The E6 Protein of the Cutaneous Human Papillomavirus Type 8 Can Stimulate the Viral Early and Late Promoters by Distinct Mechanisms (original) (raw)
Abstract
The expression of the proteins encoded by human papillomaviruses (HPVs) is tightly linked to the differentiation program of the infected keratinocytes. The late promoter, expressing the structural proteins, becomes activated in the differentiated keratinocytes, while the early promoter is also active in the basal layers. We have shown previously that the viral transcriptional regulator E2 and the cellular coactivator p300 cooperate in activation of gene expression of HPV8, which infects the skin and is associated with epidermodysplasia verruciformis. Here we demonstrate that this activation is further stimulated after overexpression of the E6 oncoprotein of HPV8 (8E6). RNase protection experiments revealed that 8E6 efficiently cooperates with 8E2 and p300 in activation of the late promoter. In addition, the early promoter, which did not respond to 8E2 and/or p300, was stimulated more than fourfold by 8E6. Our data suggest that both promoters are activated via distinct mechanisms, since the activation of the early promoter was achieved by the N-terminal moiety of 8E6; in contrast, its C-terminal half was sufficient for late promoter activation. This was markedly reduced by the deletion of amino acids 132 to 136 of 8E6, which also abolished the binding to p300, indicating that a direct interaction between 8E6 and p300 is involved. Moreover, a 45-amino-acid segment within the C/H3 region of p300 is required for 8E6 to stimulate the coactivator function of p300. Our results demonstrate for the first time that an E6 oncoprotein of HPV directly contributes to the regulation of HPV gene expression.
Human papillomaviruses (HPVs) are small double-stranded DNA viruses causing a variety of benign hyperproliferative lesions of the skin or the mucosa. Infections with certain HPV types have a high risk of undergoing malignant progression. In more than 95% of cervical cancers, the DNA of genital high-risk types such as HPV16 and HPV18 can be found, indicating that infection with these types is a prerequisite for cancer progression (34). Among the HPVs infecting the skin, HPV5 and HPV8 are classified as high-risk types. This is based on the observation that HPV-induced skin lesions in epidermodysplasia verruciformis patients, which often develop into squamous cell carcinomas after a long latency preferred in sun exposed areas, mainly contain HPV5 or HPV8 DNA (41). The oncogenic potential of HPV8 was confirmed by the ability of the early HPV8 region, encoding the regulatory proteins, to induce benign and malignant skin tumors in transgenic mice (43). Additionally, there is limited evidence for the carcinogenicity of epidermodysplasia verruciformis-associated HPVs in non-melanoma skin carcinomas of the immunocompetent population in general (40, 41).
The E6 and E7 proteins of mucosal HPVs transform cells by interfering with cell cycle regulation and counteracting apoptosis. Corresponding proteins of epidermodysplasia verruciformis-associated HPVs possess transforming activity as well. Compared to genital HPVs, little is known about the mechanisms of cutaneous E6 and E7 acting as oncoproteins. The E6 protein of HPV8 (8E6) is able to induce morphological transformation and anchorage-independent growth (18, 21). However, 8E6 does not interact with p53 (9, 22, 46). The binding of the E6 proteins of high-risk genital HPVs to p53 initiates the accelerated degradation of p53, eliminating its tumor suppressor function (44). In addition, HPV16 E6 (16E6) suppresses the transcriptional activity of p53 by targeting its coactivator, CBP (CREB-binding protein)/p300. Moreover, the interaction with CBP/p300 acting as coactivator for a variety of sequence-specific DNA binding proteins involved in numerous signaling pathways may allow 16E6 to modulate cellular transcription and probably its own transcription as well. In line with this, it has been demonstrated that 16E6 reduces activation of several cellular activators, such as NF-κB and AP1 (17, 38, 50, 52), which use CBP/p300 as coactivators (3, 11, 19, 29). Here we intended to study an interaction of 8E6 with the cellular coactivator p300 and its consequences for the viral life cycle.
