Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties (original) (raw)
Abstract
The epithelial compartment of the human breast comprises two distinct lineages: the luminal epithelial and the myoepithelial lineage. We have shown previously that a subset of the luminal epithelial cells could convert to myoepithelial cells in culture signifying the possible existence of a progenitor cell. We therefore set out to identify and isolate the putative precursor in the luminal epithelial compartment. Using cell surface markers and immunomagnetic sorting, we isolated two luminal epithelial cell populations from primary cultures of reduction mammoplasties. The major population coexpresses sialomucin (MUC+) and epithelial-specific antigen (ESA+) whereas the minor population has a suprabasal position and expresses epithelial specific antigen but no sialomucin (MUC−/ESA+). Two cell lines were further established by transduction of the E6/E7 genes from human papilloma virus type 16. Both cell lines maintained a luminal epithelial phenotype as evidenced by expression of the tight junction proteins, claudin-1 and occludin, and by generation of a high transepithelial electrical resistance on semipermeable filters. Whereas in clonal cultures, the MUC+/ESA+ epithelial cell line was luminal epithelial restricted in its differentiation repertoire, the suprabasal-derived MUC−/ESA+ epithelial cell line was able to generate itself as well as MUC+/ESA+ epithelial cells and Thy-1+/α-smooth muscle actin+ (ASMA+) myoepithelial cells. The MUC−/ESA+ epithelial cell line further differed from the MUC+/ESA+ epithelial cell line by the expression of keratin K19, a feature of a subpopulation of epithelial cells in terminal duct lobular units in vivo. Within a reconstituted basement membrane, the MUC+/ESA+ epithelial cell line formed acinus-like spheres. In contrast, the MUC−/ESA+ epithelial cell line formed elaborate branching structures resembling uncultured terminal duct lobular units both by morphology and marker expression. Similar structures were obtained by inoculating the extracellular matrix-embedded cells subcutaneously in nude mice. Thus, MUC−/ESA+ epithelial cells within the luminal epithelial lineage may function as precursor cells of terminal duct lobular units in the human breast.
Keywords: Human cell, breast, stem cell, immortal, K19
Understanding how the normal human breast develops and which cell compartment becomes neoplastic by necessity is dependent on the isolation of relevant cells as the true targets of human breast carcinogenesis and progression. More than two decades ago it was proposed that human breast cancer originates from the luminal epithelial lineage within the terminal duct lobular units (TDLU) (Wellings et al. 1975), a basic mammary structure consisting of a branching ductal-alveolar system lined by an inner layer of luminal epithelial cells and an outer layer of myoepithelial cells.
Recently, we and others have provided some evidence that the stem cells of the human and mouse mammary gland may be contained within the luminal epithelial lineage (Smith 1996; Stingl et al. 1998; Péchoux et al. 1999; Smalley et al. 1999). However, detailed ultrastructural characterizations of the rodent and human breast gland in situ have led to the widely discussed hypothesis that the stem cell is a ‘basal cell‘ with clear cytoplasm (for review, see Rudland 1987; Smith and Chepko 2001). In particular, the important work of Smith and colleagues in the mouse mammary gland has outlined an elaborate morphological tree identifying a candidate stem cell (small light cells; SLC), a first degree progenitor cell (not distinct from SLC), a second degree progenitor which is still multipotent (undifferentiated large light cell; ULLC), and then two compartments of nondividing, preluminal- and premyoepithelial cells which gradually mature into the fully differentiated lineages. At the ultrastructural level, the SLC never reaches the acinus lumen and only a fraction of the ULLC does so (Smith and Chepko 2001). Because sialomucin is the most prominent marker of luminal epithelial cells and is exclusively expressed on the apical surface of luminal epithelial cells (Petersen and van Deurs 1986), this would imply that some epithelial cells (not facing the lumen) are sialomucin negative. Nevertheless, there is reason to believe that these cells are indeed full members of the luminal epithelial lineage. The cells on the basal side of multilayered breast ducts express several luminal epithelial markers including simple epithelial keratins and epithelial-specific antigen (ESA), but no α-smooth muscle actin (ASMA) (Petersen and van Deurs 1988; Taylor-Papadimitriou et al. 1989). Cells with this phenotypic profile have in fact been observed in cultures of human reduction mammoplasties and shown to be bipotent, suggesting a stem cell potential (Stingl et al. 1998). However, further characterization of the putative stem cells to show the full potential of generating TDLU have not been pursued due to a limited growth potential in primary culture.
In the present study, we set out to identify and to define the properties of subluminal epithelial cells that express ESA but little or no sialomucin. To accomplish this, we isolated distinct cell populations using immunomagnetic sorting and immortalized them with human papilloma virus (HPV)-16 E6/E7 (Wazer et al. 1995).
Our data show that MUC−/ESA+ epithelial cells indeed exist in vivo. The luminal MUC+/ESA+ epithelial cells were differentiated, acinar, luminal epithelial-restricted without stem cell properties. The MUC−/ESA+ epithelial cells were also clearly of the luminal epithelial lineage because they expressed tight junctional proteins and exhibited a high transepithelial electrical resistance on transwell filters. However, they had a striking ability to form the entire TDLUs inside a three-dimensional reconstituted basement membrane and in nude mice and could generate myoepithelial cells. Finally, we showed that these cells have a counterpart in vivo: We localized a subpopulation of luminal epithelial cells in the normal breast in situ by the restricted expression of keratin K19. We propose that candidate stem cells of the human breast should be found within this population and that the cells we have isolated and characterized fulfill the criteria for such candidate stem cells.
Results
Identification of MUC−/ESA+ luminal epithelial cells in the breast
In culture, a putative “stem” cell of the human breast was defined based on a positive staining for the luminal epithelial marker ESA and a negative or weakly positive staining for sialomucin (MUC) (Stingl et al. 1998). In mouse mammary gland in vivo, candidate stem cells have been defined ultrastructually as, among other features, having no luminal contact but are distinct from myoepithelial cells that we refer to as suprabasal (Smith and Chepko 2001). To find a candidate stem cell in human breast in vivo, we combined these two observations and double-stained histological sections of normal human breast tissue with ESA and MUC. Most luminal epithelial cells stained as expected, that is, MUC was expressed on the apical surface and ESA at the basolateral surface (Fig. 1A).
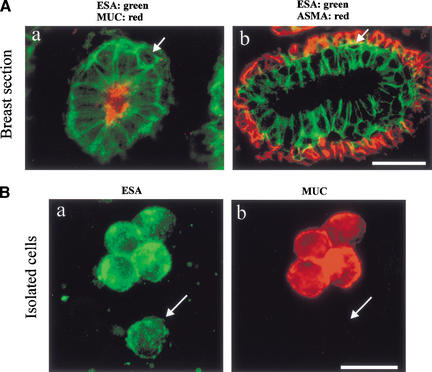
Figure 1.
Identification of MUC−/ESA+ luminal epithelial cells in the breast. (A) MUC−/ESA+ epithelial cells belong to the luminal epithelial lineage. (a) Double-labeling immunofluorescence staining of epithelial specific antigen (ESA) and sialomucin (MUC1). The arrow indicates an example of an epithelial cell that does not appear to reach the lumen and fails to express sialomucin. (b) Double-labeling immunofluorescence staining of ESA and α-smooth muscle actin (ASMA). Note that the suprabasal epithelial cells (arrow) are resting on a layer of ASMA+ myoepithelial cells. Bar, 20 μm. (B) A subset of cells within the luminal epithelial lineage is sialomucin-negative. Uncultured, trypsinized breast epithelial cells were double-stained to demonstrate ESA (green) and sialomucin (red). Whereas the majority of cells were MUC+/ESA+, a small fraction was MUC−/ESA+ (arrow). Bar, 20 μm.
However, it was also evident that a subset of ESA+ cells in occasional acinar profiles was indeed abluminal in location with no visible extensions reaching the lumen (Fig. 1A, a, arrow). To confirm that these cells were distinct from myoepithelial cells, we double-stained for ESA and ASMA. As expected, myoepithelial cells were negative for ESA, whereas a very minor population of “basal” cells were suprabasal, and did not appear to reach the lumen or stain with ASMA (Fig. 1A, b, arrow). If these cells truly never reached the lumen, not even outside the sectioned plane, a sample of smeared trypsinized, uncultured breast cells should also contain two ESA+ luminal epithelial populations: one major being MUC+ and a minor being MUC−. This was found to be the case as evidenced by double-staining of such smears for ESA and MUC (Fig. 1B, a,b). The average frequency of MUC− cells in such preparations was 8 ± 3%. It is concluded that suprabasally positioned MUC−/ESA+ epithelial cells also exist in human breast.
