi-associated small RNAs were estimated to be 25–29 nt in length. Northern blot analysis using probes specific for miRNAs (miR-1 and miR-310) clearly revealed that the Piwi-associated small RNAs are not miRNAs (Fig. 2D).
Figure 2.
(A) Protein components of the Piwi complex immunopurified from ovary lysate are visualized by silver staining. A strong discrete band appearing at ∼92 kDa on the gel is Piwi. h.c. shows where the heavy chain of the antibodies used migrated on the gel, and n.i. indicates nonimmune antibody used as a negative control. (B) Western blotting with anti-Piwi on the immunoprecipitates shown in A confirms that the 92-kDa band is indeed Piwi. (C) Visualization of small RNAs associated with Piwi in ovary. RNA molecules were isolated from the Piwi immunopurified complex shown in A, labeled with 32P-pCp, and run on a 7 M urea-containing denaturing acrylamide gel. RNAs associated with AGO1 in ovary involve molecules ∼21–23 nt in length (blue lines), which from their size are expected to be miRNAs. Piwi-associated small RNAs (red lines) migrated slightly slower than miRNAs. The size was estimated at ∼25–29 nt in length. (n.i.) Nonimmune antibody used as a negative control. (D) Northern blot analyses using specific probes for miR-1 and miR-310 show that Piwi-associated small RNAs do not include the miRNAs.
Identification of small RNAs associated with Piwi in ovary
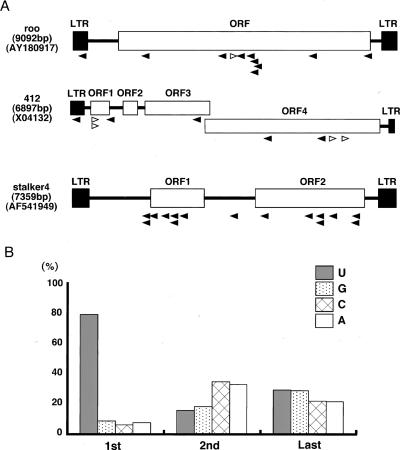
To identify the Piwi-associated small RNAs, we cloned and sequenced them. Of 800 clones sequenced, 392 hit to the Drosophila genomic sequences in databases. (Database searches were carried out by BLAST search using the databases at the National Center for Biotechnology Information [NCBI] and at FlyBase.) The compositions of the small RNAs obtained are listed in Table 1. The largest class of cloned RNAs (84.1%) was rasiRNAs, originally found in a small RNA profiling study in Drosophila melanogaster embryo and testis (Aravin et al. 2003). Breakdown products of rRNA, tRNA, mRNA, and snoRNA were also obtained (6.6%, 0.2%, 4.8%, and 4.1%, respectively). It is noteworthy that no miRNAs were obtained in this experiment. Among the rasiRNAs (330 clones), ∼40% (130 out of 330) matched to various kinds of transposable elements, including LTR (long terminal inverted repeat) retrotransposons and LINE (long interspersed nuclear elements)-like elements (Table 2). Those LINE-like elements include Het-A and TART, both known to transpose only to the chromosome ends to maintain telomere length (Mason and Biessmann 1995). Another 60% (200 out of 330) corresponded to heterochromatic regions in the genome (Table 2). By looking at the nucleotide sequences of all the clones (Supplementary Table S1), we noticed that the Piwi-associated rasiRNAs include both the sense and the anti-sense of a transposable transcript, although sense (+) orientation clones tended to be less abundant than those with an antisense (−) orientation (Table 2; Fig. 3A), and that the first nucleotide at the 5′ end was predominantly U (∼78%) (Fig. 3B). In addition, the Piwi-associated rasiRNAs were cloned by the procedure originally developed for miRNA cloning (Maniataki et al. 2005), indicating that they contain 5′ phosphates and 2′,3′-hydroxyl termini. These characteristics suggested that the Piwi-associated rasiRNAs are produced from dsRNA precursors by an RNase III domain-containing enzyme. In some cases, such as stalker4, rasiRNAs are only found in an antisense (−) orientation (Table 2; Fig. 3A). However, where the orientation bias of rasiRNA loading onto Piwi comes from is unknown and requires further investigation.