CBP and its homologue p300 (named CBP/p300) are essential components of the transcriptional machinery. CBP/p300 directly interact with components of the basal transcriptional machinery and a variety of transcription factors connecting the two major elements. Moreover, the stimulation of gene expression by CBP/p300 occurs through their inherent histone acetyltransferase (HAT) activity or by the recruitment of other HATs bound to CBP/p300, leading to the modification of the tails of the histones or of transcription factors directly by adding an acetyl group (6, 12). CBP/p300 are also implicated in HPV gene expression. As coactivators of cellular factors binding to the long control region of HPVs, such as AP1 and C/EBP (3, 19, 31), CBP/p300 are essential for the transcriptional activity of the HPV18 early promoter in position 105 that also direct the expression of E6 and E7 (4). CBP/p300 are targets of the virally encoded transcription factor E2 as well (24, 33, 39). We showed previously that coexpression of p300 strongly enhances activation of HPV8 gene expression by HPV8 E2 (8E2). Since p300 is expressed at highest levels in the most differentiated cells of the skin, we suspected that p300 contributes to the differentiation-dependent induction of the late HPV8 promoter (33).
Here we demonstrate that 8E6 is an activator of HPV8 gene expression. 8E6 stimulates the late HPV8 promoter in cooperation with 8E2 and/or p300. A detailed analysis demonstrates that amino acids 132 to 136 of 8E6, which mediate the binding to p300, are essential for this effect, indicating that a direct interaction between 8E6 and p300 is involved. In addition, the early promoter of HPV8 is activated by 8E6, which seems to occur independently of p300 and 8E2. Thus, distinct mechanisms contribute to 8E6-mediated activation of HPV8 gene expression, which may have evolved due to the different factor requirements of the respective promoters. Our data suggest that 8E6 may play an important role as coactivator of HPV8 gene expression.
MATERIALS AND METHODS
Plasmid constructs.
The expression vectors for 8E2, 18E2, p300, and the reporter constructs 4E2-Sp1-Luc and 8NCR-Luc have been described previously (33). PG5-Luc was purchased from Clontech. GST-p300 fusion proteins were obtained by cloning PCR fragments, amplified with appropriate primers, into the vector pGEX2T (Promega). 8E6 and its derivatives were expressed with an N-terminally fused FLAG epitope. Therefore, complementary oligonucleotides encoding the FLAG epitope were cloned into the BamHI-HindIII sites of pcDNA3.1+ (Invitrogen) to obtain pcDNA3.1+ FLAG. This vector was used to clone PCR fragments, obtained with appropriate primers encoding 8E6, 8E6-N (amino acids 1 to 93), and 8E6-C (amino acids 94 to 155). The expression vector for GST-8E6-C has been described previously (10). All mutations in 8E6 and p300 were introduced with the QuickChange Site-Directed Mutagenesis kit from Stratagene. To allow the expression of 8E6 in bacteria with a tag of six histidines, a PCR product containing the 8E6 open reading frame was cloned into pET32c (Novagen). To obtain a probe for RNase protection assays (RPA), an HPV fragment from nucleotides 7480 to 282 was amplified by PCR with corresponding primers and cloned into the vector pGEM4Z (Promega). 8B-Luc was obtained by changing a T at position 11 into a C to obtain a HindIII site. Subsequently, this site was used to delete the upstream HPV8 noncoding region (8NCR) sequences (the construct was kindly provided by H. Pfister).
Cell culture and transient transfections.
C33A cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. The skin keratinocyte cell line RTS3b (42) was cultivated in E medium (30). Primary foreskin keratinocytes were purchased from Cambrex and were cultivated in KGM2 medium. Transient transfections were performed using calcium phosphate precipitation (C33A cells), as described previously (33), or with FuGene reagent (RTS3b and primary keratinocytes; Roche Diagnostics). Luciferase activity was determined as described previously (33). All transient transfection experiments have been repeated at least three times. The statistical significance of the results of some experiments was calculated by a t test for paired samples with the SPSS 11.01 program (SPSS Inc., Chicago, Ill.).
Protein expression and protein-protein interaction studies.