Isolation, immortalization, and characterization of MUC+/ESA+ and MUC−/ESA+ epithelial cells
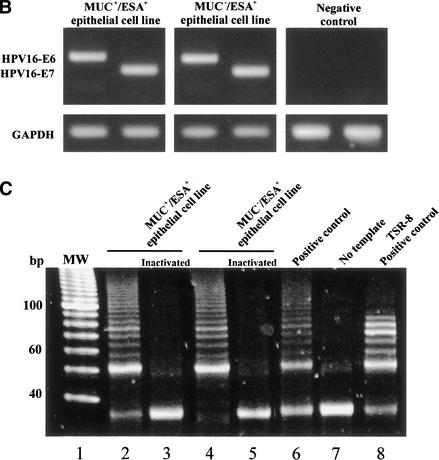
To show that MUC−/ESA+ epithelial cells were multipotent and had stem cell properties, we needed to isolate, purify, and characterize them. We accomplished this by immunomagnetic sorting. The MUC+/ESA+ epithelial cells were purified from two consecutive sialomucin-columns. The MUC−/ESA+ epithelial cells were purified as the flow-through from a sialomucin-column that was later retained in an ESA-column. To generate cell lines, we immortalized both populations with an E6/E7 construct of HPV16. The resulting established cell lines were MUC+/ESA+ and MUC−/ESA+, respectively (Fig. 2A), and are referred to below as the luminal and suprabasal-derived epithelial cells. The cell lines displayed immortalized characteristics: They have been cultured for >50 passages over two years and continue to express both E6 and E7 (Fig. 2B), and a distinct telomerase activity that is absent from finite life span breast epithelial cells (Fig. 2C) (Stampfer et al. 2001).
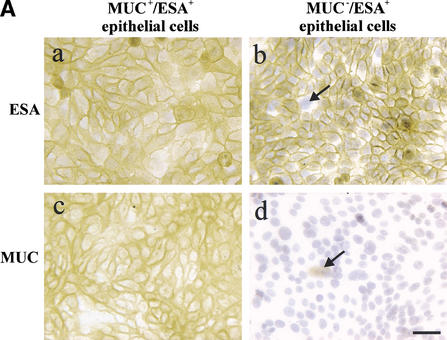
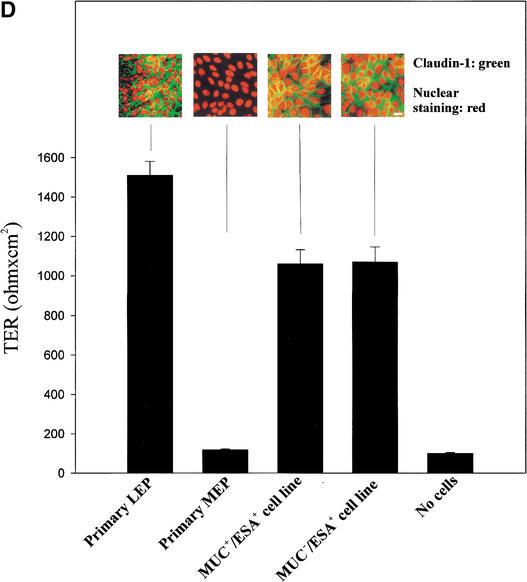
Figure 2.
Isolation, immortalization and characterization of MUC+/ESA+ and MUC−/ESA+ epithelial cells. (A) The MUC+/ESA+ and MUC−/ESA+ epithelial cells differ by sialomucin expression. Immunoperoxidase staining with ESA (a,b) and MUC1 (c,d) of the MUC+/ESA+ (a,c) and the MUC−/ESA+ epithelial cell line (b,d). Note that whereas the MUC+/ESA+ epithelial cell line is homogenous in its staining pattern and expressed sialomucin (MUC1), the MUC−/ESA+ epithelial cell line is heterogeneous (arrows) and essentially negative for sialomucin. Bar, 50 μm. (B) The cell lines express E6 and E7 stably. RT–PCR of HPV16 E6 and E7 show that both the MUC+/ESA+ and the MUC−/ESA+ epithelial cells are stably transduced. (C) The cell lines exhibit telomerase activity. TRAP assay of equal numbers of MUC+/ESA+ and MUC−/ESA+ epithelial cell lines showed telomerase activity in transduced cells. (Lane 1) Molecular weight markers; (lane 2) MUC+/ESA+ epithelial cell line; (lane 4) MUC−/ESA+ epithelial cell line; (lanes 3 and 5) heat inactivated negative control of the cell lines; (lane 6) positive control pellet; (lane 7) negative control without cell lysate; (lane 8) positive control TSR8. (D) Both the MUC+/ESA+ and MUC−/ESA+ epithelial cell lines belong to the luminal epithelial lineage. Confluent cultures were plated on Transwell filters and assayed for transepithelial resistance (TER), and parallel monolayer cultures were double-stained for claudin-1 as well as propidium iodide to visualize the nuclei (insets). Primary luminal epithelial cells (LEP) and myoepithelial cells (MEP) are readily discriminated by TER and claudin-1 expression. Bar, 20 μm.
Whereas the MUC+/ESA+ epithelial cell line continued to be homogeneous (Fig. 2A, a,c), the MUC−/ESA+ suprabasal-derived epithelial cell line contained occasional subpopulations of ESA− cells and MUC+ cells (Fig. 2A, b,d, arrows). Double immunofluorescence staining for ESA and MUC (data not shown) revealed that this cell line contained three cellular subtypes: The majority population was MUC−/ESA+ and two minor populations were either MUC−/ESA− or MUC+/ESA+. To substantiate that both cell lines belonged to the luminal epithelial lineage, even though one of them was essentially devoid of luminal epithelial MUC expression, we tested for a marker that is a hallmark of glandular epithelial phenotype: that of functional tight junctions. This was carried out by staining for the tight junction proteins claudin and occludin, and by measuring the level of transepithelial electrical resistance (TER) in confluent cultures on transwell filters. By these criteria, primary luminal epithelial cells were readily distinguished from primary myoepithelial cells (Fig. 2D). As also seen in Figure 2D, the newly established cell lines resembled luminal epithelial cells both by staining at the cell boundaries for tight junction proteins (insets) and exhibiting a high TER comparable to what has been published for other simple epithelia with barrier function such as kidney and thyroid cells (Jaeger et al. 1997). We conclude that both cell lines belong to the luminal epithelial lineage.
Clonal cell lines of the MUC−/ESA+ epithelial cell line are multipotent
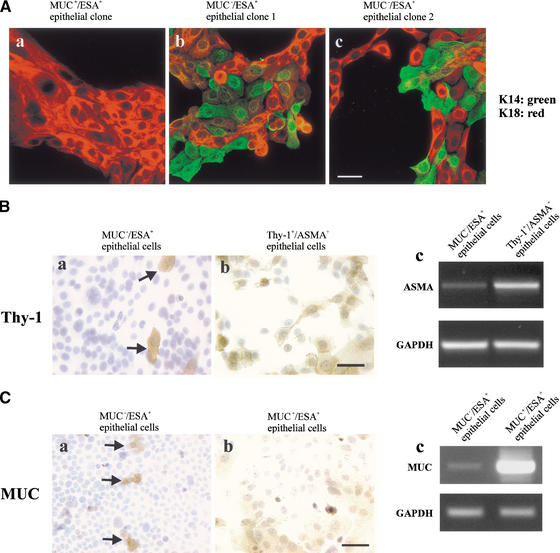
The fact that the MUC−/ESA+ suprabasal-derived epithelial cells continued for >50 passages to generate subpopulations of MUC+/ESA+ and MUC−/ESA− cells as well as MUC−/ESA+ cells make them strong candidates for multipotent progenitor or stem-like cells of the breast, and the MUC+/ESA+ cells could represent differentiated luminal epithelial cells, and the MUC−/ESA− cells could represent myoepithelial cells, which are MUC−/ESA− in vivo. To test this, clonal cultures (limited dilution) were established and double-stained for keratin K18 (luminal marker) and K14 (myoepithelial marker). Although the MUC+/ESA+ epithelial cell line did not generate any K18− myoepithelial cells, the MUC−/ESA+ suprabasal-derived epithelial cell line readily formed mixed clones of K18+/K14+, K18+/K14−, and K18−/K14+ epithelial cells (Fig. 3A). We then explored whether the K14+ epithelial cells were precursor of a myoepithelial progenies or whether they already contained all of the myoepithelial markers. We found that these cells represented a primitive level of myoepithelial differentiation because <1% of the cells expressed other myoepithelial markers such as Thy-1 (Fig. 3B, a). However, if the cells were retained in a Thy-1 column, a myoepithelial subline could be generated that also expressed α-smooth muscle actin referred to as Thy-1+/ASMA+, which is restricted to postmitotic myoepithelial cells in vivo (Fig. 3A, b,c) (Sapino et al. 1990). We reasoned that, if K18+ epithelial cells were also precursor cells of a progeny within the luminal epithelial compartment, they could further mature within this compartment to differentiated cells. Sialomucin-expressing cells were essentially eliminated (0.04% weakly positive) by retention on a sialomucin-retaining column, but evidence for spontaneous maturation into sialomucin-positive cells was provided by the reoccurrence (5%) of these cells after 6 d (Fig. 3C, a). These cells in turn could be retained in a similar column and maintained MUC+ in high-density cultures in the presence of serum (Fig. 3C, b,c). Taken together, these observations provide evidence for the existence of a MUC−/ESA+, multipotent progenitor cell belonging to the luminal epithelial lineage that can give rise to differentiated myoepithelial and luminal epithelial cells, and their precursors.
