Sex and virulence in Escherichia coli: an evolutionary perspective (original) (raw)
Abstract
Pathogenic Escherichia coli cause over 160 million cases of dysentery and one million deaths per year, whereas non-pathogenic E. coli constitute part of the normal intestinal flora of healthy mammals and birds. The evolutionary pathways underlying this dichotomy in bacterial lifestyle were investigated by multilocus sequence typing of a global collection of isolates. Specific pathogen types [enterohaemorrhagic E. coli, enteropathogenic E. coli, enteroinvasive E. coli, K1 and _Shigella_] have arisen independently and repeatedly in several lineages, whereas other lineages contain only few pathogens. Rates of evolution have accelerated in pathogenic lineages, culminating in highly virulent organisms whose genomic contents are altered frequently by increased rates of homologous recombination; thus, the evolution of virulence is linked to bacterial sex. This long-term pattern of evolution was observed in genes distributed throughout the genome, and thereby is the likely result of episodic selection for strains that can escape the host immune response.
Introduction
Pathogenic bacteria present ongoing challenges to human and animal health but the processes by which virulence has evolved remain incompletely understood, even for bacteria as well studied as Escherichia coli. E. coli is ubiquitous, asymptomatically colonizing the intestines of mammals and birds, and is widely distributed in the environment. E. coli also includes pathogens of global significance that are responsible for epidemic dysentery (e.g. Shigella), neonatal meningitis (associated with the K1 capsular polysaccharide), haemolytic uraemic syndrome (O157:H7) as well as a variety of other diseases. Virulence is often associated with the presence of specific gene clusters, termed ‘pathogenicity islands’ (Groisman and Ochman, 1996; Hacker and Kaper, 2000). However, it remains unclear how virulence affects the overall patterns of evolution within a genome. Addressing such questions requires a global overview of how diversity has evolved within the species at large.
E. coli was the first bacterium for which population genetic techniques were introduced. Results of multilocus enzyme electrophoresis (MLEE) indicated that certain combinations of alleles occurred multiple times, which was interpreted as indicating a clonal population structure with infrequent recombination (Selander and Levin, 1980). Further support for this conclusion was provided by subsequent MLEE analyses of 1000s of natural and clinical isolates from humans and other sources; 72 of these isolates, the ECOR collection, were chosen to represent the known genetic diversity at that time (Ochman and Selander, 1984). The ECOR collection was subdivided into four groups, designated A, B1, B2 and D, plus a minor group E that has largely been ignored because it clustered inconsistently in subsequent analyses. Phylogenetic trees of housekeeping gene sequences from the ECOR collection indicated that group D diverged first and that groups A and B1 are sister groups that separated later (Nelson et al., 1991; 1997; Nelson and Selander, 1992; Boyd et al., 1994; Wang et al., 1997). More recent analyses suggest that perhaps B2, rather than D, is ancestral (Lecointre et al., 1998; Escobar-Paramo et al., 2004a).
Based on the examination of MLEE data from a variety of sources, Maynard Smith et al. (1993) questioned whether most species of bacteria were truly clonal. Indeed, recombination has largely destroyed statistical support for trees of housekeeping genes within Streptococcus pneumoniae and Neisseria meningitidis (Feil et al., 2001), and the limited genetic diversity among five closely related members of the ECOR collection was attributed to recombination, rather than to mutation (Guttman and Dykhuizen, 1994). Furthermore, hyper-mutable and hyper-recombinant phenotypes in E. coli have been detected in laboratory experiments (Sniegowski et al., 1997; Vulic et al., 1999; Cooper and Lenski, 2000) and among natural isolates (Denamur et al., 2000). If recombination were frequent in E. coli, it would blur phylogenetic signals (Holmes et al., 1999; Schierup and Hein, 2000), reduce the degree of differentiation between groups and erase statistical support for any inferred branching patterns of trees. However, trees based on different E. coli genes were congruent and seemed to differ statistically from random trees (Reid et al., 2000; Feil et al., 2001). Therefore, the contribution of recombination to the population structure of E. coli, though controversial, seems to be minor.
Here, we describe a publicly available database of extensive sequence data from housekeeping gene fragments for a global sample of E. coli. These sequences were used to estimate the population structure of E. coli and to demonstrate that recombination is widespread within this species. As a result, the ancestry of numerous hybrid isolates is derived from multiple sources. Furthermore, it is exactly these hybrid strains that tend to be highly virulent, whereas genetic admixture is less frequent among avirulent isolates, suggesting that sex and virulence are causally related.
Results
Fragments of seven housekeeping genes distributed around the E. coli chromosome were sequenced from 462 isolates from diverse sources (Fig. 1A, Table S1). These strains were isolated in Europe, Africa, North America and the Pacific Rim from humans and 41 species of domesticated, captive and wild mammals, birds and reptiles (Table S2). The isolates derive from a variety of extraintestinal and intestinal diseases and from the faeces of healthy individuals. To facilitate retrospective analyses, the strains tested include the complete ECOR reference collection. Strain and sequence data are freely available for interrogation and data submission via a public website (http://web.mpiib-berlin.mpg.de/mlst), to which data for several hundred additional strains has been submitted since this analysis was completed.
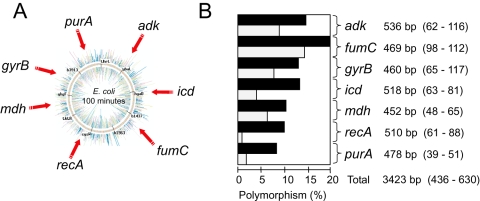
Fig. 1.
(A) Genomic locations and (B) genetic diversity of seven housekeeping genes. (B) Polymorphism levels for each gene are indicated in the histogram in which black bars reflect nucleotide polymorphisms and grey bars indicate amino-acid polymorphisms. Each gene symbol is followed by the length of the sequenced gene fragment (informative sites – polymorphic sites).
There is no clear phylogeographic component to the sources of diversity, nor is there a strong correlation between genetic groups and the host species from which bacteria were isolated. The following analyses therefore focus on phylogenetic and population genetic aspects of the entire dataset.
Population expansions and contractions
Eight to 20% of the nucleotides were polymorphic within each of the seven gene fragments (Fig. 1B), for a total of 630 single nucleotide polymorphisms over the 3423 bp sequenced from each strain. Neighbour-joining trees of the concatenated sequences showed that two (0.4%) of the isolates, from a dog and a parrot, differed strongly from the main group of isolates, which were much more closely related (Fig. 2A). Thus, E. coli is considerably more diverse than had been previously appreciated and the ECOR collection contains only a subset of the full genetic diversity within E. coli. Second, the tree topology for the 460 isolates in the main group is star-like, as expected if the vast majority of modern E. coli represented the results of population expansion subsequent to a major historical bottleneck or selective sweep that removed most of the extant diversity (Cohan, 2002).