Table 1.
Compostitions of RNAs found in the Piwi-associated small RNA identification experiment
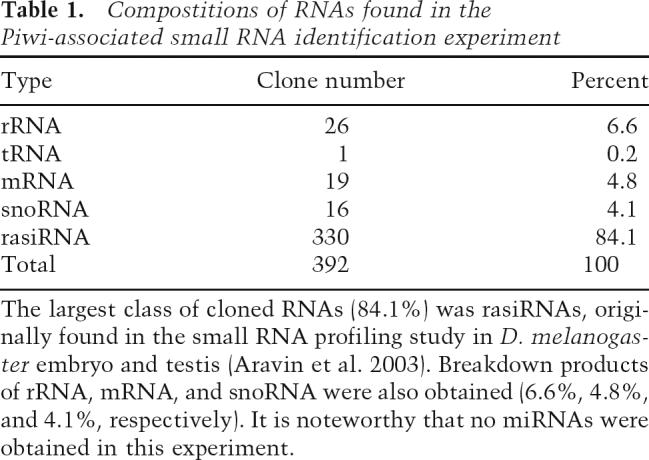
Table 2.
Identification of rasiRNAs associated with Piwi in ovary
Figure 3.
Identification of small RNAs associated with Piwi in ovary. (A) The regions where roo, 412, and stalker4 rasiRNAs are identified as Piwi-associated rasiRNAs corresponding to the transcripts are shown by arrowheads. rasiRNAs in sense and antisense orientations are indicated in white and black, respectively. (B) The first nucleotide of the rasiRNA associated with Piwi is predominantly U (1st). This tendency was not found to be obvious in the second and last nucleotides of the rasiRNAs.
Specific association of Piwi with rasiRNAs
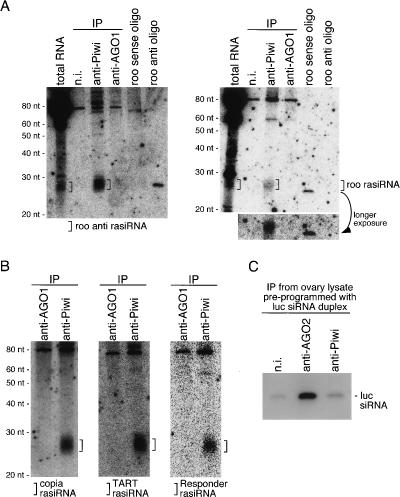
To confirm that rasiRNAs associated with Piwi indeed involve both the sense and antisense of a transcript, Northern blotting analysis was performed. roo rasiRNA was chosen for examination because it was one of most abundant clones obtained (11 out of 300) and in both sense and antisense orientations (Table 2). Both sense and antisense roo rasiRNAs were detected in the immunopurified Piwi complex as expected (Fig. 4A). Neither was found in the AGO1 complex, indicating that the Piwi association with rasiRNAs is specific (Fig. 4A). roo rasiRNA that is derived from the roo antisense transcript (hereinafter, we refer to it as roo anti-rasiRNA) seems more abundant in the Piwi complex (Fig. 4A) than the roo rasiRNA that was derived from the roo transcript in the sense orientation. This was determined from the fact that the signal of roo anti-rasiRNA in the anti-Piwi lane is about threefold stronger than that of roo anti-oligo (10 fmol) (Fig. 4A, left panel), whereas the signal of roo rasiRNA is a little less (∼85%) compared with that of roo sense oligo (10 fmol) (Fig. 4A, right panel). This observation correlated well with the fact that we obtained less roo rasiRNA than roo anti-rasiRNA in the cloning experiment (Table 2). The Piwi–rasiRNA association was further verified by Northern blotting on total RNAs isolated from Piwi and AGO1 immunopurified complexes in ovary. rasiRNAs originating from copia and TART retrotransposon transcripts, and one of the repetitive elements located in the centromeric heterochromatin region, Responder (Supplementary Table S1; Cabot et al. 1993), were detected only in Piwi but not in AGO1 complexes (Fig. 4B), demonstrating that the association with rasiRNAs is specific to Piwi. Whether Piwi was able to associate with siRNA was also investigated. luc siRNA duplex was first incubated in ovary lysate. Piwi and AGO2 complexes were then immunopurified from the lysate using specific antibodies to each protein. Northern blotting revealed that guide siRNA was specifically loaded onto AGO2 but not onto Piwi (Fig. 4C). These data, together with those shown in Figure 2D, strongly indicate the high specificity of Piwi association with rasiRNAs. It is suggested that a complex for loading rasiRNAs specifically onto Piwi exists; as the Dicer1/ Loquacious (R3D1) and Dicer2/R2D2 complexes load, after processing, mature miRNA and guide siRNA onto AGO1 and AGO2, respectively.