Figure 3.
Evidence for multipotency in the MUC−/ESA+ epithelial cell line. (A) Double-staining with luminal epithelial K18 and myoepithelial K14 in clones of the MUC+/ESA+ epithelial cell line and the MUC−/ESA+ epithelial cell line. Clonal cultures of the MUC+/ESA+ (a) and the MUC−/ESA+ epithelial cells (b,c) were double-stained with keratin K18 and K14. No evidence for myoepithelial cells was found in any of the MUC+/ESA+ clones. Conversely, a mixture of cells was always present in the MUC−/ESA+ epithelial clones. Bar, 40 μm. (B) Evidence of spontaneous maturation to Thy-1+/ASMA+ myoepithelial cells. Immunoperoxidase staining of Thy-1, a marker for myoepithelial cells, in cultures of MUC−/ESA+ epithelial cells before (a) and after (b) purification in a Thy-1 retaining column. The spontaneous occurrence of Thy-1 stained cells is limited to <1% (arrows). However, on purification, a myoepithelial subline can be obtained that also expresses ASMA (c). Bar, 50 μm. (C) Evidence for maturation to MUC+/ESA+ epithelial cells. MUC−/ESA+ epithelial cells were cleared of sialomucin-positive cells and stained for sialomucin after 2 wk (a, arrows), and after further sorting of the newly formed sialomucin-positive cells (b). The MUC stainings was confirmed by RT–PCR (c). Bar, 50 μm.
MUC−/ESA+ epithelial cells give rise to TDLU
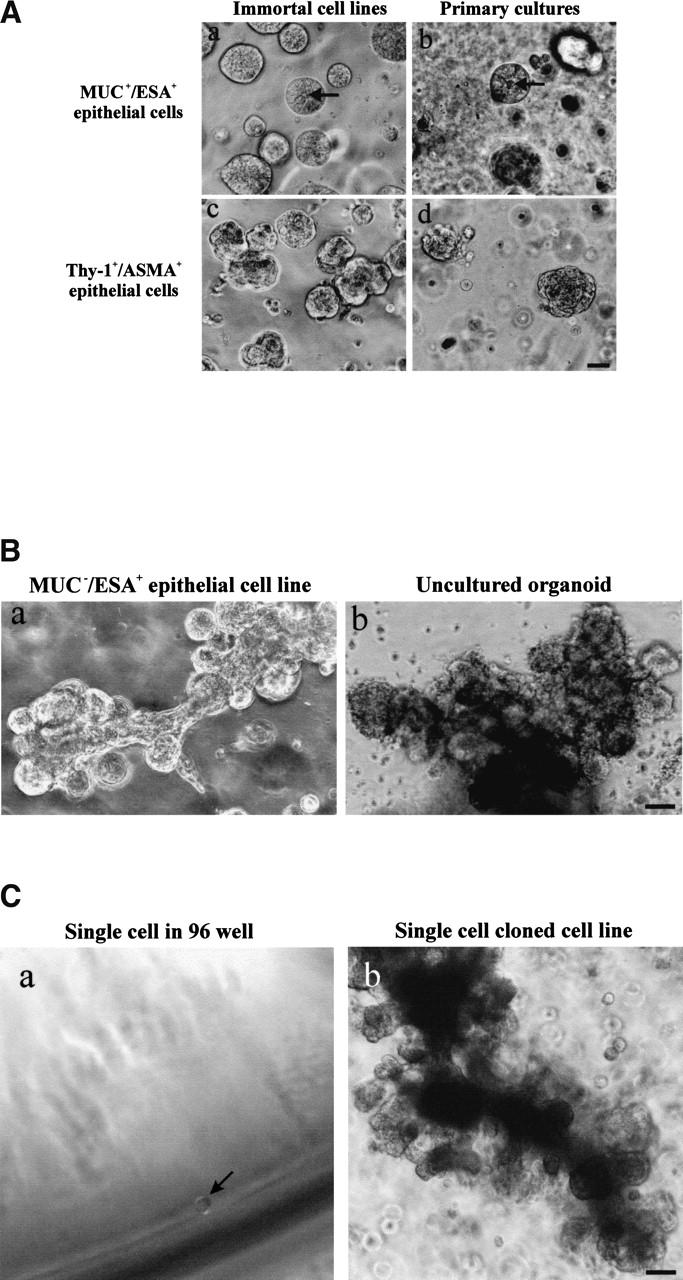
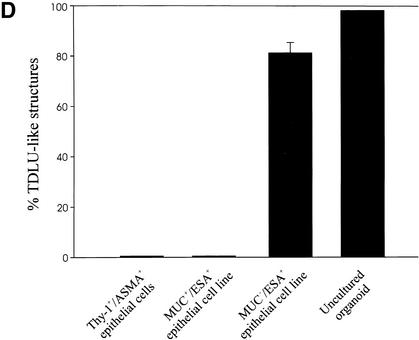
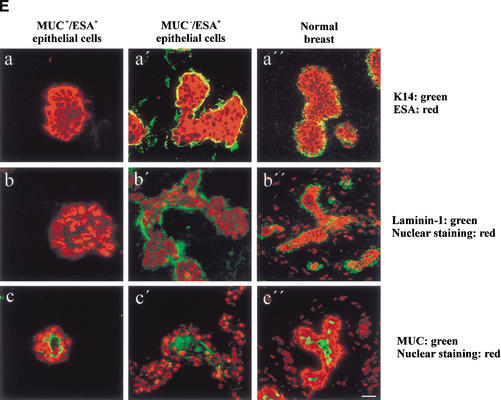
In the mouse and rat, the standard criteria for the presence of stem cells has been: (1) self-renewal and (2) the ability to regenerate the entire mammary epithelium from a single cell on reimplantation of cells in syngeneic gland-free fat pads (Smith and Medina 1988). We tested MUC−/ESA+ cells for these properties in a laminin-rich gel, conditions that have been shown previously to provide physiological cues that allow mammary morphogenesis for both rodent and human cells (for review, see Bissell et al. 1999). Previously, two prominent cell morphologies were reported for primary human breast cells in laminin-rich gels: small acinus-like structures of luminal epithelial origin and solid colonies for myoepithelial cells (Petersen et al. 1992). Embedding the MUC+/ESA+ epithelial cell line or the Thy-1+/ASMA+ cells yielded morphologies very similar to those already described for primary breast cells (Fig. 4A). In contrast, the MUC−/ESA+ epithelial cells gave rise to formation of larger, more elaborate morphologies resembling the entire functional unit of the breast gland, that is, the TDLU (Fig. 4B,D). A similar frequency of TDLU morphological structures (73%), albeit not as elaborate, was recorded if MUC−/ESA+ epithelial cells were freshly prepared from primary cultures. To verify the multipotency of MUC−/ESA+ epithelial cells, we further made clonal cultures from single cells and identified a clone that was able to generate the TDLU-like structures as described above for the parent line (Fig. 4C). Sections of MUC−/ESA+ epithelial cells embedded in laminin-rich gels and stained for differentiation markers of normal breast revealed a remarkable similarity to TDLUs in vivo (Fig. 4E). Nevertheless, the experimentally generated TDLUs showed a subtle difference from normal breast by residual expression of K14 in the central cells and less sialomucin staining. The MUC+/ESA+ acini also showed some acini that were inside-out with regards to polarization of sialomucin (data not shown). Finally, to provide evidence that the ability to generate a MUC−/ESA+ epithelial cell line from the human breast with all the above mentioned progenitor cell characteristics was not an accident, we repeated the entire protocol including immortalization of cells from two additional biopsies, and reproduced the TDLU morphology both times. Collectively, these data are in strong support of MUC−/ESA+ epithelial cells as precursors of the TDLU, the functional unit in the human breast.
Figure 4.