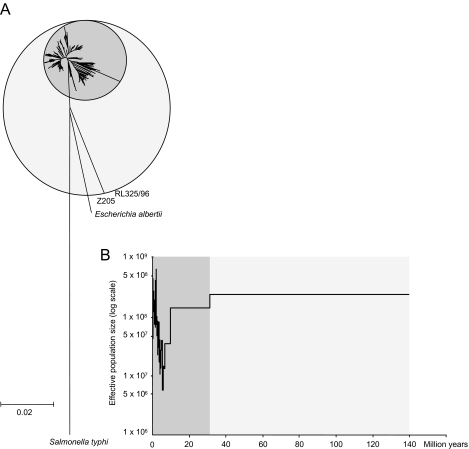
Fig. 2. Diversity and population size within E. coli.
A. Neighbour-joining (NJ) tree of 462 E. coli, three E. albertii and one Salmonella typhi (outgroup) based on concatenated sequences from the seven loci using pairwise genetic distances based on the GTR+G+I evolutionary model. The dark grey circle represents the main group of 460 isolates within E. coli whereas the light grey circle also encompasses the two divergent isolates.
B. Generalized skyline plot showing changes in effective E. coli population size over the last 140 million years. The ultrametric tree used was a UPGMA tree calculated from the genetic distances in part A and calibrated to a molecular clock rate of 7.6 × 10−10 per year.
Generalized skyline plots can be used to calculate the statistical probability of different demographic scenarios and to estimate when changes in population size occurred (Strimmer and Pybus, 2001). Although they are based on a coalescent approach that is appropriate for non-recombining sequences, such as mitochondrial DNA, they have also been used for the analysis of HIV phylogenies, wherein recombination is frequent (Strimmer and Pybus, 2001). When applied to the genetic distances estimated for the E. coli data, this algorithm indicated that models of population structure which included piece-wise demographic expansion or exponential growth were much more likely than a model of constant population size (log likelihood = −4430.94; P < 0.01). The skyline plots also indicated that a major population expansion had occurred within the last 5 million years during which the effective population size increased by an estimated 50-fold (1 × 107 − 5 × 108), following a prior contraction that began 10–30 million years ago (Fig. 2B). The two exceptional strains, which are almost as distant from most E. coli as are isolates of a distinct species, Escherichia albertii, are likely to be remnants of the original diversity that existed in E. coli prior to the population contraction event. Hereafter, we concentrate on genetic diversity within the main modern group of E. coli isolates.
Limitations of phylogenetic inferences within modern E. coli
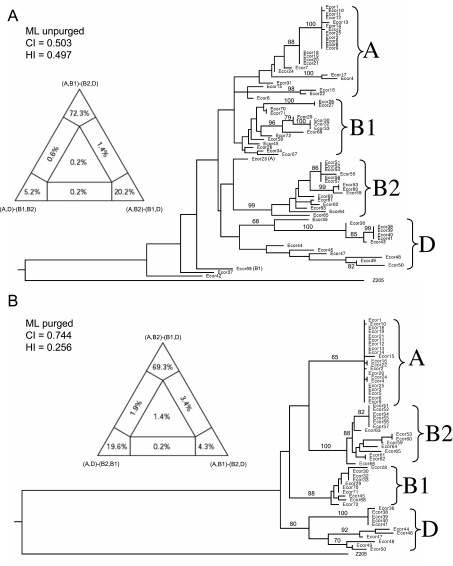
Concatenated sequences from the ECOR collection fell into four phylogenetic clades whose composition was largely concordant with MLEE groups A, B1, B2 and D (Fig. S1). In agreement with previous nucleotide sequence-based analyses, the neighbour-joining trees of the concatenated sequences place group D as ancestral. However, as is commonly the case for star-like phylogenies, bootstrap support values for this tree (Fig. S1) were very low (data not shown) and the branch lengths to the four clades were very short, both suggesting that the branch order is not reliable. To resolve such ambiguities, we performed heuristic maximum likelihood analysis, which provides statistical measures of reliability of tree topology. This algorithm yielded an alternate tree topology (Fig. 3A) in which the A/B1 (A plus B1) groups branched contemporaneously from the root with B2/D.
Fig. 3.
Phylogenies of concatenated sequences from the ECOR collection. Left: Four-cluster likelihood mapping analysis (TREE-PUZZLE) represented as triangles showing likelihood supports for each of three alternative topologies (at tips of diagram), as well as support for unresolved quartets (centre) and for partly resolved quartets (edges). Right: Heuristic maximum likelihood trees based on neighbour-joining (NJ) starting trees with NNI branch swapping. The group labels reflect the original groupings based on MLEE and the numbers at the nodes are bootstrap confidence values above 70%.
A. Original sequences. ML settings = GTR+G+I; G = 0.4846; I = 0.8042.
B. Sequences purged of recombinant sites. ML settings = GTR+G+I; G = 0.7066; I = 0.7245. CI, Consistency index; HI, Homoplasy index.
Due to these contradictory results, we attempted to deduce the true branching order by purging recombinant sites (see Experimental procedures) and then testing all possible topologies of both purged and unpurged maximum likelihood trees by the Shimodaira–Hasegawa likelihood test (S–H test), and by likelihood mapping according to quartet puzzling (Strimmer and von Haeseler, 1997). According to the S–H test, the unpurged topology where A/B1 branched at the same time as B2/D received the strongest statistical support, but it was not significantly better than a tree in which D was ancestral (Table 1). The likelihood of this topology was only approximately 70% according to quartet puzzling versus 20% for (A,B2)-(B1,D) (Fig. 3A, left). After purging recombinant sites, the data yielded a higher consistency index and a lower Homoplasy ratio (see below), and the major groups were more clearly defined (Fig. 3B and Fig. S1). In this case, D is ancestral and A branched contemporaneously with B1; but again, the most likely branching order was not significantly better than other topologies (Table 1, Fig. 3B, left). We were also unable to resolve the branching order of the groups by comparing two-group clades with an outgroup, either in purged or unpurged trees (data not shown), and phylogenetic trees of sequences from all 463 strains yielded results comparable to those limited to the 72 strains from the ECOR collection.
Table 1. Shimodaira–Hasegawa tests of constrained maximum likelihood trees using the GTR+G+I model.
| Tree | Topology | –Ln L | Diff – Ln L | P |
|---|---|---|---|---|
| 1 | (Z205,(A,(B1,(D,B2)))) | 9205.15 | 61.82 | 0.000* |
| 2 | (Z205,(B1,(A,(D,B2)))) | 9194.72 | 51.39 | 0.001* |
| 3 | (Z205,(B2,(D,(A,B1)))) | 9167.37 | 24.04 | 0.043* |
| 4 | (Z205,(D,(B2,(A,B1)))) | 9157.41 | 14.08 | 0.299 |
| 5 | (Z205,((A,B1),(D,B2))) | 9143.33 | Best | |
| 1 | (Z205,(A,(B1,(D,B2)))) | 7353.18 | 7.90 | 0.222 |
| 2 | (Z205,(B1,(A,(D,B2)))) | 7345.61 | 0.33 | 0.778 |
| 3 | (Z205,(B2,(D,(A,B1)))) | 7353.39 | 8.11 | 0.190 |
| 4 | (Z205,(D,(B2,(A,B1)))) | 7345.28 | Best | |
| 5 | (Z205,((A,B1),(D,B2))) | 7345.55 | 0.27 | 0.802 |
We conclude that, despite claims that E. coli is largely clonal (Feil et al., 2001), it is not possible to convincingly deduce the ancestral relationships among the major modern groups by phylogenetic reconstructions. This phenomenon may be due to a fast radiation of the groups after the population bottlenecks and/or to frequent recombination within E. coli, as described below.