Figure 4.
Specificities of the Piwi–rasiRNA association. (A) RNAs associated with Piwi in ovary were isolated and probed with sense and antisense roo transcripts internally labeled with 32P in vitro. The left panel shows that Piwi associates with roo anti-rasiRNAs originating from roo antisense transcript. The right panel shows the Piwi association with roo rasiRNAs that is derived from the roo transcript in sense orientation. roo sense and antisense DNA oligos (10 fmol each) were also loaded on gels to show the probe specificities. roo anti-rasiRNA signal in the anti-Piwi lane was several-fold stronger than that of roo antisense oligo (left panel), whereas roo rasiRNA signal in the anti-Piwi lane (right) is a little less (∼85%) compared with that of the roo sense oligo, indicating that Piwi likely associates preferentially with roo anti-rasiRNA. Note that the AGO1 immunopurified complex (both panels) does not contain roo rasiRNAs, showing the specificity of the Piwi–rasiRNA association. (B) Blots containing RNAs associated with Piwi and AGO1 in ovary were probed with copia and TART transcripts and one of repetitive elements, Responder. Small RNAs corresponding to copia and TART rasiRNAs are detected only in the anti-Piwi lanes, indicating that the association with both rasiRNAs is specific for Piwi. (C) siRNA preprogrammed in ovary lysate was loaded onto AGO2, but not onto Piwi. Ovary lysate was prepared first and luc siRNA duplex was added to the lysate. After incubation, AGO2 and Piwi were immunopurified from the mixture using the specific antibodies. RNA molecules were then isolated from the complexes and probed with luc siRNA passenger strand to visualize the luc guide siRNA. In the anti-AGO2 lane, the luc guide siRNA strongly appeared, but not in the anti-Piwi and n.i. (nonimmune) lanes, indicating that luc guide siRNA was specifically loaded onto AGO2.
Piwi exhibits Slicer activity
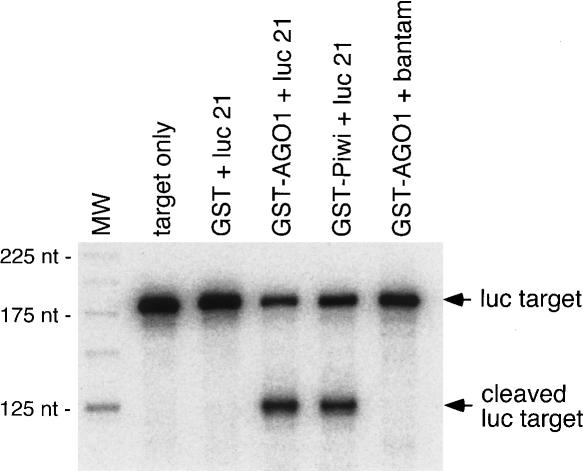
Previously, using GST-tagged recombinant full-length AGO1 (GST-AGO1), we showed that AGO1 is able to cleave target RNA completely complementary to miRNA (Miyoshi et al. 2005). In this study, we produced full-length GST-Piwi and examined the activity in an in vitro target RNA cleavage assay. Interestingly, we noticed that in the alignment of the Piwi domains of Piwi, AGO1, and AGO2, the D–D–H (Asp–Asp–His) motif, shown to be essential for Slicer activity of human Ago2 (Rivas et al. 2005), was not well conserved, and that the third residue was replaced with Lys (Supplementary Fig. S2). Thus, we suspected that Piwi may not exhibit Slicer activity. However, we observed that GST-Piwi was able to cleave luc target RNA when it was preincubated with luc guide siRNA (21 nt; luc 21) as well as GST-AGO1 (Fig. 5A). In this assay, it should be noted that we used single-stranded luc guide siRNA, in contrast to the siRNA duplex used in Figure 4C. GST-Piwi bound with another single-stranded siRNA, luc 30, which contains an additional 9 nt at the 3′ end of luc 21, also cleaved the luc target (data not shown). By searching for peptide sequence similarity over species, we noticed that the D–D–K triad is conserved in a member of the Argonautes (EAL29401) in Drosophila pseudoobscura and one of the Piwi domain-containing proteins (EAR93067) in Tetrahymena thermophila.