MUC−/ESA+ epithelial cells give rise to terminal duct lobular units (TDLU). (A) MUC+/ESA+ epithelial- and Thy-1+/ASMA+ myoepithelial cells make colonies with distinct morphologies in a laminin-rich gel. Immortalized (a,c) and primary (b,d) MUC+/ESA+ epithelial cells (a,b) and Thy-1+/ASMA+ myoepithelial cells (c,d) were embedded as single cells in a laminin rich gel. Both MUC+/ESA+ epithelial and Thy-1+/ASMA+ myoepithelial cell lines resembled the corresponding primary cells. Whereas the luminal epithelial cells formed acinus-like spheres with a central lumen, the myoepithelial cells formed irregular solid clusters of cells. Bar, 25 μm. (B) MUC−/ESA+ epithelial cells make an elaborate TDLU-like structure in a laminin-rich gel. MUC−/ESA+ epithelial cells (a) were embedded as single cells in a laminin-rich gel and compared with the morphology of freshly isolated, uncultured TDLU organoids (b). Both consist of small branching ductules terminating in globular acinus-like structures. Bar, 50 μm. (C) Single cell cloned MUC−/ESA+ cells maintain the ability to make TDLU-like structures in a laminin-rich gel. Single cell cloning was verified by the demonstration of only one cell in a 96 well using a 10× objective (Ca; arrow). Single cell clone ‘TH123‘ embedded as single cells in Matrigel could give rise to TDLU-like structures by 6 d (b). Bar, 50 μm. (D) Quantification of TDLU-like structures in laminin-rich gels. MUC+/ESA+ and MUC−/ESA+ epithelial cells and Thy-1+/ASMA+ myoepithelial cells were embedded inside laminin-rich gels and allowed to grow for 12 d and compared to uncultured organoids. The number of TDLU-like structures was quantified by phase contrast microscopy. (E) MUC−/ESA+ epithelial colonies in laminin-rich gels resemble TDLU in vivo. Sections of laminin-rich gels containing MUC+/ESA+ (left column) and MUC−/ESA+ epithelial cells (middle column) were compared with sections of normal breast tissue (right column) and double-stained for ESA and keratin K14 (a–a"), propidium iodide and laminin-1 (b–b") and propidium iodide and sialomucin (c–c"). Only the MUC−/ESA+ epithelial cells showed a differentiation pattern reminiscent of normal breast tissue. Bar, 15 μm.
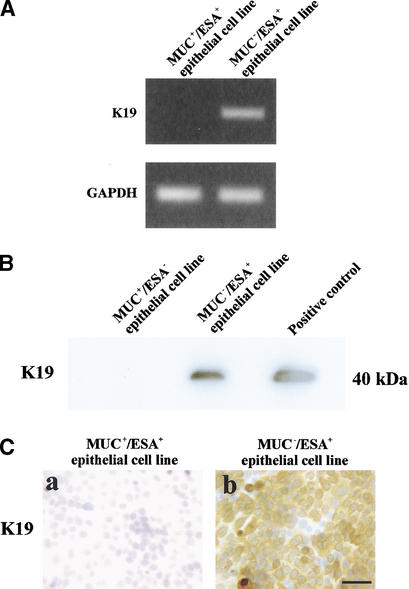
The MUC−/ESA+ epithelial cells are keratin K19+, similar to a subpopulation of cells in TDLU and neoplastic breast epithelial cells in vivo
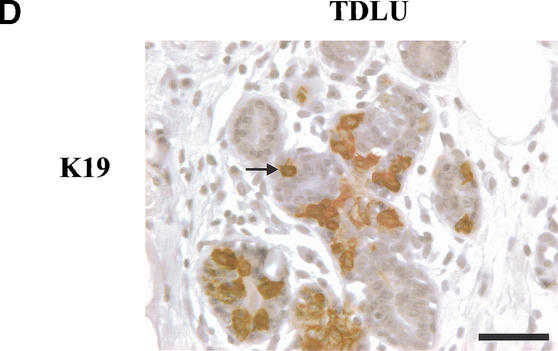
To identify a candidate subpopulation within TDLU in which the progenitor cell could reside, we performed an analysis of the markers expressed by the cell lines we had established. Keratin K19 was identified as a distinctive trait expressed by the MUC−/ESA+ as opposed to the MUC+/ESA+ epithelial cells (Table 1). This difference in phenotype was confirmed further by RT–PCR, immunoblotting, and immunostaining (Fig. 5A–C). It was reported 16 years ago (Bartek et al. 1985) that keratin K19 is expressed in only a subpopulation of luminal epithelial cells within TDLU. We confirmed the limited ex-pression of keratin K19 in vivo but also showed that some K19+ cells are suprabasal (Fig. 5D, arrow). It should be noted that in the majority of breast carcinomas, the neoplastic epithelial cells stain positive for keratin K19 (Bartek et al. 1985).
Table 1.
Keratin K19 is a distinctive trait expressed by the MUC − /ESA + epithelial cell line as revealed by immunocyto-chemical staining
| Differentiated trait | MUC+/ESA+ epithelial cell line | MUC−/ESA+ epithelial cell line | Thy-1+/ASMA+ myoepithelial cells |
|---|---|---|---|
| Claudin-1 | + | +/− | − |
| Occludin | + | −/+ | − |
| ESA | + | +/− | −/+a |
| Keratin K18 | + | +/− | −/+ |
| Keratin K19 | − | +/−b | −/+ |
| MUC | + | −/+ | − |
| E-cadherin | + | + | ND |
| Keratin 14 | −c | +/− | +/− |
| Thy-1 | − | −/+ | +/− |
| ASMA | − | −/+ | +/−d |
Figure 5.
The MUC−/ESA+ cells are keratin K19-positive similar to a subpopulation of cells in TDLU in vivo. (A) MUC+/ESA+ epithelial- and MUC−/ESA+ epithelial cells differ by expression of mRNA for keratin K19. RT–PCR of keratin K19 in MUC+/ESA+ epithelial- and MUC−/ESA+ epithelial cells. (B) MUC+/ESA+ epithelial- and MUC−/ESA+ epithelial cells differ by expression of keratin K19. Immunoblot of keratin K19 of protein lysates from MUC+/ESA+ epithelial- and MUC−/ESA+ epithelial cells. (C) Keratin K19 staining in cultures of MUC+/ESA+- and MUC−/ESA+-epithelial cells. Cultures were stained for keratin K19 by immunoperoxidase and counterstained with hematoxylin. Whereas the MUC+/ESA+ epithelial cells were completely negative, the other cell line was heterogeneous with a large contribution from keratin K19-positive cells (after passage 27). Bar, 50 μm. (D) Keratin K19 staining in TDLU in situ. Section of breast tissue showing a TDLU stained for keratin K19 and counterstained with hematoxylin. Note the heterogeneous staining and the presence of several stained suprabasal epithelial cells in the TDLU (arrow indicates an example). Bar, 50 μm.
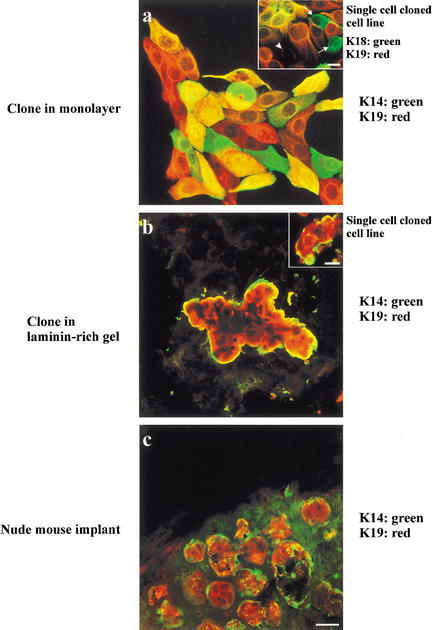
If the keratin K19+ cells were indeed potential candidate stem cells, then the MUC−/ESA+ epithelial cell line should show evidence of multipotency with regard to keratin K19 expression as well. This was tested in clonal cultures (limited dilution) double-stained for keratin K19 and the myoepithelial keratin K14. As seen in Figure 6a, clones could be identified that diversified into cells that were K14+ and K19+ alone or were positive for both. In addition, dual staining for K18 and K19 indicated the expected mix of markers (Fig. 6a, inset). Similarly, cloning in laminin-rich gels also resulted in formation of TDLU structures, which showed correct segregation of cells into suprabasally/luminally positioned K19+ cells and basally located K14+ cells (Fig. 6b). Finally, to provide yet further evidence for the morphogenic potential of these cells in vivo, we inoculated nude mice subcutaneously after preembedding the cells in a mixture of collagen gel and rBM. Using the entire population derived from reduction mammoplasties in primary culture, it was shown previously that human breast histology (TDLU) could be recapitulated (Yang et al. 1994). We used this assay to show that the MUC−/ESA+ epithelial cell line segregated into central keratin K19+ cells and basal keratin K14+ cells (Fig. 6c).