Reticulate evolution rather than clonality
Population genetic tools can be more appropriate than phylogenetic trees for deducing deep evolutionary splits in species where homologous recombination is common. To this end, we analysed the polymorphisms in all seven gene fragments with STRUCTURE (Pritchard et al., 2000; Falush et al., 2003a), which employs a Bayesian method to discern groupings among recombining organisms. The linkage model of STRUCTURE assigns probabilities of derivation from ancestral source groups for each polymorphic nucleotide. The ancestry of each strain is then estimated as the summed probability of derivation from each group over all polymorphic nucleotides. STRUCTURE recognized four ancestral sources of polymorphisms within E. coli, and separated most ECOR isolates into groups whose membership was consistent with their original assignments to groups A, B1, B2 and D. However, within the entire dataset, numerous strains fell into hybrid groups that contain significant ancestry from multiple sources (Fig. 4A, Table S2).
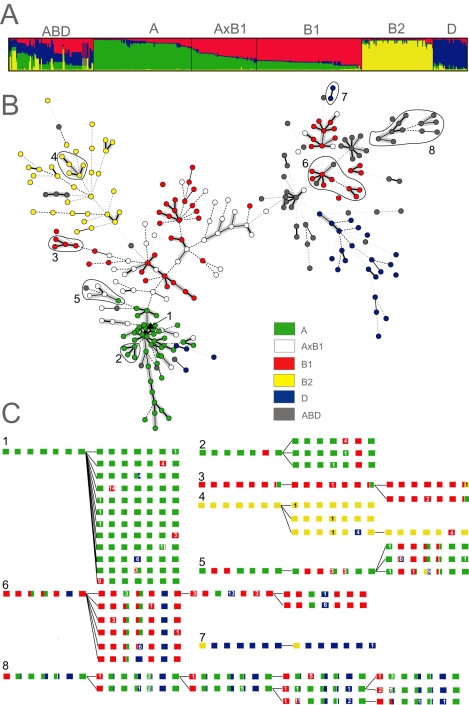
Fig. 4. Ancestry of 460 E. coli isolates.
A. Proportions of ancestry from groups A, B1, B2 and D as inferred by STRUCTURE and their assignment to six groups as displayed with DISTRUCT (Rosenberg, 2004). The plot shows one vertical line for each isolate indicating the proportions of ancestry from the four groups, colour-coded as in part B.
B. Distribution of A, B1, B2, D, AxB1 and ABD within a minimal spanning tree (MSTREE) of 275 E. coli STs based on the degree of allele sharing. Dots are coloured according to group and lines connecting ST complexes (see Fig. 6) are shaded in grey. Clades that are illustrated in greater detail in part c are indicated by numbers and encircled by lines.
C. Details of microevolution within eight selected clades. The branch order within each clade and the ancestral ST were taken from the MSTREE. Boxes are colour-coded as in panel B and indicate stretches of nucleotides that are derived from the four ancestral groups A, B1, B2 and D within each of the seven loci, shown in the order: adk, fumC, gyrB, icd, mdh, purA and recA. Numbers in contrasting colours indicate the numbers of nucleotides that differ from the allele that was previously present.
We assigned two-thirds of the strains (312/460), whose proportion of nucleotides from one of the four ancestral sources exceeded a threshold value of 2/3, to groups A, B1, B2 and D, and one-third (148) to hybrid groups. (This 2/3 threshold resulted in 96% concordance between our assignments of ECOR strains and their original groupings based on MLEE.) In addition to A, B1, B2 and D, we defined one hybrid group called AxB1, containing strains which derive most of their ancestry from A and B1, and a second group called ABD, where extensive recombination has yielded a highly heterogeneous set of isolates with multiple sources of ancestry.
Groups ABD and AxB1 contain isolates where recombination has been particularly frequent, but recombination is not exclusive to these hybrid groups. Virtually all E. coli possess some imported nucleotides (Fig. 4A), and many of the allelic differences between related isolates reflect mosaics due to homologous recombination with other groups (Fig. 4C), thereby augmenting the genetic diversity provided by mutation.
We sought to quantify the frequency of homologous recombination within groups by two independent measures, the Homoplasy ratio (Maynard Smith and Smith, 1998), which is applicable to sequences that differ by up to 2% (Posada et al., 2002), and the composite likelihood of r/μ (recombination rate/mutation rate) (McVean et al., 2002). Both tests confirm that significant levels of recombination had occurred within each of the E. coli groups (Table 2 and Table S3), and that the levels of recombination differ between the groups.
Table 2. Estimates of mutation (θ) and recombination (ρ) rates.
| θ | ρ | r/μ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B2 | B1 | D | A | B2 | B1 | D | A | B2 | B1 | D | |
| adk | 0.011 | 0.008 | 0.004 | 0.012 | 0.000 | 0.013 | 0.008 | 0.015 | 0.000 | 1.554 | 2.050 | 1.282 |
| fumC | 0.009 | 0.009 | 0.006 | 0.027 | 0.003 | 0.010 | 0.021 | 0.043 | 0.303 | 1.100 | 3.621 | 1.578 |
| gyrB | 0.004 | 0.007 | 0.007 | 0.012 | 0.204 | 0.009 | 0.008 | 0.004 | 47.37 | 1.290 | 1.147 | 0.304 |
| icd | 0.004 | 0.009 | 0.010 | 0.019 | 0.011 | 0.013 | 0.010 | 0.031 | 2.683 | 1.523 | 1.020 | 1.602 |
| mdh | 0.005 | 0.006 | 0.004 | 0.009 | 0.002 | 0.022 | 0.007 | 0.034 | 0.308 | 3.633 | 1.581 | 3.574 |
| purA | 0.004 | 0.006 | 0.004 | 0.007 | 0.000 | 0.024 | 0.012 | 0.016 | 0.000 | 3.918 | 2.733 | 2.219 |
| recA | 0.002 | 0.007 | 0.005 | 0.010 | 0.000 | 0.017 | 0.007 | 0.003 | 0.263 | 2.297 | 1.478 | 0.337 |
| Mean | 0.006 | 0.007 | 0.006 | 0.014 | 0.002 | 0.015 | 0.009 | 0.029 | 0.321 | 2.053 | 1.614 | 2.139 |
The lowest Homoplasy ratio (0.30) was observed in group A, consistent with the estimates of r/μ, which are close to zero for five of the seven genes in group A (Table 2). Higher Homoplasy ratios were detected in the other groups and r/μ values were more frequently significant in B1 and D. Furthermore, mean r/μ ratios were about five to six times higher in the B1, B2 and D groups than in the A group, showing that mutation is the main driving evolutionary force in the A group whereas recombination is more prevalent in the others (Table 2). We also note that the mutation rate within group D is higher than within the other groups.