Protein expression, purification, glutathione _S_-transferase (GST) pulldown assays, and direct interaction studies have been performed as described previously (33). To detect the FLAG-8E6 or coprecipitated p300 proteins in vivo, transfected C33A cells were resuspended in 100 mM LSDB (50 mM Tris Cl, pH 7.9, 20% glycerol, 1 mM dithiothreitol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 100 mM KCl), and whole-cell extracts were prepared by sonication. One milligram of these extracts was incubated with FLAG-M2 antibody coupled to Sepharose for 2 h at 4°C, followed by three washes in 100 mM LSDB. Bound 8E6 proteins were detected by a Western blot developed with the FLAG-M5 monoclonal antibody (Sigma). Coprecipitated endogenous p300 and hemagglutinin (HA)-tagged p300 were detected in a Western blot developed with the p300 antibody (C20; Santa Cruz) or the HA antibody (3F10; Roche), respectively. For precipitation of endogenous p300 from HeLa nuclear extracts, about 200 ng of purified GST fusion protein or GST alone was incubated with 500 μg HeLa nuclear extracts for 3 h at 4°C, prepared according to a previously described method (8). After three washes in 100 mM LSDB, bound proteins were detected in a Western blot with the p300 antibody (C20; Santa Cruz).
RNase protection assays.
Total cellular RNA was isolated from transfected RTS3b cells 48 h after transfection with the RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions, including an additional treatment with RNase-free DNase. Ten micrograms of RNA was hybridized to 32P-radiolabeled cRNA encompassing the HPV sequences from positions 282 to 7480. The RNase protection assay was performed with reagents of the Ribo Quant Multi Probe RNase protection system (Pharmingen) according to the protocol of the manufacturer.
RESULTS
8E6 stimulates activation by the viral E2 protein.
The E6 protein of HPV16 inhibits coactivation of transcription mediated by the cellular coactivators CBP and p300. We thought to investigate whether the E6 protein of the cutaneous high-risk type HPV8 affects coactivator function of p300. To analyze the effect of 8E6 on p300-mediated transcription, we coexpressed 8E6 along with an activator, using p300 as coactivator. Since it has been shown that activation by the E2 proteins of HPV16 (16E2), HPV18 (18E2), and HPV8 (8E2) and of the bovine papillomavirus type 1 (BPV1E2) requires CBP/p300 (24, 33, 39), we tested the influence of 8E6 on activation by E2. In transient transfections of C33A cells, 8E2 activated an E2-responsive synthetic promoter (Fig. 1A) 42-fold, which was further stimulated up to 49-fold after coexpression of 8E6. The stimulating effect of 8E6 on activation by E2 was also observed with 18E2 (Fig. 1A). Thus, 8E6 further increases activation mediated by E2. A similar effect could be observed with p53 (data not shown).
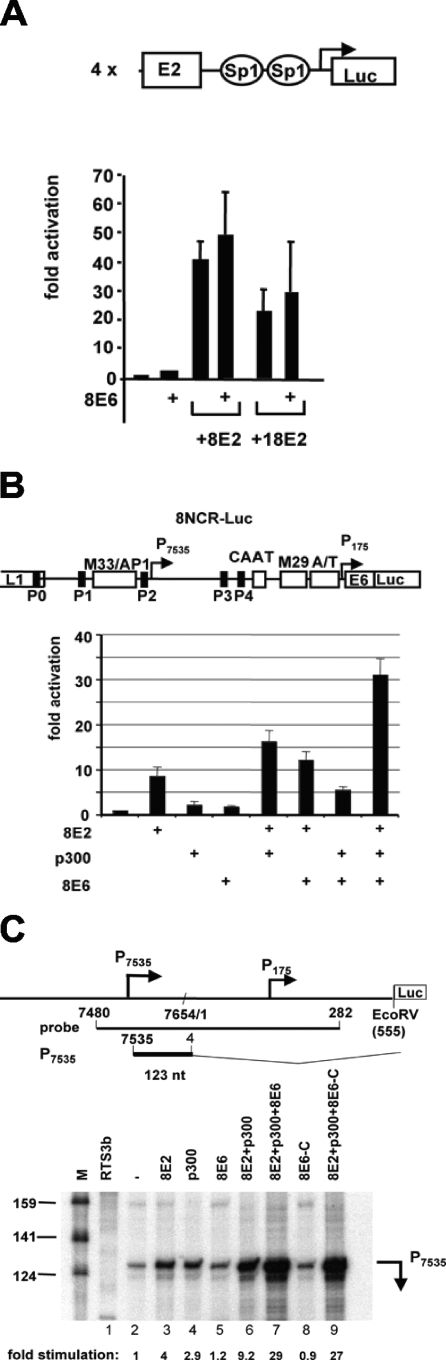
FIG. 1.