Figure 6.
Clonal segregation of keratin K19-positive and K14-positive cells in two- and three-dimensional culture and mouse implants of MUC−/ESA+ cells. Clonal culture of MUC−/ESA+ epithelial cells on monolayer collagen coated plastic (a), in a laminin-rich gel (b), and implanted orthotopically in the nude mouse (c) double-labeled with keratin K19 and keratin K14. Inset in a shows staining of the single cell cloned cell line, ‘TH123’, double-stained with keratin K19 and K18. Arrows indicate the presence of K19−/K18+ cells and arrowhead indicates a K19−/K18− cell. Collectively, the monolayer cultures show distinct evidence of bipotency. Inset in b shows an example of a colony from the single cell cloned cell line. In three-dimension organization resembles TDLU-like structures including terminal ducts and acini. (Bars, 20 μm).
We conclude that the MUC−/ESA+ epithelial cells are a plausible candidate for multipotent progenitors in the human breast. Their easily identifiable equivalent in vivo could therefore contain the candidate stem cells of the human breast.
Discussion
The experiments described here establish both the existence of multipotent progenitor cells in the human breast and the fact that they can be immortalized without loss of stem cell-like potential. The experiments also outline a method for their isolation and further characterization. The multipotent cell line belongs to the luminal epithelial lineage as evidenced by expression of ESA, claudin-1, keratins K18 and K19, and by the ability to form monolayers that display a high transepithelial electrical resistance TER. Embedding clonal populations in a three-dimensional basement membrane gel or in mammary fat pads of mice reveal that the stem cell-like MUC−/ESA+ epithelial cell line recapitulates an elaborate morphology closely reminiscent of TDLU in vivo. We propose that MUC−/ESA+ cells within the luminal epithelial lineage are precursors to human breast TDLUs and candidate breast stem cells.
Molecular characterization of the evolution of the cellular lineage in the human breast requires the ability to amplify the lineages to be studied. To achieve this goal, we immortalized the purified cells by transduction with HPV16–E6/E7 (Band et al. 1990; Band 1998; Garbe et al. 1999). The major concern with this technique is that although it has been shown that immortality is achieved by inactivation of p53 and retinoblastoma protein (RB), these may not be the only affected molecules and other cellular functions may also be affected (Garbe et al. 1999; for review, see Zwerschke and Jansen-Durr 2000). Evidence suggests, however, that human cells derived from E6/E7 immortalization retain much of their original phenotype. In organotypic cultures of endocervical cells, which originate from the target organ in vivo for HPV-16 infection, the cells appeared normal with ordinary stratification and production of a cornified layer (Halbert et al. 1992). Also, normal adult human pancreatic epithelial cells transfected with E6/E7 remained polarized on collagen gels, did not grow in soft agar, and expressed typical simple keratins (Furukawa et al. 1996). Thus, whereas E6 and E7 readily transform rodent cells, human cells appear to be significantly more robust. Infected human cells do not form tumors in nude mice even after >100 passages in culture (Willey et al. 1991; Band 1995; Furukawa et al. 1996). Immortalization of normal human breast cells with either E6 or E7 did not lead to aberrant functional behavior in luminal or myoepithelial cells tested (Wazer et al. 1995). It has been reported that breast cells may lose keratin K19 expression as a consequence of E7-immortalization (Spancake et al. 1999), but this is not necessarily the case as shown here (see below) and by others (Wazer et al. 1995). However, we cannot at present exclude some loss of K19+ cells in the MUC+/ESA+ cell line during immmortalization. Finally, human salivary gland cells transduced with E6/E7 were reported to remain diploid or near-diploid without a general destabilization of the karyotype (Queimado et al. 1999). We have confirmed these studies, and have found that the transduced breast cells are nontumorigenic and have a diploid karyotype (46, XX; T. Gudjonsson and C. Jin, unpubl.) even after more than one year in culture and 25 passages. It should be pointed out here that the immortalized clones are most robust before passage 40. Once the cells have reached passages beyond 50 they retain some stem-like properties but begin to behave more erratically in our hands.
The strategy for isolating progenitor cells in the human breast was based on two assumptions. First, recent data from a number of different groups including ours have implicated the luminal epithelial lineage as the source of bipotent cells in the human breast (Smith 1996; Stingl et al. 1998, 2001; Péchoux et al. 1999; Smalley et al. 1999). Second, ultrastructural studies in rodent mammary glands, but also to some extent in the human breast, have led to the identification of a basal ‘clear‘ cell as the candidate stem cell or first-degree progenitor cell (Rudland 1987; Smith and Chepko 2001). The finding that the candidate stem cells in the mouse mammary gland are ‘undifferentiated‘ (Smith and Chepko 2001) agrees with our finding that in the human breast these cells do not express sialomucin. That the MUC−/ESA+ putative stem cells, nevertheless, express large amounts of ESA, also known as the 40-kD cell adhesion molecule Ep-CAM, is also compatible with the reported expression of this molecule in putative stem cells of other tissues. Thus, in contrast to what is the general pattern of adhesion molecule expression such as cadherins, Ep-CAM is expressed highly in those parts of epithelial tissues that express low levels of other differentiation markers (Balzar et al. 1999).
One major and novel conclusion from our study is that the multipotent progenitor cells of the human breast reside in a predominantly keratin K19+ compartment. This was somewhat surprising because keratin K19− cells have been widely thought to be the potential stem cells of the breast. This belief has been based on earlier assumptions that stem cells should proliferate rapidly and that highly proliferative breast cell lines and to some extent benign proliferative lesions were keratin K19−, and that keratin K19+ cells proliferate poorly in culture (Bartek et al. 1985; Paine et al. 1992; Stampfer and Yaswen 1993; Rønnov-Jessen et al. 1996). However, more recent evidence supports our observation of a keratin K19+ precursor cell compartment. First, keratin K19 is one of the earliest keratins expressed in the embryo (Tamai et al. 2000), and whereas the fetal breast contains a homogenously keratin K19+ luminal epithelial compartment, keratin K19− luminal cells arise only in adulthood (Anbazhagan et al. 1998). Second, >90% of breast carcinomas are K19+. Although it could be argued that K19− luminal epithelial cells could express K19 along with malignant transformation, so far all reported transformations of nonmalignant K19− breast cell lines have resulted in K19− tumor cells (Petersen et al. 1998; Santner et al. 2001). Third, studies of other organs, including liver, pancreas, skin, testes, and prostate have revealed that the stem cell compartment expresses keratin K19 (Stosiek et al. 1990; Fridmacher et al. 1995; Michel et al. 1996; Bouwens 1998; Hudson et al. 2001). This does not imply that all keratin K19+ cells are progenitor cells because, for instance, the entire basal layer of the skin is K19+ and this by far exceeds the expected number of stem cells. It should be pointed out also that we can not at present rule out the possibility that the MUC−/K19+ cells may be downstream of the ultimate stem cell of the breast. Regardless, however, these cells are capable of generating a fundamental epithelial component of the breast.
Finally, it remains to be seen whether the ultrastructurally defined stem cell in the mouse mammary gland may also resemble the keratin K19+ cells of the human breast. The two species are not directly comparable because of the difference in the levels of glandular maturation in the virgin gland of mice versus the resting gland of humans (for review, see Rønnov-Jessen et al. 1996). In particular, the mouse mammary gland from nulliparous mice is less developed than the human breast and contains no TDLUs. Accordingly, keratin K19 staining of the virgin mouse mammary gland resembles staining of the fetal human breast, which also lacks TDLUs, that is, a more widespread staining of essentially all luminal epithelial cells (cf. Anbazhagan et al. 1998 and Smalley et al. 1998). An indication that these cells are progenitor cells for the entire mouse mammary gland comes from studies of primary cultured clones of sorted luminal epithelial cells (K19+/K18+) from 10-week-old virgin mice (Smalley et al. 1998, 1999). Such clones rapidly diversify (within a week) to generate K19−/K18+ indicating maturation towards heterogeneity reminiscent of that of adult human breast cells (Smalley et al. 1999). Although no information on staining of pregnant and lactating mouse mammary glands yet exists to confirm maturation-induced heterogeneity in vivo, additional evidence for profound similarities in the lineage evolution in mouse and human comes from studies of mouse cells on rBM gels. In the three-dimensional assays, a proportion of clones can form what is referred to as “mammospheres” with segregation of luminal epithelial (K19+/K18+/K14−) and myoepithelial (K19−/K18−/K14+) cells into a central and peripheral position, respectively. Such behavior was interpreted in favor of the presence of pluripotent stem cells (Smalley et al. 1999). It is possible that the mammospheres recorded in these cultures of mouse mammary epithelial cells, although morphologically less elaborate, are nevertheless the counterparts to the TDLUs seen in rBM cultures of human MUC−/ESA+ epithelial cells. As such they may be the result of the existence of similar progenitor cells in these mouse cultures. It is interesting to note that we estimated the frequency of human MUC−/ESA+ epithelial cells to be ∼8%, and the crude preparations of mouse luminal epithelial cells yielded multipotent clones with a frequency of ∼10% cells (Smalley et al. 1999). This also agrees well with previous published data for mice that the estimated frequency of candidate mammary stem cells and first-degree progenitor cells is ∼8%–12% (Smith and Chepko 2001). Taken together, it is likely that the mouse mammary stem cell is also both suprabasal and K19+ positive.