Each of the four major groups of E. coli contains both pathogens and non-pathogens but their relative proportions differ widely among groups (Fig. 5A). For the following, we focus on the five pathogen types that are most frequently represented in the database (Table S2): EHEC (enterohaemorrhagic E. coli; one of the primary food-borne pathogens), EPEC (enteropathogenic E. coli associated with infantile and traveller's diarrhoea), EIEC (enteroinvasive E. coli), Shigella and K1 E. coli. Most striking is the fact that highly virulent pathogens are rare in group A, which contains only a single EPEC isolate and no K1, EHEC, EIEC or Shigella. In contrast, the hybrid groups are particularly rich in pathogens, suggesting a link between virulence and homologous recombination. Of the 61 Shigella strains, which cause epidemic disease, 34 were assigned to the hybrid groups ABD and AxB1, 27 were assigned to B1 and D, and none were assigned to A or B2 (Table S2). Similarly, 24 EIEC were in the hybrid ABD and AxB1 groups versus 14 in B1 and D and genetic mosaicism within the housekeeping genes was quite frequent even among pathogens that were assigned to B1 and D (Fig. 4C).
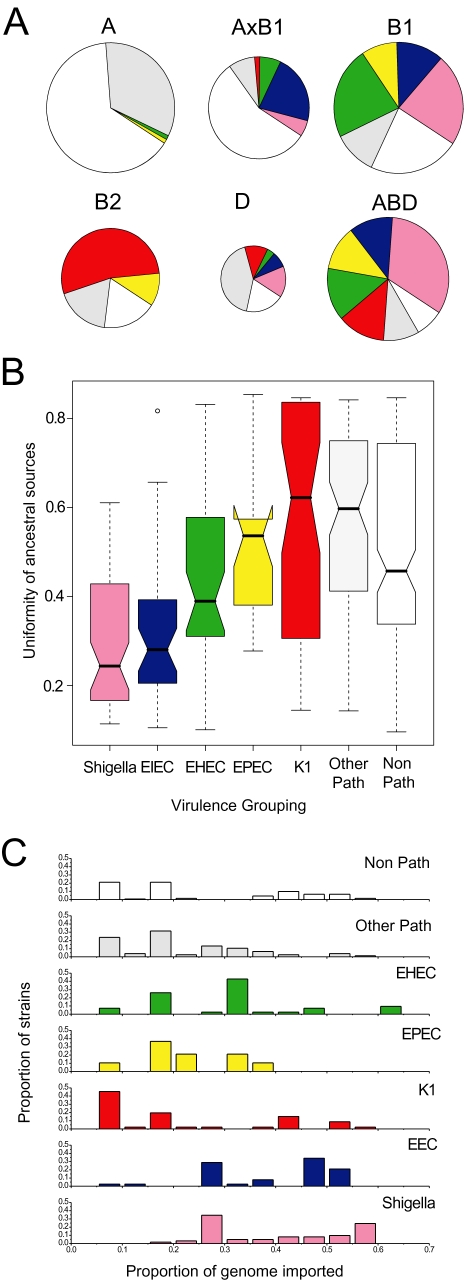
Fig. 5. Sources of ancestry versus pathogenicity.
A. Proportion of pathogens per group. The six groups are represented by circles whose areas are proportional to the numbers of isolates. Arcs are colour-coded as in panel B and indicate the frequencies of pathogen types.
B. Notched box and whisker plots of the uniformity of ancestral sources by pathogen group. Within each plot, the central box indicates the two central quartiles, separated by a horizontal line indicating the median value. The 95% confidence limits of the median are indicated by a notch; pairs of boxes whose notches do not overlap possess significantly different median values (P < 0.05). Lines above and below each box indicate the upper and lower quartiles, and outliers are indicated by small circles. Uniformity was calculated for each strain as the sum of its squared ancestries from the A, B1, B2 and D groups according to STRUCTURE, which ranges from 0.25 to 1.0, followed by normalization to a range from 0 to 1.
C. Frequency of isolates versus proportion of genome that has been imported from other groups by pathogen type. For each strain, the proportion of imported DNA was estimated conservatively by subtracting its maximal ancestry from any single group (A, B1, B2 or D) from 1.0. Other Path: other pathogens; Non-Path: non-pathogens.
These observations indicate that different groups of pathogens may be associated with different frequencies of homologous recombination. We therefore calculated the genetic uniformity of ancestral sources by pathogen group (Fig. 5B). The results showed a statistically significant trend of increasing uniformity from Shigella and EIEC through EPEC through EHEC, K1 and other pathogens. In general, non-pathogens have a less uniform ancestry than do EHEC, K1 and other pathogens, but this may reflect assignment errors because non-pathogens consist of a mixture of strains whose pathogenicity determinants have not been determined and/or which were isolated from healthy individuals. Similar results were obtained with histograms of the frequency of strains versus the proportion of imported nucleotides (Fig. 5C). Thus, the most virulent strains of E. coli have also undergone the greatest degree of homologous recombination between ancestral sources.
Allele-based population genetic structure of pathogens and non-pathogens
The population structure of microbial species with intermediate levels of recombination can be revealed by allele-based analyses (Maiden et al., 1998; Feil, 2004). The 630 sequence polymorphisms defined 50–82 unique sequences for each of the seven gene loci, which are referred to as alleles. Each unique combination of alleles was assigned a sequence type (ST) number, e.g. ST1, ST2, and many of the resulting 278 STs fall into groups of related STs. Related STs were assigned to 23 so-called ST complexes, using the principles of the eBurst algorithm (Feil et al., 2004): each ST complex includes at least three STs that differ from their nearest neighbour by no more than two of the seven loci while ST complexes differ from each other by three or more loci. STs that did not match the criteria for inclusion within an ST complex are simply referred to by their ST designation.
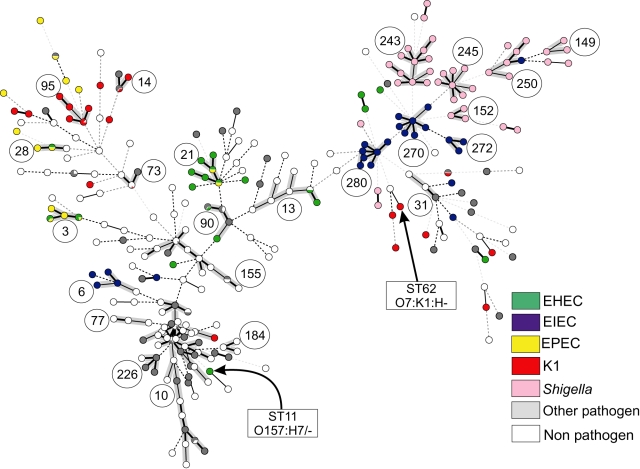
The allele-based relationships within a minimal spanning tree, referred to as an MSTREE, correlate strongly with the assignments to groups by STRUCTURE (Fig. 4B). STs that had been assigned to A, B1, B2 or D cluster together with others assigned to the same group within the MSTREE. Almost all group A isolates correspond to ST complex 10 (compare Figs 4 and 6), whereas the other groups each contained multiple ST complexes and individual STs. STs containing AxB1 isolates are distributed in the centre of the MSTREE, intermingled among A and B1 strains, while ABD strains are distributed throughout the MSTREE. Second, many ST complexes are associated with particular virulence phenotypes (Fig. 6). Almost all isolates within the ST complexes 149, 152, 243, 245 and 250 were Shigella, and almost all Shigella were found within these ST complexes. Similarly, ST complexes 270, 272 and 280 are specific for and contain most EIECs. Other well-known groups of pathogens are also associated with specific STs or ST complexes (Table S4), e.g. ST95 complex contains the related bacteria of serogroups O1, O2 and O18 that express the K1 polysaccharide (Achtman and Pluschke, 1986; Weissman et al., 2006), ST62 contains O7:K1 bacteria and all O157:H7 bacteria are in ST11 (Fig. 6).