8E6 enhances activation by the HPV E2 protein. (A) C33A cells have been cotransfected with a synthetic E2-responsive reporter construct containing four E2 and two Sp1 binding sites in front of a minimal promoter. Expression vectors for E2 proteins of HPV8 and HPV18 have been cotransfected either alone or together with an expression vector for 8E6. The mean of the fold activations of three independent experiments and the error bars are shown. (B) RTS3b cells transfected with 500 ng of a luciferase reporter construct containing the noncoding region (NCR) of HPV8. The schematic drawing of the HPV8 NCR above the graph indicates the position of the early (P175) and the late (P7535) promoters, the five E2 binding sites P0, P1, P2, P3, and P4, a CAAT element, an A- and T-rich (A/T) region, the AP1 binding site, and two motifs of either 33 or 29 nucleotides in length, M33 and M29, which are conserved among epidermodysplasia verruciformis-associated HPV types. This reporter construct was cotransfected with expression vectors for 8E2 (10 ng), p300 (500 ng), and 8E6 (500 ng), as indicated. The bars represent the means of the fold activations calculated from four independent experiments. (C) RNase protection experiment with RNA isolated from RTS3b cells transfected with the 8NCR-Luc construct alone (−) or together with the expression vectors for 8E2, p300, 8E6, or 8E6-C, encoding the C-terminal half of 8E6, in different combinations as indicated. Total RNAs have been hybridized with radioactively labeled cRNA encompassing the HPV8 sequence from positions 282 to 7480, as given in the schematic drawing. The position of the protected fragment of 123 nucleotides corresponding to transcripts initiating from P7535 is indicated, as are positions of the marker bands. The fold stimulations of P7535 have been calculated after quantifying the signals with a PhosphorImager. M, molecular size marker. nt, nucleotide.
These observations imply that 8E6, in contrast to 16E6, fails to repress transcriptional activation by activators using p300 (38, 50, 52). Previously, we have demonstrated a direct interaction between 8E2 and p300, which we correlated with a cooperative activation of HPV8 gene expression (33). To analyze the role of 8E6 on the functional interaction between 8E2 and p300 in activation of HPV8 gene expression, we transiently cotransfected RTS3b cells, an HPV-negative immortalized keratinocyte cell line (42). Transcription from the reporter construct 8NCR-Luc, expressing the luciferase gene under control of the entire regulatory region of HPV8 (noncoding region 8NCR), was enhanced by about twofold after overexpression of 8E6 (Fig. 1B). 8E6 also slightly stimulated the activations obtained by overexpressed 8E2 and p300 alone (1.4- and 2.5-fold, respectively). As observed previously, both 8E2 and p300 cooperated, since they stimulated promoter activity 16.4-fold, compared to 8.7-fold and 2.3-fold by 8E2 and p300 alone, respectively (Fig. 1B). The effect of 8E2 and p300 is reduced compared to that seen in previous observations, which is due to the increased amount of transfected pcDNA3.1 vector responsible for the expression of 8E6 (33). The addition of 8E6 to 8E2 and p300 resulted in a 31.2-fold activation, which is far beyond the sum of the effects of the single components (Fig. 1B). Thus, 8E6 can activate HPV8 gene expression in cooperation with 8E2 and p300. A dose-dependent regulation by 8E2 has been demonstrated to concern the late promoter (47-49). However, in addition to the late promoter at position 7535 (P7535), the reporter construct used here harbors the early promoter at position 175 (P175), which is rather weak compared to that at P7535. In order to test which promoter is stimulated by 8E6, 8E2, and p300 of HPV8 (noncoding region 8NCR), we performed RNase protection assays (RPA) with RNA isolated from transiently transfected RTS3b cells. As a probe we used in vitro-synthesized, radioactively labeled cRNA encompassing HPV8 sequences from nucleotides 7480 to 282 in antisense. Figure 1C, lane 2, shows that a major fragment of about 123 nucleotides was protected with RNA isolated from cells which have been transfected with the reporter construct 8NCR-Luc only. This corresponds to transcripts initiated at P7535. As revealed in Fig. 1C, it has been demonstrated that the P7535-derived transcripts are spliced at a splice donor at position 4. In reporter constructs, the splice acceptor was mapped to the boundary of the viral insert and the multiple cloning site of the vector (48). The results obtained by RPA perfectly correlated with the data obtained by reporter gene assays shown in Fig. 1B and demonstrate that 8E6 stimulates the 8E2- and p300-mediated activation of the HPV8 late promoter. Furthermore, the C-terminal moiety of 8E6 was sufficient for the effect. Transcripts initiating at P175 should have a size of 107 nucleotides. A corresponding fragment was not detected, which may be due to the low level of activity of P175 (47-49). As a result of technical reasons, we were not able to analyze a specific transcript in the presence of 8E6, which makes it impossible to study the effect of 8E6 on P175 by this assay. (The 8E6 RNA expressed from the expression vector hybridized with the probe giving rise to an identical signal; data not shown.)