The isolation, immortalization, and characterization of the putative stem cells of the adult human breast, and the parallel between the rodent and the human breast stem cells may now make it possible to isolate and immortalize mammary stem cells from other species and other organs as well. Once we have pure populations of stem cells and progenitor cells, we can then start to analyze which genes regulate their behavior and see how these genes are disrupted during malignant progression. This information will be useful both for the treatment of breast cancer as well as possible tissue regeneration.
Materials and methods
Cell culture
Breast luminal epithelial cells were generated from primary cultures of biopsies from patients undergoing reduction mammoplasty for cosmetic reasons. The use of human material has been reviewed by the Regional Scientific–Ethical Committees for Copenhagen and Frederiksberg, Denmark and approved with reference to (KF) 01-161/98. The tissue was prepared as described previously (Péchoux et al. 1999). Briefly, it was mechanically disaggregated followed by enzymatic digestion with collagenase to release epithelial organoids. The organoids were plated in CDM3 medium (Petersen and van Deurs 1987, 1988) on collagen-coated (Vitrogen-100, Cohesion) T-25 flasks (Nunc). Cells were kept at 37°C in a humidified incubator with 5% CO2 and the medium was changed three times a week. In some instances organoids were trypsinized directly after collagenase digestion to obtain uncultured single cells for smears, which were fixed in methanol and further analyzed.
Cell separation
Cells were separated from six biopsies (D382, D490, D492, TH69 for immortalizations and TH82 and TH95 for short-term cultures). Luminal epithelial cells were purified after organoids had spread out to monolayers in primary culture. Cells were trypsinized and filtered as described previously (Péchoux et al. 1999). The luminal MUC+/ESA+ epithelial cells (D382) were separated immunomagnetically using two anti-sialomucin (115D8, Biogenesis Ltd.) columns and expanded for two passages on CDM6 medium (Péchoux et al. 1999) prior to immortalization. The MUC−/ESA+ epithelial cells were collected as the flow through population from an anti-sialomucin column and later (passage 3, 10, or 27) retention of cells by an anti-ESA (VU-1D9, NovoCastra) column. The time point of ESA retention did not affect the ability to generate TDLU’s. The MUC−/ESA+ cells were kept on DME-F12 supplemented with either choleratoxin (10 ng/mL; Sigma-Aldrich), epidermal growth factor (100 ng/mL; PreproTech EC LTD), and keratinocyte growth factor (10 μg/mL; PreproTech), or insulin (3 ug/mL; Boehringer Mannheim, Roche), hydrocortisone (1.4 μM; Sigma-Aldrich), epidermal growth factor (100 ng/mL), and 10% FCS or CDM3 prior to immortalization (Péchoux et al. 1999). All cultured breast epithelial cells expressed β4-integrin in the presence of choleratoxin or serum in contrast to fibroblasts (Gudjonsson et al. 2002; T. Gudjonsson, unpubl.). Thus, when fibroblasts were present they were removed by retaining epithelial cells in a β4-integrin column using MAB1964 antibody (Chemicon International). This step was optional. For isolation and purification of myoepithelial-derived cells we used an antibody against Thy-1 (AS02, Dianova). All cell separations were carried out by use of the MiniMACS magnetic cell separation system according to the manufacturer's instructions (Miltenyi Biotech).
Establishment of immortalized cell lines and clonal cultures
The MUC+/ESA+ epithelial and MUC−/ESA+ epithelial cells were transduced in passage 3 with sterile filtered retrovirus-containing supernatant from the PA317 LXSN HPV16E6E7 packaging cell line (CRL-2203, ATCC) in the presence of 8 μg/mL polybrene (Sigma-Aldrich) (Band et al. 1990; Wazer et al. 1995). Transduced cells were selected in the presence of 100 μg/mL G418 (Life Technologies). Established cell lines were kept routinely in H14 medium (Blaschke et al. 1994). Clonal cultures were prepared by limited dilution according to a protocol for prostate epithelial cells (Hudson et al. 2000). Briefly, 500, 1000, or 5 × 103 cells (5 × 104 in three-dimensional laminin rich gels) were plated onto T25-flasks in serum-free H14 medium and kept for 2 wk prior to staining. For single-cell cloning, cells were collected with a pipette and dispensed to a 96-well dish. The presence of only one single cell was verified visually under the phase contrast microscope. Cells were expanded and transferred to a 24-well dish or a T25 flask before Matrigel experiments. One clone (TH123) was used for further experimentation.
TER
For TER measurements, cells were plated on polycarbonate filters with a pore size of 0.4 μm (Corning Costar Corporation) and allowed to reach confluency. A Millicell-ERS volt-ohm meter (Millipore) was used to determine the TER value. All TER values were normalized for the area of the filter and were obtained after background subtraction. All experiments were done in triplicate.
RNA isolation and reverse transcription PCR
Total RNA was extracted from monolayer cultures with Trizol according to the manufacturer's instructions (Life Technologies). DNase-treated (DNase I Amp Grade, Life Technologies) total RNA (1.3 μg) was used as template for first strand synthesis with oligo dT primers (SuperScript First-Strand Synthesis System for RT–PCR, Life Technologies) in a 20 μL volume. A volume of 1 μL from this cDNA served as template for the subsequent PCR amplifications, using primers specific for HPV16 E6 and E7 (HPV16 E6 and HPV16 E7, respectively), keratin K19 (K19), sialomucin (MUC1), α smooth muscle actin (ASMA), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primer sequences, annealing temperature (TA), and number of amplification cycles for each reaction, as well as the resulting product size are listed below (Table 2). Each PCR reaction was initiated with a 15 min incubation step at 95°C, followed by the specified number of cycles with denaturation at 94°C, annealing at the specified TA, and extension at 72°C, for 1 min each, followed by a final extension step at 72°C for 7 min. Each reaction was performed in a 50 μL volume containing 2.5 U HotStar taq polymerase (QIAGEN, KEBO Lab A/S), 10× PCR buffer including MgCl2 (QIAGEN), 200 μM dNTP, and 200 nM of forward and reverse primers.
Table 2.
Primer sequences
| Primer | Sequence (5′-3′ direction) | TA (C) | Amplification cycles | Product size (bp) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV16 E6-FW | G | C | A | A | C | A | G | T | T | A | C | T | G | C | G | A | C | G | T | G | 55° | 30 | 234 | ||
| HPV16 E6-RV | G | G | A | C | A | C | A | G | T | G | G | C | T | T | T | T | G | A | C | A | |||||
| HPV16 E7-FW | G | A | T | G | G | T | C | C | A | G | C | T | G | G | A | C | A | A | G | C | 55° | 30 | 143 | ||
| HPV16 E7-RV | G | T | G | C | C | C | A | T | T | A | A | C | A | G | G | T | C | T | T | C | |||||
| K19-FW | G | A | G | G | T | G | G | A | T | T | C | C | G | C | T | C | C | G | G | G | C | A | 58° | 25 | 462 |
| K19-RV | A | T | C | T | T | C | C | T | G | T | C | C | C | T | C | G | A | G | C | A | G | ||||
| MUC1-FW | G | T | A | C | C | A | T | C | A | A | T | G | T | C | C | A | C | G | A | C | 60° | 30 | 351 | ||
| MUC1-RV | C | T | A | C | G | A | T | C | G | G | T | A | C | T | G | C | T | A | G | G | |||||
| αSM Actin-FW | G | G | A | A | T | C | C | T | G | T | G | A | A | G | C | A | G | C | T | C | 56° | 32 | 1200 | ||
| αSM Actin-RV | C | A | C | A | G | T | T | G | T | G | T | G | C | T | A | G | A | G | A | C | A | G | A | G | |
| GAPDH-FW | G | A | A | G | G | T | G | A | A | G | G | T | C | G | G | A | G | T | 54° | 25 | 226 | ||||
| GAPDH-RV | G | A | A | G | A | T | G | G | T | G | A | T | G | G | G | A | T | T | T | C |
Control amplification was performed on RNA samples not subjected to reverse transcription to verify that no contaminating genomic DNA was present (data not shown). The PCR products were analyzed by electrophoresis in 1.5% agarose gels.