Fig. 6.
Pathogenic types within an MSTREE. Each ST is represented by a dot. Dots with uniform colours indicate that all isolates were of the same pathogen type (see legend) while the small pie charts indicate the fraction of isolates belonging to each pathogen type. Circled numbers indicate ST complexes, whereas arrows indicate STs 11 and 62. Black lines connecting pairs of STs indicate that they share six (thick lines), five (thin) or four alleles (dotted). Grey, dotted lines connecting pairs of STs of increasing line length indicate that they share three to one alleles respectively. In addition, the lines connecting the STs within an ST complex are shaded in grey.
Repeated and independent evolution of pathogenic strains
Allele-based relationships within the MSTREE also allowed us to address whether virulent phenotypes evolved once or on multiple occasions, as previously suggested for EHEC isolates (Reid et al., 2000).
None of the five pathogen types is restricted to a single grouping (Fig. 6 and Table S3) and each pathogenic type occurs in multiple, unrelated ST complexes, indicating that virulence has been acquired independently on multiple occasions. EIEC strains are present in seven ST complexes and Shigella in five, whose locations and relationships within the MSTREE (Fig. 6) confirm that they have evolved on multiple occasions (Lan et al., 2004). As noted above, STs containing Shigella or EIEC are found in the hybrid AxB1 and ABD groups as well as in groups B1 and D (Table S2). K1 E. coli are found in six ST complexes, most of which are in groups B2 and ABD with a minor proportion in D and AxB1.
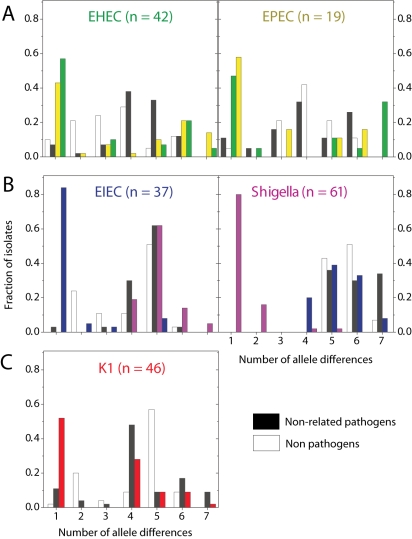
The situation with EHEC and EPEC is more complex. Not only do these pathogens occur in multiple, distinct ST complexes, but they do not seem to represent distinct genetic entities and are occasionally found within a single ST. On average, EHEC strains were almost as closely related to EPECs as they were to other EHECs (Fig. 7A), and vice versa. Other data (Donnenberg and Whittam, 2001) indicate that EHEC1 (strains DEC3 and DEC4; O157:H7) evolved recently from an EPEC strain (DEC5, O55:H7) and are closely related to ECOR37. We find that DEC3 through DEC5 are all in ST11 and ECOR37 is in ST61, both of which are members of the ST11 Complex (group ABD). Similarly, it was thought that EHEC2 (strains DEC9 and DEC10; O26:H11) evolved recently from EPEC strains of serotype O111:H8 (DEC8). In agreement, each of these three strains belong to a distinct ST within the ST29 complex. EHEC and EPEC strains are also found together in the ST20 Complex, which includes DEC11 (EPEC2), as well as in three other ST complexes for a total of six ST complexes containing both EHEC and EPEC versus only three ST complexes that contain either EHEC or EPEC strains but not both (data not shown). We note that a primary distinction between EHEC and EPEC is that the former secrete a Shiga toxin (Nataro and Kaper, 1998). Lysogenization with a bacteriophage can convert EPEC to EHEC (Schmidt et al., 1999), and EHEC can readily lose the ability to express Shiga toxin (Karch et al., 1992), providing a facile mechanism for repeated conversions between these virulence profiles.
Fig. 7.
Allele sharing between pathogen types. Histograms show the distributions of the number of allele differences between strains from a given pathogen group to other strains from different STs, colour-coded by class: same pathogen group (colour of label), related pathogen group (colour of label in neighbouring plot), different pathogen group (black) and non-pathogens (white).
Discussion
We attempted to sample all natural diversity in E. coli by examining isolates from diverse geographic sources, including Africa and Australia, from healthy and diseased individuals, from wild animals, including reptiles and birds, as well as from domesticated animals and humans. We also deliberately included Shigella and EIEC bacteria, which have only rarely been compared with other E. coli pathogens. The resulting sample is unique in its diversity of sources and provided insights not achievable with the more limited sampling strategy of former analyses.
Demographic changes
The global sample was so extensive that we identified two strains that are very distinct from the 460 other isolates (Fig. 2A). The existence of these strains indicates that E. coli was originally much more diverse than had been previously appreciated. This diversity was reduced an estimated 10–30 million years ago by population contractions and bottlenecks, and most of the genetic diversity observed in contemporary populations has accumulated during extensive expansions in the last 5 million years. During these population expansions, the descendants of four major lineages, A, B1, B2 and D, have become predominant and represent the majority of modern isolates. Rare representatives of the originally greater diversity continue to exist and additional sampling of non-pathogens from wild animals will probably identify more isolates that do not belong to the four main groups. The additional identification of such ‘living fossils’ is important because they can be used to date genomic events, such as the import of genes and gene clusters that facilitated the evolution of virulence and other specific environmental adaptations.
Are phylogenetic approaches appropriate for E. coli?
Phylogenetic approaches can be used to reconstruct the time scale and branching order of evolutionary events among distinct groups of organisms that rarely interbreed, such as multiple species. They are not necessarily suitable for evolutionary analyses within breeding populations, e.g. within a sexual species, because recombination both distorts branch lengths and obscures branch order. Initial analyses of the population structure of E. coli focused on the existence of long-lasting clones (Selander and Levin, 1980; Achtman et al., 1983), which would be suitable for phylogenetic analyses. And the original phylogenetic analyses within a clonal context identified four groups, A, B1, B2 and D, which correspond well with the predominant groups described here.
Unfortunately, the existence of discrete groups does not necessarily indicate clonality, nor does it justify the use of phylogenetic approaches. Tree-building algorithms always produce trees, whether they are valid or not, and tests are needed to determine whether branch orders have strong statistical support. Distinct groups can be separated by phylogenetic approaches even when numerous individuals are hybrids due to homologous recombination (Falush et al., 2003b), and other tests are needed to determine the frequencies of hybrid individuals.