Amino acids 132 to 136 within 8E6 are required for functional interaction with p300.
To elaborate the function of 8E6 in transcriptional activation, we mapped the amino acids in 8E6 involved in stimulation of 8E2- and p300-mediated transcription. All 8E6-derived proteins were expressed with an N-terminal FLAG tag in order to detect the proteins in the cell extracts to verify their expression. Initially, these functional analyses were addressed by transient transfections of primary keratinocytes, the cells naturally infected by these HPVs. Here the fold activation of HPV8 transcription by coexpressed E2 and p300 varied from experiment to experiment, but regardless 8E6 strongly stimulated transcription obtained by coexpression of E2 and p300 by 4.1-fold (Fig. 2A). Like other E6 proteins, 8E6 contains two zinc-binding domains, which are involved in protein-protein interactions. It has been shown that both can fold and bind to zinc independently from each other (27). Figure 2A reveals that overexpression of 8E6-N (from amino acids 1 to 93, expressing the N-terminal zinc binding domain) reduced 8E2- and p300-mediated transcription by 40%, whereas 8E6-C (from amino acids 94 to 155, expressing the C-terminal zinc binding domain) retained the effect of full-length 8E6 (see also Fig. 1C), indicating that its coactivator function resides within the C-terminal half. The inhibitory function of 16E6 on CBP has also been mapped to the C-terminal zinc finger (38, 52). A small deletion within the second zinc finger of 16E6 (16E6Δ123-127) resulted in the loss of transcriptional repression, which correlated with a markedly reduced affinity to CBP in GST pulldown experiments (17, 38, and data not shown). A mutant of 8E6 lacking amino acids 132 to 136 (8E6Δ132-136), which are homologous to amino acids 123 to 127 in 16E6, was significantly reduced in its capacity to stimulate the activation by 8E2 and p300 compared to wild-type 8E6 (P = 0.013) (Fig. 2B). To narrow down the amino acids involved in p300 stimulation, we changed each of these amino acids into alanine. The 8E6H135A mutant revealed reduced capacity to activate transcription by E2 and p300 compared to 8E6; however, it seemed more functional than 8E6Δ132-136 (Fig. 2B). An immunoprecipitation with extracts from C33A cells transfected with the corresponding expression vectors was performed to confirm the expression of the E6 proteins (Fig. 2B).
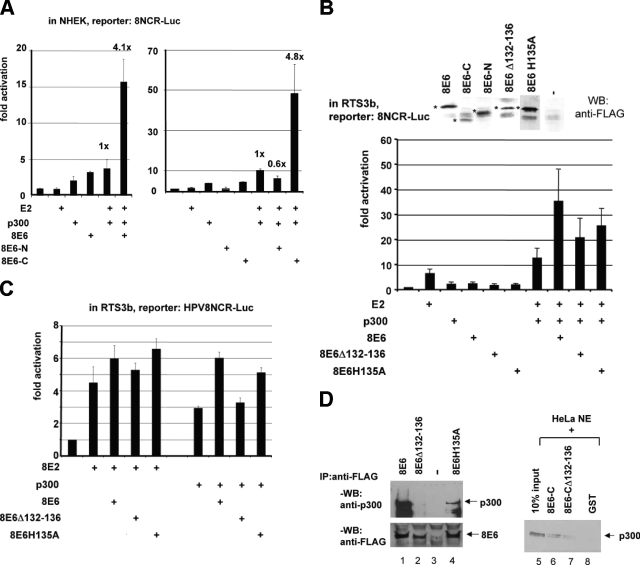
FIG. 2.