TRAP-assay
The telomerase activity was determined with the TRAP assay (Kim et al. 1994) using the TRAPeze Telomerase Detection Kit (Intergen) according to the manufacturer's instructions. Cells were grown to 70%–80% confluence, trypsinized and counted. A lysate volume equal to 1000 cells was used for each reaction, and electrophoresed on a 12% nondenaturing acrylamide gel (Bio-Rad), stained in SYBR green 1 (Molecular Probes), and visualized by UV transillumination and image recording in a Gel Doc 1000 (Bio-Rad).
Matrigel experiments
For three-dimensional cultures, 2.5 × 105 MUC+/ESA+, MUC−/ESA+, or Thy-1+/ASMA+ cell lines or short-term cultures were plated separately inside rBM (Matrigel, lot no. 40230A, Becton Dickinson). Experiments were carried out in 24-well dishes (Nunc) using 300 μL Matrigel in which single cells were suspended. In some experiments the MUC−/ESA+ cells were re-retained in an ESA column prior to embedding in Matrigel to obtain optimal morphogenesis. Primary luminal epithelial cells, myoepithelial cells, and uncultured TDLU organoids from the breast were used as control (Petersen et al. 1992). The percentage of TDLU formation defined by branching of cell clusters on Matrigel was quantified after no longer than 6–14 d of culture by phase contrast microscopy using a 10× objective and a 10× eye piece.
Immunocytochemistry and confocal microscopy
Breast biopsies and cells cultured within rBM were frozen in n-hexan (Merck) and mounted in Tissue Freezing Medium (Leica Instruments) for sectioning. Frozen biopsies and rBM gels were sectioned at an 8-μm setting in a cryostat. The sections and cell cultures were dried for 15 min at room temperature and fixed in methanol as described previously (Petersen and van Deurs 1988). Primary antibodies were directed against keratin K18 (F3006; Trichem Aps), keratin K19 (BA17 and RCK108, DAKO, or Ab1 Neomarkers), sialomucin (MAM6, clone 115D8, Biogenesis), occludin (OC-3F10, Zymed Laboratories), polyclonal claudin-1 (Zymed), ESA (VU-1D9, Novocastra), E-cadherin (HECD-1, kindly provided by Dr. Atsushi Ochiai, Tokio, Japan), Thy-1 (AS0-2, Dianova), ASMA (1A4, Sigma-Aldrich), α1 chain of laminin-1 (EB7, kindly provided by I. Virtanen, University of Helsinki), and keratin K14 (LL002, Novocastra). Rabbit anti-mouse immunoglobulins (Z259, DAKO) were used as secondary antibodies and a peroxidase conjugated anti-peroxidase mouse mAb was used as tertiary antibody (P850, DAKO). The peroxidase reactions were performed using 0.5 mg/mL 3,3-diaminobenzidine (Sigma) and 0.5 μL/mL of 30% H2O2 (Merck) for 10 min. The cultures were counterstained with hematoxylin. For double-labeling experiments we used iso-type specific antibodies, all from Southern Biotechnology as described previously (Rønnov-Jessen et al. 1995). Antibody incubations were carried out for 30 min, and specimens were rinsed twice for 5 min each, all at room temperature. Some sections received a nuclear counter stain with 1 μg/mL propidium iodide (Molecular Probes). Afterwards sections were mounted with coverslips by use of Fluoromount-G (Southern Biotechnology) supplemented with 2.5 mg/mL _n_-propyl gallate (Sigma-Aldrich) as described previously (Rønnov-Jessen et al. 1992). For keratin K19 staining, biopsies were fixed routinely in formalin, parafin embedded, and sectioned at a 4 μm thickness. The antibodies were visualized by Streptavidin-Biotin (DAKO 5004; kindly performed by Dr. F. Rank, Rigshospitalet). Immunofluorescence was visualized using a Zeiss LSM 510 laser-scanning microscope (Carl Zeiss). Sections were observed by use of a 20×, 40×, 63×, or 100× objective and slicing in the z-plane into 0.25 μm-thick focal planes was indispensable for high quality FITC, Texas red, or propidium iodide exposures. Smeared cells double-stained for ESA and MUC were quantified by counting 100 cells in five different fields.
Immunoblotting
Semiconfluent T-25 flasks of MUC+/ESA+ and MUC−/ESA+ epithelial cells, and T47D breast cancer cells (positive control) were lysed for 30 min at 4°C in buffer containing 1% Triton X-100, 1% sodium deoxycholate, 10% glycerol, 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5 mM EDTA, 1 mM PMSF, 1% Trasylol, 100 μM NaVO3. The lysates were centrifuged and samples were subjected to 12% SDS-PAGE and run at 35 mV for 4 h. The loading of lanes was equilibrated based on protein determinations by the Bio-Rad protein assay (Bio-Rad Laboratories). The samples were electrophoretically transferred to Immun-Blot PVDF Membrane (Bio-Rad) at 400 mA for 3–4 h at 4°C in 20% methanol, 0.2 M glycine, and 25 mM Tris-HCl. Blots were blocked in phosphate-buffered saline containing 5% dried milk and 0.05% Tween-20 for 1 h at room temperature before probing with anti-keratin K19 (BA17). The blots were washed three times for 10 min in PBS containing 0.05% Tween-20 and then incubated with the anti-mouse IgG, horseradish peroxidase linked whole antibody (NA931, Amersham Pharmacia Biotech). After washing, bound antibodies were visualized using the ECL immunoblotting detection system (Amersham).
Transplantation into nude mice
The MUC+/ESA+ and MUC−/ESA+ epithelial cell lines were inoculated subcutaneously into BALB/C nude mice after preembedding 106 cells in 500 μL of a mixture of collagen and rBM (20% Matrigel/ 80% collagen) (Yang et al. 1994). The mice were sacrificed after one week and the implants were sectioned and stained (see above).
Acknowledgments
We thank Tove Marianne Lund for expert technical assistance and Dr. Derek Radisky for critical reading of the manuscript. The Aasted Clinic, the Private Clinic, and the Søllerød Plastic Surgery Clinic and Rigshospitalet (Dr. Fritz Rank) are gratefully acknowledged for providing the biopsy material. This work was supported by grants from the Icelandic Research Fund for Graduate Students, Dansk Kræftforskningsfond, the Dagmar Marshalls fond (to T.G.), The Danish Research Council, The Novo Nordisk Foundation, The Thaysen Foundation, Friis Fonden, The Meyer Foundation, The Danish Cancer Society, The Danish Research Council, The Danish Medical Association Research Fund (to O.W.P. and L.R.-J.), and The National Institutes of Health (grant CA-64786-02 to M.J.B. and O.W.P.) and United States Department of Energy Office of Biological and Environmental Research (contract DE-AC03-76SF00098 to M.J.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL o.w.petersen@mai.ku.dk; FAX 45-35327285.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.952602.