The analyses presented here show that phylogenetic approaches are largely unsuitable for most modern E. coli. Different branching orders were obtained with different algorithms, and no single branching order had strong statistical support. As a result, we were unable to elucidate which of the four groups is ancestral and anticipate that the assignment of individual isolates to these groups by phylogenetic approaches will often be inconsistent or incorrect. In support of this argument, the branching order, numbers of groups and assignments of individual strains to groups have differed between different analyses (Boyd et al., 1994; Pupo et al., 1997; Wang et al., 1997; Lecointre et al., 1998; Arnold et al., 1999; Escobar-Paramo et al., 2003; 2004a,b). These difficulties partially reflect a lack of signal due to the star-shaped phylogeny of most modern isolates (Fig. 2), probably reflecting not only rapid diversification but also frequent homologous recombination within E. coli.
It has long been recognized that recombination within E. coli can occur in nature and is an important source of the diversity among modern isolates (Guttman and Dykhuizen, 1994). However, recombination has generally been thought to be rare and the population structure of E. coli continues to be treated as if it were more or less clonal (Reid et al., 2000; Feil et al., 2001) despite some contradictory inferences about the ancestry of modern groupings (Lecointre et al., 1998; Escobar-Paramo et al., 2004a). Our data show that homologous recombination in E. coli has been so frequent that one-third of all isolates, and an even higher proportion of pathogens, were assigned to the hybrid groups AxB1 and ABD (Figs 4A and 5A). And recombination has been a more important source of genetic diversity within groups B1, B2 and D than has mutation (Table 2 and Table S3). As a result, the genetic structure of E. coli housekeeping genes does not fit a classical clonal framework.
Why then have phylogenetic analyses often correctly identified the existence of the four groups and found congruent signals between sequences from different genes (Reid et al., 2000). First, prior analyses rarely included Shigella and EIEC, which contain particularly high frequencies of hybrids. Second, the analysis presented here encompasses a much wider range of global diversity than has been previously tested. Finally, we do not dispute the existence of the four groups but rather interpret them as reflecting the descent from four ancestral sources, with subsequent partial admixture. This interpretation provides a useful framework for investigating the evolution of virulence within E. coli, but attempts (Hommais et al., 2005) to assign all modern pathogenic isolates to one of the classical four groups are not appropriate and may be misleading. Instead, we substitute a finer-grained structure based on allele sharing, and suggest that the association of pathogens with ST complexes is more informative than with groups or clades whose boundaries are fluid.
Sex and virulence
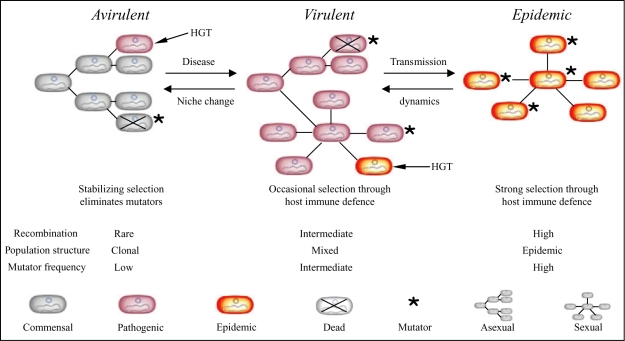
Striking parallel patterns were observed between the frequencies of recombination and pathogens within the population structure of E. coli. Recombination is rare within group A (ST10 complex), which contains very few pathogens, more frequent within groups B1, B2 and D, and most common within the hybrid AxB1 and ABD groups whose very existence reflects extensive recombination (Fig. 4A). Furthermore, the most virulent pathogen types, EIEC and Shigella, have recombined more than any other pathogen type or the non-pathogens (Fig. 5), and Shigella share the fewest alleles with other E. coli (Fig. 7). These observations point to a link between sex, i.e. homologous recombination, and virulence. This leads us to postulate that virulence is the driving force for more frequent recombination, occurring by the mechanism outlined in Fig. 8. According to this model, commensal E. coli maintain low frequencies of homologous recombination, but can occasionally acquire novel genes that result in virulence by horizontal genetic exchange (HGT) (Hacker and Kaper, 2000). The resulting pathogenic lifestyle results in greater exposure to host immune defences, which in turn selects for variants that can evade those defences. Selection for such variants results in higher mutation and recombination rates. Finally, certain virulent organisms, such as Shigella, EIEC and EHEC, which are referred to as epidemic in the model, arise through secondary HGT events. These epidemic organisms are exposed to even stronger selection by host immune defences, resulting in even higher levels of mutation and recombination.
Fig. 8.
Model of evolutionary mechanisms that link sex and virulence. Virulence determinants are assumed to be introduced initially to a species by HGT, indicated by short arrows. We deduce that (at least) two events are needed to convert an ancestral avirulent and commensal group of bacteria to virulent, pathogenic organisms and then to highly virulent organisms that cause epidemic disease. Within each population, mutators arise at low frequencies by random mutations. Mutator strains are eliminated due to their lower fitness (crosses), and are only transient within each population. But because virulent and epidemic organisms face selection pressures for more rapid diversification in response to host immune defences, there will be higher frequencies of those mutators that have not yet been eliminated among virulent bacteria and still higher frequencies among epidemic bacteria. The frequency of transient mutators determines the population structure. At low frequencies, populations are largely asexual (clonal), whereas sex becomes more frequent with the frequency of transient mutators. As a result, virulent bacteria are expected to possess patchy sexual population structures within a generally asexual framework while epidemic bacteria are largely sexual.
How could selection pressure result in higher frequencies of mutation and homologous recombination? Reduced mismatch repair (MMR) increases both mutation and recombination rates, reducing genetic barriers between distantly related bacteria (Taddei et al., 1997; Cooper and Lenski, 2000; Denamur et al., 2000; Lenski et al., 2003). It has been suggested that MMR-deficient E. coli (mutators) were responsible for the rapid emergence of antibiotic resistance and the import of virulence genes by lateral gene transfer (LeClerc et al., 1996). Thus, the occurrence of transient mutators within the B1, B2, D and hybrid groups might explain the uneven distribution of recombination between different E. coli populations, and we predict that mutators should be even more frequent within epidemic E. coli than in other pathogens. To date, surveys within E. coli have examined only very few mutators, and have not included either Shigella or EIEC, so it not surprising that no significant association between mutators and either virulence or group has been reported (Matic et al., 1997; Picard et al., 2001). Note that even if mutators are responsible for the different rates of recombination and mutation, their frequencies would be expected to be low because mutators are strongly selected against due to their reduced long-term fitness. Our model predicts that they would be transient in all groups but would simply not be as rapidly eliminated in pathogens as in non-pathogens due to selection for genetic change imposed by the host.
If an increased frequency of transient mutators accompanied the evolution of virulence, is it possible that transient mutators also played a role in more frequent HGT by reducing barriers to HGT with unrelated organisms? In that case, a single factor would be responsible for the link between sex and virulence. The only support for this speculation that we could find was the average genome size, which reflects the number of imports of foreign DNA. The average genome size of groups B1, B2 and D is larger than for group A (5.07 ± 0.09 Mb versus 4.74 ± 0.06 Mb) (Bergthorsson and Ochman, 1998). However, other correlates of HGT, such as the copy numbers of IS elements, do not correlate well with either a split between A versus B1, B2 and D or with virulence according to the limited data that are available (see Supplementary material). This question should therefore be left open until multiple complete genomes from strains of pathogenic and non-pathogenic E. coli are available.