Amino acids 132 to 136 of 8E6 are involved in activation and are required for binding to p300. (A) Primary keratinocytes (NHEK) were cotransfected with the 8NCR-Luc reporter construct and vectors expressing wild-type 8E6, 8E6-N (from amino acids 1 to 93), or 8E6-C (encoding amino acids 94 to 155) (500 ng) either alone or in combination with E2 and/or p300, as indicated. The values represent the means of at least three independent experiments, and the error bars are given. (B and C) Transient transfections similar to those in panel A but in RTS3b cells and with an 8E6 with a deletion of amino acids 132 to 136 (E6Δ132-136) or with a mutation of the H at position 135 to an A (8E6H135A). To monitor the expression of the various E6 proteins, extracts from C33A cells transiently transfected with the corresponding vectors expressing the various E6 proteins with an N-terminal FLAG tag or the empty vector were incubated with M2-FLAG-Sepharose beads, and the bound proteins were detected by a Western blot developed with the FLAG antibody. The positions of the various E6 proteins are indicated by an asterisk. (D) Extracts from C33A cells which were transfected with expression vectors for FLAG-8E6, FLAG-8E6Δ132-136, FLAG8E6H135A, or the empty vector (lanes 1 to 4) were incubated with M2-FLAG-Sepharose. Bound endogenous p300 was detected by a Western blot developed with an antibody against p300. The expression of the FLAG-tagged 8E6 proteins was confirmed by a Western blot developed with the FLAG antibody, shown beneath. In lanes 5 to 8, HeLa nuclear extracts have been incubated with GST-8E6-C, GST-8E6-CΔ132-136, or GST to detect bound endogenous p300 by a Western blot. In lane 5, 10% of the input HeLa nuclear extracts was loaded onto the gel. WB, Western blot; IP, immunoprecipitation.
These results suggest that 8E6 targets p300 and/or 8E2 to superstimulate the respective activations. To analyze this, we studied the effect of the mutations in 8E6 on the activations of HPV8 transcription obtained by 8E2 or p300 alone (see Fig. 1B). Figure 2C shows that while the deletion of amino acids 132 to 136 within 8E6 hardly affected its capacity to stimulate the transcriptional activation by 8E2, the deletion abolished the twofold 8E6-specific enhancement of HPV8 transcription by overexpressed p300, indicating that amino acids 132 to 136 of 8E6 are relevant for stimulation of p300-mediated activation of HPV8 transcription.
To correlate the activation of 8E6 with binding to p300, we performed coimmunoprecipitation experiments. After transient transfections of C33A cells with an expression vector for FLAG-8E6, endogenous p300 was coprecipitated by the FLAG antibody, while in the presence of FLAG-8E6Δ132-136 it failed to do so (Fig. 2D). This indicates that 8E6 binds to cellular p300, which is mediated by the amino acids 132 to 136. A faint band corresponding to endogenous p300 was present when FLAG-8E6H135A was overexpressed (Fig. 2D, lane 4), reflecting a weaker interaction with p300. In these experiments, 8E6-C was unable to precipitate p300 (data not shown), which may be due to the low expression level of 8E6-C (Fig. 2B). However, GST-8E6-C bound p300 present in HeLa nuclear extracts, as indicated in Fig. 2D. In contrast, the p300-specific band was much weaker when GST-8E6-CΔ132-136 was applied. Thus, the stimulation of the coactivator function of p300 by 8E6 correlates with an interaction of both proteins in vivo requiring amino acids 132 to 136 of 8E6.
Binding of 8E6 to the C/H3 region of p300 is involved in activation of P7535.