References
- Anbazhagan R, Osin PP, Bartkova J, Nathan B, Lane EB, Gusterson BA. The development of epithelial phenotypes in the human fetal and infant breast. J Pathol. 1998;184:197–206. doi: 10.1002/(SICI)1096-9896(199802)184:2<197::AID-PATH992>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17-1A antigen (Ep-CAM) J Mol Med. 1999;77:699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- Band V. Preneoplastic transformation of human mammary epithelial cells. Semin Cancer Biol. 1995;6:185–192. doi: 10.1006/scbi.1995.0015. [DOI] [PubMed] [Google Scholar]
- ————— The role of retinoblastoma and p53 tumor suppressor pathways in human mammary epithelial cell immortalization. Int J Oncol. 1998;12:499–507. doi: 10.3892/ijo.12.3.499. [DOI] [PubMed] [Google Scholar]
- Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc Natl Acad Sci. 1990;87:463–467. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Taylor-Papadimitriou J, Miller N, Millis R. Patterns of expression of keratin 19 as detected with monoclonal antibodies in human breast tissues and tumours. Int J Cancer. 1985;36:299–306. [PubMed] [Google Scholar]
- Bissell MJ, Weaver VM, Lelièvre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757s–1764s. [PubMed] [Google Scholar]
- Blaschke RJ, Howlett AR, Deprez P-Y, Petersen OW, Bissell MJ. Cell differentiation by extracellular matrix components. Meth Enzymol. 1994;245:535–556. doi: 10.1016/0076-6879(94)45027-7. [DOI] [PubMed] [Google Scholar]
- Bouwens L. Cytokeratins and cell differentiation in the pancreas. J Pathol. 1998;184:234–239. doi: 10.1002/(SICI)1096-9896(199803)184:3<234::AID-PATH28>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Fridmacher V, Le Bert M, Guillou F, Magre S. Switch in the expression of the K19/K18 keratin genes as a very early evidence of testicular differentiation in the rat. Mech Dev. 1995;52:199–207. doi: 10.1016/0925-4773(95)00401-l. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- Garbe J, Wong M, Wigington D, Yaswen P, Stampfer MR. Viral oncogenes accelerate conversion to immortality of cultured conditionally immortal human mammary epithelial cells. Oncogene. 1999;18:2169–2180. doi: 10.1038/sj.onc.1202523. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Rønnov-Jessen L, Villadsen R, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DL, O'Hare M, Watt FM, Masters JR. Proliferative heterogeneity in the human prostate: Evidence for epithelial stem cells. Lab Invest. 2000;80:1243–1250. doi: 10.1038/labinvest.3780132. [DOI] [PubMed] [Google Scholar]
- Hudson DL, Guy AT, Fry P, O'Hare MJ, Watt FM, Masters JR. Epithelial cell differentiation pathways in the human prostate: Identification of intermediate phenotypes by keratin expression. J Histochem Cytochem. 2001;49:271–278. doi: 10.1177/002215540104900214. [DOI] [PubMed] [Google Scholar]
- Jaeger MM, Kalinec G, Dodane V, Kachar B. A collagen substrate enhances the sealing capacity of tight junctions of A6 cell monolayers. J Membr Biol. 1997;159:263–270. doi: 10.1007/s002329900289. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;265:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Michel M, Torok N, Godbout MJ, Lussier M, Gaudreau P, Royal A, Germain L. Keratin 19 as a biochemical marker of skin stem cells in vivo and in vitro: Keratin 19 expressing cells are differentially localized in function of anatomic sites, and their number varies with donor age and culture stage. J Cell Sci. 1996;109:1017–1028. doi: 10.1242/jcs.109.5.1017. [DOI] [PubMed] [Google Scholar]
- Paine TM, Soule HD, Pauley RJ, Dawson PJ. Characterization of epithelial phenotypes in mortal and immortal human breast cells. Int J Cancer. 1992;50:463–473. doi: 10.1002/ijc.2910500323. [DOI] [PubMed] [Google Scholar]
- Péchoux C, Gudjonsson T, Rønnov-Jessen L, Bissell MJ, Petersen OW. Human mammary luminal epithelial cells contain progenitors to myoepithelial cells. Dev Biol. 1999;206:88–99. doi: 10.1006/dbio.1998.9133. [DOI] [PubMed] [Google Scholar]
- Petersen OW, van Deurs B. Characterization of epithelial membrane antigen expression in human mammary epithelium by ultrastructural immunoperoxidase cytochemistry. J Histochem Cytochem. 1986;34:801–809. doi: 10.1177/34.6.3009605. [DOI] [PubMed] [Google Scholar]
- ————— Preservation of defined phenotypic traits in short-time cultured human breast carcinoma derived epithelial cells. Cancer Res. 1987;47:856–866. [PubMed] [Google Scholar]
- ————— Growth factor control of myoepithelial-cell differentiation in cultures of human mammary gland. Differentiation. 1988;39:197–215. doi: 10.1111/j.1432-0436.1988.tb00094.x. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Rønnov-Jessen L, Weaver VM, Bissell MJ. Differentiation and cancer in the mammary gland: Shedding new light on an old dichotomy. Adv Cancer Res. 1998;75:135–161. doi: 10.1016/s0065-230x(08)60741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queimado L, Lopes C, Du F, Martins C, Fonseca I, Bowcock AM, Soares J, Lovett M. In vitro transformation of cell lines from human salivary gland tumors. Int J Cancer. 1999;81:793–798. doi: 10.1002/(sici)1097-0215(19990531)81:5<793::aid-ijc21>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Rudland PS. Stem cells and the development of mammary cancers in experimental rats and humans. Cancer Met Rev. 1987;6:55–83. doi: 10.1007/BF00047609. [DOI] [PubMed] [Google Scholar]
- Rønnov-Jessen L, Petersen OW, Koteliansky VE, Bissell MJ. The origin of the myofibroblasts in breast cancer: Recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J Clin Invest. 1995;95:859–873. doi: 10.1172/JCI117736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: The importance of the stromal reaction. Physiol Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- Sapino A, Macri L, Gugliotta P, Bussolati G. Evaluation of proliferating cell types in human and mouse mammary gland by a double immunostaining procedure. Acta Histochem. 1990;Suppl.-Band XL:81–84. [PubMed] [Google Scholar]
- Smalley MJ, Titley J, O'Hare MJ. Clonal characterization of mouse mammary luminal epithelial and myoepithelial cells separated by fluorescence-activated cell sorting. In Vitro Cell Dev Biol Anim. 1998;34:711–721. doi: 10.1007/s11626-998-0067-0. [DOI] [PubMed] [Google Scholar]
- Smalley MJ, Titley J, Paterson H, Perusinghe N, Clarke C, O'Hare MJ. Differentiation of separated mouse mammary luminal epithelial and myoepithelial cells cultured on EHS matrix analyzed by indirect immunofluorescence of cytoskeletal antigens. J Histochem Cytochem. 1999;47:1513–1524. doi: 10.1177/002215549904701203. [DOI] [PubMed] [Google Scholar]
- Smith GH. Experimental mammary epithelial morphogenesis in an in vivo model: Evidence for distinct cellular progenitors of the ductal and lobular phenotype. Breast Cancer Res Treat. 1996;39:21–31. doi: 10.1007/BF01806075. [DOI] [PubMed] [Google Scholar]
- Smith GH, Medina D. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J Cell Sci. 1988;89:173–183. doi: 10.1242/jcs.90.1.173. [DOI] [PubMed] [Google Scholar]
- Smith GH, Chepko G. Mammary epithelial stem cells. Microscopy Res Techn. 2001;52:190–203. doi: 10.1002/1097-0029(20010115)52:2<190::AID-JEMT1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Spancake KM, Anderson CB, Weaver VM, Matsunami N, Bissell MJ, White RL. E7-transduced human breast epithelial cells show partial differentiation in three-dimensional culture. Cancer Res. 1999;59:6042–6045. [PubMed] [Google Scholar]
- Stampfer MR, Yaswen P. Culture systems for study of human mammary epithelial cell proliferation, differentiation, and transformation. Cancer Surveys. 1993;18:7–34. [PubMed] [Google Scholar]
- Stampfer MR, Garbe J, Levine G, Lichtsteiner S, Vasserot AP, Yaswen P. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor beta growth inhibition in p16INK4A(−) human mammary epithelial cells. Proc Natl Acad Sci. 2001;98:4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67:93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- Stosiek P, Kasper M, Karsten U. Expression of cytokeratin 19 during human liver organogenesis. Liver. 1990;10:59–63. doi: 10.1111/j.1600-0676.1990.tb00436.x. [DOI] [PubMed] [Google Scholar]
- Tamai Y, Ishikawa T, Bosl MR, Mori M, Nozaki M, Baribault H, Oshima RG, Taketo MM. Cytokeratins 8 and 19 in the mouse placental development. J Cell Biol. 2000;151:563–572. doi: 10.1083/jcb.151.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane EB, Leigh IM. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: Relation to in vivo phenotypes ad influence of medium. J Cell Sci. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- Wazer DE, Liu X-L, Chu C, Gao Q, Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci. 1995;92:3687–3691. doi: 10.1073/pnas.92.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55:231–273. [PubMed] [Google Scholar]
- Willey JC, Broussoud A, Sleemi A, Bennett WP, Cerutti P, Harris CC. Immortalization of normal human bronchial epithelial cells by human papillomaviruses 16 or 18. Cancer Res. 1991;51:5370–5377. [PubMed] [Google Scholar]
- Yang J, Guzman RC, Popnikolov N, Bandyopadhyay GK, Christov K, Collins G, Nandi S. Phenotypic characterization of collagen gel embedded primary human breast epithelial cells in athymic nude mice. Cancer Lett. 1994;81:117–127. doi: 10.1016/0304-3835(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Zwerschke W, Jansen-Durr P. Cell transformation by the E7 oncoprotein of human papillomavirus type 16: Interactions with nuclear and cytoplasmic target proteins. Adv Cancer Res. 2000;78:1–29. doi: 10.1016/s0065-230x(08)61022-2. [DOI] [PubMed] [Google Scholar]