Concluding remarks
Based on genetic variation within E. coli, we find that pathogenic strains have accelerated rates of mutation and recombination. Mutation and recombination each leave recognizable signatures in the genome, and their influence can be extracted from analyses of contemporary populations, even if they are transient properties of particular strains (Taddei et al., 1997). Because these forces will operate independently of strain background and the specific virulence determinants present within the genome, extending such studies to additional bacterial pathogens could lend credence to a new paradigm in which sex and virulence are intimately related across a broad range of microbes.
Experimental procedures
Bacterial strains
To cover a large portion of the known bacterial diversity within this species (Table S2), a total of 462 E. coli strains from multiple healthy and diseased sources were investigated. We scored as pathogenic those bacteria isolated from diseased hosts or with known virulence determinants (see bottom of Table S2) and all others as non-pathogens. One focus of the collection consisted of pathogens from both humans and domesticated animals that had been classified as EHEC (41 isolates), EPEC (20), EAEC (9), or ETEC (20) on the basis of virulence determinants (Nataro and Kaper, 1998) or APEC (13) on the basis of typical disease in domesticated animals. To add geographical as well as host diversity, and to expand the numbers of non-pathogens, the collection included all 72 isolates from the ECOR collection (Ochman and Selander, 1984), 15 isolates that represent the known diversity of E. coli from healthy wild mammals in Australia (Gordon et al., 2002) and 114 isolates from patients with diarrhoea in Ghana plus their close contacts including food handlers. We also included 61 Shigella from all known serotypes and species, 38 EIEC of different serotypes and 46 isolates from a variety of clonal groupings that express the K1 capsular polysaccharide (Achtman and Pluschke, 1986). Additional details including geographic origin are in Table S2.
Sequence-based phylogenetic analysis showed that two E. coli isolates (isolates RL325/96 and Z205 from a dog and a parrot respectively) differed markedly from the remaining isolates (Fig. 2). These strains clearly belong to E. coli according to biochemical, serological and metabolic typing schemes and by 16S rDNA sequences. Based on the MLST data, they represent the deepest known evolutionary lineages in this species. Because of their extensive sequence divergence from the vast majority of E. coli strains, they were excluded from subsequent analysis.
Nucleotide sequencing of gene fragments
Fragments of seven gene fragments (Fig. 1) were amplified and sequenced from all isolates using the primers in Table S1. PCR reactions were as follows: denaturation, 94°C for 1 min; annealing 56°C for 1 min; extension, 72°C for 1 min; 35 cycles. Independent amplicons were used to sequence both strands by an Applied Biosystems Prism 3700 automated sequencer with dRhodamine-labelled terminators (PE applied Biosystems).
STRUCTURE analysis
We used the linkage model in STRUCTURE (Falush et al., 2003a) to identify groups with distinct allele frequencies as described (Falush et al., 2003b). This procedure assigns a probability of ancestry for each polymorphic nucleotide for a given number of groups, K, and also estimates q, the combined probability of ancestry from each of the K groups for each individual isolate. We chose four groups for this report because repeated analyses (20 000 iterations, following a burn-in period of 10 000 iterations) with K between 2 and 7 showed that the model probability increased dramatically until K = 4 and only slowly thereafter. A cut-off value of q ≥ 0.67 was then used to assign individual isolates to one of the four groups, which were named A, B1, B2 and D on the basis of the assignments for the ECOR collection. Unassigned isolates were designated as AxB1 if the combined _q_-values for A and B1 were ≥ 0.8 and all other isolates were designated as ABD.
Minimum spanning tree
MSTREE, a graphical tool that links allele designations within an MLST database to a minimal spanning tree, was implemented as part of BIONUMERICS V3.5 (Applied Maths BVBA, Sint-Martens-Latem, Belgium). The minimal spanning tree is calculated by Prim's algorithm, modified to choose between otherwise equivalent, alternative subtrees at each step by implementing priority rules that incorporate aspects of the BURST algorithm (Feil and Spratt, 2001). The highest priority is given to STs with the largest numbers of single locus variants. Any ties were resolved by choosing the ST (or a random ST) with the largest number of isolates. The first node in the network is the ST with the highest priority according to these rules and subsequent links are chosen by a recursive strategy. ST complexes were defined as containing at least three STs, with links of one or two shared alleles. Identical results were obtained by an independent implementation of these rules written in Python (data not shown). The graphical representation displays the quantitative relationships between STs and ST complexes, measured as the number of shared alleles, by lines of different thickness and type (Figs 4B and 6).
Recombination and mutation
Population-based recombination and mutation rates were estimated by a composite-likelihood method using LDHAT (McVean et al., 2002). LDHAT employs a parametric approach, based on the neutral coalescent, to estimate the scaled parameter ρ= 2_N_e_r_, where N_e is the effective population size, and r is the rate at which recombination events separate adjacent nucleotides. LDHAT also estimates θ = 2_N_e_μ, where μ is the mutation rate per nucleotide. The ratio between these two estimates is r/μ. Homoplasy ratios were calculated as described (Suerbaum et al., 1998). Homoplasy ratios range from 0.0, which indicates a clonal population, to 1.0, which would be expected under free recombination.
Phylogenetic and demographic analyses
Sequences were aligned and trimmed to a uniform size by using SEQLAB and PILEUP (Wisconsin Package 9.1, GCG, Madison, WI), and then concatenated. Concatenated sequences from the seven loci are referred to as ‘unpurged’ data. Purged data consisted of sequences taken exclusively from isolates assigned to groups A, B1, B2, and D, i.e. excluding isolates in AxB1 and ABD. Within these sequences, stretches of DNA that had a different ancestry, as illustrated in Fig. 4C, were replaced by stretches of N's and were treated as missing data. Phylogenetic trees were performed with both unpurged and purged concatenated sequences as follows. The best-fit model of DNA substitution and the parameter estimates used for tree reconstruction were chosen by performing hierarchical likelihood ratio tests that are implemented in PAUP* (Swofford, 2003) and MODELTEST 1.05 (Posada and Crandall, 1998). Phylogenetic trees were estimated for each dataset with PAUP* incorporating the best-fit maximum-likelihood model of evolution (Figs 2A and 3). Additional analyses were also performed using MrBayes 2.01 (Huelsenbeck and Ronquist, 2000). This program was used to sample phylogenies according to their posterior probabilities using Metropolis-coupled Markov Chain Monte-Carlo methods and to determine clade credibility values across a consensus phylogeny. These procedures yielded comparable results to those shown in Fig. 3 (data not shown). We used the non-parametric S–H test (Shimodaira and Hasegawa, 1999) to compare likelihood scores of trees directly derived from the merged sequences, and we applied the S–H test to all potential topologies. Finally, we also used quartet puzzling (Schmidt et al., 2002) to assign estimations of support to each internal branch.
Changes in the effective population size of E. coli over time were evaluated with a generalized skyline plot based on the distribution of coalescence events within a phylogenetic tree using the Tamura-Nei model of evolution (gamma = 0.312; invariable sites = 0.642) with GENIE (Pybus and Rambaut, 2002). GENIE was also used to calculate the likelihoods according to the coalescent probability distribution on the basis of different demographic models such as exponential or logistic growth. The age of events was calculated based on a molecular clock rate of 7.6 × 10−10 per year. This is based on the divergence of 21.3% between the concatenated sequences of E. coli and Salmonella enterica, which diverged 140 million years ago (Ochman and Wilson, 1987).