Figure 5.
Target RNA cleavage assay with GST-Piwi and GSTAGO1. Recombinant full-length Piwi and AGO1 fused to GST were produced in and purified from Escherichia coli. GST was used as a control. As guide siRNA, luc 21 (luc guide siRNA, 21 nt; Miyoshi et al. 2005) and bantam miRNA (21 nt; Miyoshi et al. 2005) were used. GST fusion proteins were first incubated with guide siRNA, and then the luc target labeled at the 5′ end (Miyoshi et al. 2005) was added to the reaction mixture. After incubation, resultant RNAs were loaded on a denaturing acrylamide gel. The cleaved product was observed not only with GST-AGO1, but also with GST-Piwi. GST-AGO1 bound with bantam did not cleave the target, as expected.
Discussion
For several years, it has been thought that Piwi is involved in gene silencing of LTR retrotransposons since (1) Piwi is one of the members of the fly Argonaute family of proteins, and (2) piwi mutations cause higher expression levels of retrotransposon transcripts in vivo. Links between protein factors involved in RNAi and the silencing of endogenous transposable elements have also been made in other species like Caenorhabditis elegans (Ketting et al. 1999; Tabara et al. 1999) and Chlamydomonas reinhardtii (Wu-Scharf et al. 2000). However, evidence at molecular levels to support such a Piwi function has not hitherto been reported. This study is the first to show that Piwi is specifically associated with rasiRNAs in vivo and that Piwi has Slicer activity. Our data suggest a novel, third kind of gene silencing pathway in Drosophila, following two distinct gene silencing pathways mediated by AGO1-miRNAs and AGO2-siRNAs (Okamura et al. 2004). The existence of rasiRNAs in Drosophila has been demonstrated previously through investigation of small RNA expression profiling in testis and embryo (Aravin et al. 2003). In that study, it was mentioned that rasiRNAs show some characteristics that suggest involvement of an RNase III domain enzyme in rasiRNA processing (Aravin et al. 2003). In this study, we observed that Piwi-associated rasiRNAs also show the same characteristics as those found in the exhaustive study of Aravin et al. (2003). Which protein factor(s) then produces rasiRNAs? Dicer1, Dicer2, and Drosha were not detected in Piwi complex immunopurified from ovary (data not shown). Although it may be due to the epitope hindering by Piwi and processing factor association, it is possible that a yet-to-be unidentified protein(s) other than Dicer1, Dicer2, and Drosha might be the crutial factor(s) for the rasiRNA-producing process.
We found that in fly testis, Piwi seems not to be expressed in GSCs, but its expression was clearly observed in the hub, the somatic cell cluster that functions to maintain GSC identity and influence its behavior. It is known that piwi1 causes male infertility due to severe defects in spermatogenesis; thus, the necessity of the Piwi function in the hub for such processes is quite apparent. Although it also will be important to determine if Piwi is associated with rasiRNAs in the testis, our findings have set a new stage for understanding how Piwi functions in the formation and maturation of GSCs in both ovary and testis.
Materials and methods
Western blot analysis
Two hundred amino acids at the N terminus of Piwi fused with GST were used as the antigen to immunize mice. Anti-Piwi monoclonal antibodies were produced essentially as described previously (Ishizuka et al. 2002). Western blotting was performed as described previously (Miyoshi et al. 2005). Ten micrograms of proteins from each sample were loaded on gels (Fig. 1A). Anti-tubulin was from the Developmental Studies Hybridoma Bank (1:1000 dilution).
Immunofluorescence
Testes and ovaries were dissected manually from adult flies in 1× PBS. Embryos were collected and dechorionated. Immunostainings were performed under standard procedures. Culture supernatants of anti-Piwi hybridoma cells were used without dilution. A rat monoclonal antibody to _D_E-cadherin (DCAD2) (1:20 dilution) was a gift from S. Hayashi (Kobe-RIKEN CDB). Alexa-488 anti-mouse and FITC-conjugated anti-rat IgGs (Jackson ImmunoResearch) were used as the secondary antibodies. All images were collected using a Zeiss LSM510 laser-scanning microscope.
Immunoprecipitation from ovary lysate
Immunoprecipitation was performed essentially as described previously (Miyoshi et al. 2005). Immunoprecipitation buffer contained 30 mM HEPES-KOH (pH 7.3), 150 mM KOAc, 2 mM MgOAc, 5 mM DTT, 0.1% NP40, 2 μg/mL Pepstatin, 2 μg/mL Leupeptin, and 0.5% Aprotinin. About 300 ovaries were used per immunoprecipitation. Silver staining of the protein components in the immunoprecipitates was performed using Silver-Quest (Invitrogen).
Visualization of small RNAs associated with Piwi and AGO1
After immunoprecipitation, total RNAs were isolated from the immunopurified complexes with phenol:chloroform and precipitated with ethanol. RNAs were labeled with 32P-pCp with T4 RNA ligase (TaKaRa) in a cold room overnight. After the reaction, RNAs were extracted with phenol:chloroform and precipitated with ethanol. RNAs labeled with pCp were loaded on a 7 M urea-containing denaturing 12% polyacrylamide gel and run. Signals were visualized using BAS-2500 (Fuji).
Small RNA cloning and sequence analysis
Small RNA cloning was carried out essentially as described (Maniataki et al. 2005). The sequences of the adapters were 3′ adapter, 5′-pUUUaaccgcatccttctctmdT-3′ and 5′ adapter, 5′-tact aatacgactcactAAA-3′ (p, phosphate; capital letters, RNA; small letters, DNA; mdT, 3′ amino-modified deoxythymidine). First-strand cDNA synthesis was performed with Stratascript RT and RT primer (5′-ACTAGCTGGAATTCAAGGATGCGGTTAA AG-3′). For PCR, an RT primer and PCR primer (5′-CAGCCA ACGGAATTCATACGACTCACTAAA-3′) were used after 5′-end phosphorylation with T4 polynucleotide kinase (TAKARA). KOD plus (TOYOBO) was used as a polymerase. PCR products were cloned into the EcoRV site of pBS SK+ vector followed by sequencing. RNA sequences between 5′ and 3′ adapters were analyzed by performing searches in annotated genomic databases (NCBI [http://www.ncbi.nlm.nih.gov/blast] and FlyBase [http://flybase.bio.indiana.edu/blast]) to determine whether the cloned RNAs mapped to the genome. To avoid confusion, we only accepted sequences that perfectly matched the databases and categorized sequences that did not match to the D. melanogaster genome as “others.”