Acknowledgments
We thank T. Cheasty for independent confirmation that RL325/96 and Z205 are true E. coli, J. Hacker and D. Gordon for providing isolates, G. McVean and Edward C. Holmes for assistance with algorithms, K. Donohoe and B. Kusecek for technical assistance. We also would like to thank P. Bunje, Vincent Daubin and A. Meyer for comments on the manuscript. Support was by grants from the Deutsche Forschungsgemeinschaft, the University of Konstanz, the Bundesministerium für Bildung und Forschung, the National Health and Medical Research Council (Australia), the National Institutes of Health (USA) (Grant GM56120), and the Department of Energy (USA).
Supplementary material
The following supplementary material is available for this article online:
Fig. S1. Neighbour-joining trees of the ECOR collection based on the seven concatenated housekeeping genes using the GTR+G+I model.
Table S1. Oligonucleotide primers used for E. coli MLST.
Table S2. Sources of 460 E. coli by group.
Table S3. Nucleotide diversity (π), Tajima's _D_-test and Homoplasy (H) ratio test for the four E. coli populations, as well as for the two major hybrid groups.
Table S4. Properties of STs and ST complexes.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Achtman M, Pluschke G. Clonal analysis of descent and virulence among selected Escherichia coli. Annu Rev Microbiol. 1986;40:185–210. doi: 10.1146/annurev.mi.40.100186.001153. [DOI] [PubMed] [Google Scholar]
- Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, et al. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C, Metherell L, Willshaw G, Maggs A, Stanley J. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J Clin Microbiol. 1999;37:1274–1279. doi: 10.1128/jcm.37.5.1274-1279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- Boyd EF, Nelson K, Wang FS, Whittam TS, Selander RK. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C, et al. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Whittam TS. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Invest. 2001;107:539–548. doi: 10.1172/JCI12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Páramo P, Giudicelli C, Parsot C, Denamur E. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J Mol Evol. 2003;57:140–148. doi: 10.1007/s00239-003-2460-3. [DOI] [PubMed] [Google Scholar]
- Escobar-Paramo P, Sabbagh A, Darlu P, Pradillon O, Vaury C, Denamur E, Lecointre G. Decreasing the effects of horizontal gene transfer on bacterial phylogeny: the Escherichia coli case study. Mol Phylogenet Evol. 2004a;30:243–250. doi: 10.1016/s1055-7903(03)00181-7. [DOI] [PubMed] [Google Scholar]
- Escobar-Páramo P, Grenet K, Le Menac'h A, Rode L, Salgado E, Amorin C, Gouriou S, et al. Large-scale population structure of human commensal Escherichia coli isolates. Appl Environ Microbiol. 2004b;70:5698–5700. doi: 10.1128/AEM.70.9.5698-5700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003a;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003b;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Feil EJ. Small change: keeping pace with microevolution. Nat Rev Microbiol. 2004;2:483–495. doi: 10.1038/nrmicro904. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DM, Bauer S, Johnson JR. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology. 2002;148:1513–1522. doi: 10.1099/00221287-148-5-1513. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- Guttman DS, Dykhuizen DE. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Urwin R, Maiden MC. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol Biol Evol. 1999;16:741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- Hommais F, Pereira S, Acquaviva C, Escobar-Paramo P, Denamur E. Single-nucleotide polymorphism phylotyping of Escherichia coli. Appl Environ Microbiol. 2005;71:4784–4792. doi: 10.1128/AEM.71.8.4784-4792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: bayesian inferences of phylogeny. Bioinformatics. 2000;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Karch H, Meyer T, Russmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect Immun. 2004;72:5080–5088. doi: 10.1128/IAI.72.9.5080-5088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc JE, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Winkworth CL, Riley MA. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20 000 generations. J Mol Evol. 2003;56:498–508. doi: 10.1007/s00239-002-2423-0. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, et al. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Smith NH. Detecting recombination from gene trees. Mol Biol Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R, Raleigh EA, McKane M, Cryderman D, Bilodeau P, McWeeny K. Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics. 1999;153:539–554. doi: 10.1093/genetics/153.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K, Selander RK. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992;174:6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K, Whittam TS, Selander RK. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K, Wang FS, Boyd EF, Selander RK. Size and sequence polymorphism in the isocitrate dehydrogenase kinase/phosphatase gene (aceK) and flanking regions in Salmonella enterica and Escherichia coli. Genetics. 1997;147:1509–1520. doi: 10.1093/genetics/147.4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Wilson AC. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Picard B, Duriez P, Gouriou S, Matic I, Denamur E, Taddei F. Mutator natural Escherichia coli isolates have an unusual virulence phenotype. Infect Immun. 2001;69:9–14. doi: 10.1128/IAI.69.1.9-14.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Holmes EC. Recombination in evolutionary genomics. Annu Rev Genet. 2002;36:75–97. doi: 10.1146/annurev.genet.36.040202.111115. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Karaolis DKR, Lan RT, Reeves PR. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A. genie: estimating demographic history from molecular phylogenies. Bioinformatics. 2002;18:1404–1405. doi: 10.1093/bioinformatics/18.10.1404. [DOI] [PubMed] [Google Scholar]
- Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Schierup MH, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Bielaszewska M, Karch H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage phi3538 isolated from Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:3855–3861. doi: 10.1128/aem.65.9.3855-3861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Selander RK, Levin BR. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210:545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of Log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. [Google Scholar]
- Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, Pybus OG. Exploring the demographic history of DNA sequences using the generalized skyline plot. Mol Biol Evol. 2001;18:2298–2305. doi: 10.1093/oxfordjournals.molbev.a003776. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. paup*– Phylogenetic Analyses Using Parsinomy and Other Methods Version 4.0. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- Taddei F, Matic I, Godelle B, Radman M. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 1997;5:427–428. doi: 10.1016/S0966-842X(97)01157-8. discussion 428–429. [DOI] [PubMed] [Google Scholar]
- Vulic M, Lenski RE, Radman M. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proc Natl Acad Sci USA. 1999;96:7348–7351. doi: 10.1073/pnas.96.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FS, Whittam TS, Selander RK. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J Bacteriol. 1997;179:6551–6559. doi: 10.1128/jb.179.21.6551-6559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman SJ, Chattopadhyay S, Aprikian P, Obata-Yasuoka M, Yarova-Yarovaya Y, Stapleton A, et al. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol Microbiol. 2006;59:975–988. doi: 10.1111/j.1365-2958.2005.04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Neighbour-joining trees of the ECOR collection based on the seven concatenated housekeeping genes using the GTR+G+I model.
Table S1. Oligonucleotide primers used for E. coli MLST.
Table S2. Sources of 460 E. coli by group.
Table S3. Nucleotide diversity (π), Tajima's _D_-test and Homoplasy (H) ratio test for the four E. coli populations, as well as for the two major hybrid groups.
Table S4. Properties of STs and ST complexes.
This material is available as part of the online article from http://www.blackwell-synergy.com