Northern blot analysis
Total RNA was isolated from 50 ovaries from flies using Isogen (Invitrogen) according to the manufacturer's instructions. Five micrograms of total RNA were resolved by electrophoresis. Northern blotting for miRNA (miR-1 and miR-310) and luc siRNA was performed as reported previously (Miyoshi et al. 2005; Saito et al. 2005). The following probes were used for detection: 5′-CTCCATACTTCTTTACATTCCA-3′ for miR-1, and 5′-AAAGGCCGGGAAGTGTGCAATA-3′ for miR-310. Gene fragments for detecting rasiRNAs were cloned into pBS SK+ vector using primers 5′-TTTGGTCAATCGAGCCTTTAA TCGACTC-3′ and 5′-CACAATTGCTGATGTATGGCCTCA AG-3′ corresponding to GenBank sequence AY180917 for roo; 5′-AAGCAAGAAAGTTATGAACGCG-3′ and 5′-CCACAGA CATCTGAGTGTAC-3′, corresponding to GenBank sequence X04456 for copia; 5′-CGCCTGTGCAGCAGCTATTTAA-3′ and 5′-CAAATAAAGGATCTGGCAATATGGTAT-3′, corresponding to GenBank sequence AY600955 for TART; 5′-TCTG GAGATTCTGATCAACTGG-3′ and 5′-AGGTGTCTTCTTT TCGCCTGG-3′ corresponding to GenBank sequence M21213 for the Responder element. PCR products carrying T7 or T3 promoters containing the cloned fragments of retrotransposon or Responder sequences were used as transcription templates. RNA probes were synthesized in vitro using MaxiScript T7 and T3 kits (Ambion) in the presence of 32P-UTP. The transcribed RNAs were extracted with phenol:chloroform and precipitated with ethanol, then partially hydrolyzed by incubating at 60°Cin a solution containing 40 mM NaHCO3 and 60 mM Na2CO3. Hybridization and washing were performed as indicated previously (Saito et al. 2005). To test the specificity for roo rasiRNA hybridization, 10 fmol of roo sense (5′-TCCTTTAAGCATCT TACAGCTAAAGG-3′) and antisense (5′-TTTAGCTGTAAG ATGCTTAAAGGAGCT-3′) DNA oligos were loaded on gels.
siRNA loading assay
Three hundred ovaries were homogenized in a hypotonic buffer (30 mM HEPES at pH 7.4, 2 mM MgOAc, 5 mM DTT, and 1 mg/mL Pefablock SC) to prepare ovary lysate. luc siRNA duplex (Miyoshi et al. 2005) was incubated in the lysate for1hat26°C. Piwi and AGO2 complexes were then immunopurified from the mixture using anti-AGO2 and anti-PIWI antibodies. Sodium chloride was added to the lysates to 800 mM just before immunoprecipitation was begun. Reaction mixtures were rocked for 1 h at 4°C and the beads washed extensively with a washing buffer (30 mM HEPES at pH 7.4, 800 mM NaCl, 2 mM MgOAc, 5 mM DTT, and 1 mg/mL Pefablock SC). RNAs were isolated from the immunoprecipitates and separated on a denaturing polyacrylamide gel. Northern blotting was performed for detecting luc guide siRNA as described previously (Miyoshi et al. 2005).
Production of recombinant proteins and target RNA cleavage assay
To obtain cDNA encoding PIWI, poly(A)+ RNAs were purified from ovaries and RT–PCR was carried out using a ProSTAR Ultra-HF RT–PCR system (Stratagene). To produce a GST-PIWI fusion protein, full-length PIWI cDNA was subcloned into a pGEX-5X-1 expression vector (GE Healthcare Bioscience). GST-tagged protein was expressed in BL21 (DE3) and purified according to the manufacturer's instructions (GE Healthcare Bioscience). Preparation of cap-labeled luc target RNA (_luc_180) and the cleavage assay were carried out essentially as described (Miyoshi et al. 2005). In brief, 1 μg of GST fusion proteins was preincubated with luc siRNA 21mer (5′-UCGAAGUAUUCC GCGUACGUG-3′) in a cleavage reaction buffer (25 mM HEPES at pH 7.4, 50 mM KOAc, 5 mM MgOAc, 5 mM DTT, 0.1 U/μL RNasin and 0.1 μg/μL yeast RNA) for 1.5 h at 26°C. Then 5000 cpm of luc target RNA (_luc_180) was added and incubated for another 1.5 h at 26°C. The cleavage products of the reactions were analyzed by electrophoresis on 6% denaturing polyacryl-amide gels.
Accession numbers
The GenBank accession numbers for the transposons and chromosome sequences used in small RNA mapping and orientation determination are as follows: BS(X77571), F-element (AY183919), G-element (X06950), HeT-A (AJ635224 and U06920), TART (U14101 and AY600955), TAHRE (AJ542581), X-element (AF237761), R1Dm (X51968), R2Dm (X51967), pilger (AJ278684), strider (AF456125), 1731 (X07656), 412 (X04132), blood (AY180916), gypsy (M12927 and Z31368), springer (AF364549), H. M. S. Beagle (AF365402 and AY183918), Max-element (AJ487856), mdg1 (X59545), mdg3 (X95908), opus (AY180918), roo (AY180917), stalker (AF420242), ninja (AF420242), 297 (X03431), cruiser (AF364550), circe (AJ132547 and X98424), BEL (U23420), Tirant (AY928610), Micropia-Dm11 (X14037), rover (AF492764), Hoppel (AF533772), copia (X04456).
Acknowledgments
We thank members of the Siomi laboratory for discussions and comments on the manuscript. We also thank S. Hayashi for anti-_D_E-cadherin, and T. Suzuki and H. Kose for discussions and technical advice. K.S. is a postdoctoral fellow of the 21st century COE Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), and K.M is a research fellow supported by the Japan Society for the Promotion of Science (JSPS). This work was supported in part by grants to M.C.S and H.S from MEXT and the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Klenov M.S., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabot E.L., Doshi P., Wu M.L., Wu C.I. Population genetics of tandem repeats in centromeric heterochromatin: Unequal crossing over and chromosomal divergence at the Responder locus of Drosophila melanogaster . Genetics. 1993;135:477–487. doi: 10.1093/genetics/135.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes& Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Cox D.N., Chao A., Baker J., Chang L., Qiao D., Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.N., Chao A., Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Du T., Zamore P.D. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M.2005Dicing and slicing: The core machinery of the RNA interference pathway FEBS Lett. 579:5822–5829 [DOI] [PubMed] [Google Scholar]
- Harris A.N., Macdonald P.M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128::2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hutvagner G. Small RNA asymmetry in RNAi: Function in RISC assembly and gene regulation. FEBS Lett. 2005;579::5850–5857. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi M.C., Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila . Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova A.I., Klenov M.S., Gvozdev V.A. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33::2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J.R., Yamaguchi S., Carthew R.W. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes & Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp T.H., van Luenen H.G., Plasterk R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Leuschner P.J., Ameres S.L., Kueng S., Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301::1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Maniataki E., De Planell Saguer M.D., Mourelatos Z. Immunoprecipitation of microRNPs and directional cloning of microRNAs. Methods Mol. Biol. 2005;309:283–294. doi: 10.1385/1-59259-935-4:283. [DOI] [PubMed] [Google Scholar]
- Mason J.M., Biessmann H. The unusual telomeres of Drosophila . Trends Genet. 1995;11:58–62. doi: 10.1016/s0168-9525(00)88998-2. [DOI] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C.2005Slicer function of Drosophila Argonautes and its involvement in RISC formation Genes& Dev.19:2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Kai T., Decotto E., Spradling A. The stem cell niche: Theme and variations. Curr. Opin. Cell Biol. 2004;16:693–699. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M.C. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol. 2005;3: doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A., Cox D.N., Wang Z., Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans . Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D. Perspective: Machines for RNAi. Genes& Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Klenov M.S., Kalmykova A.I., Stolyarenko A.D., Kotelnikov R.N., Gvozdev V.A. The RNA Interference proteins and Vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster . RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- Williams R.W., Rubin G.M. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf D., Jeong B., Zhang C., Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290::1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]