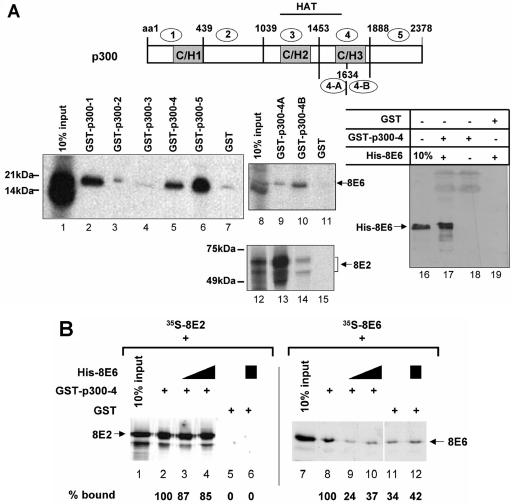
16E6 has been shown to bind to three regions within CBP, the C/H1 region (amino acids 340 to 413), the C-terminal region (amino acids 1970 to 2220), and the C/H3 region (amino acids 1621 to 1877) (38). Within this domain, a small motif from amino acids 1808 to 1826 of CBP, which corresponds to amino acids 1770 to 1788 in p300, has been determined to be the 16E6-interacting region and has been suggested to be involved in repression of p53-mediated transcription and that of other factors, such as c-fos (35, 52). Moreover, the binding of 16E6 to segments within the C terminus of p300 has been correlated with repression of activation by NF-κB and p53 (17, 50). To elaborate the domains within p300 which are involved in the 8E6-mediated activation, we initially used a set of five GST-p300 fusion proteins covering the entire p300 (Fig. 3A). GST-p300-1, GST-p300-4, and GST-p300-5 were bound by 35S-labeled 8E6 (Fig. 3A). Thus, 8E6 binding to p300 is similar to that of 16E6 and BPV1E6, as well as the E2 proteins of HPV18 and BPV1 (33, 38, 53, and data not shown). Previously, we were able to show that 8E2 only interacts with GST-p300-4 in vitro and that this interaction may be relevant for the functional cooperativity (33). The binding of 8E6 to this fragment of p300 is direct, since GST-p300-4 precipitated bacterially expressed purified, His-tagged 8E6, in contrast to GST alone (Fig. 3A, lanes 17 and 19). A simultaneous binding of 8E2 and 8E6 to p300 should be a prerequisite for the cooperation observed here. To confirm this, we divided p300-4 into two parts, p300-4A, from amino acids 1453 to 1634, and p300-4B, from amino acids 1635 to 1887. Figure 3A shows that 8E6 was precipitated by GST-p300-4B, while GST-p300-4A only precipitated trace amounts of 8E6. The reverse was observed with 35S-labeled 8E2. It strongly interacted with GST-p300-4A and only weakly with GST-p300-4B (Fig. 3A). Thus, the major binding motifs of 8E6 and 8E2 are not overlapping, and both proteins may bind simultaneously to p300 to form a ternary complex. This was confirmed by an in vitro competition experiment (Fig. 3B). Preincubation of GST-p300-4 with increasing amounts of nonlabeled His-tagged 8E6 only marginally affected the binding of 35S-labeled 8E2 to GST-p300-4 (10% reduction). In contrast, the interaction of 35S-labeled 8E6 with GST-p300-4 was reduced by 75% when GST-p300-4 was preincubated with the same amounts of purified His-8E6, which served as a control. This is in line with the notion that the 8E6 binding sites within p300 were occupied by the nonlabeled 8E6 protein, competing with the binding of labeled 8E6. In contrast, the access of 8E2 was not inhibited after His-8E6 had bound.
FIG. 3.
8E2 and 8E6 can bind simultaneously to the central part of p300. (A) 8E6 and 8E2 bind to distinct domains of p300. Shown is a schematic drawing of p300 with the positions of the three cysteine-histidine-rich regions C/H1 to C/H3 being conserved to CBP and the domain with histone acetyltransferase (HAT) activity. The binding of 35S-labeled 8E6, obtained by in vitro translation, was examined with five GST-p300-1 to GST-p300-5 fusion proteins, covering the open reading frame of p300 (lanes 1 to 7). The amino acids (aa) comprising the five p300 fragments fused to GST are given above the schematic drawing of p300. To analyze the binding of 8E6 (lanes 8 to 11) and 8E2 (lanes 12 to 15) in more detail, GST pulldown assays with GST-p300-4A and GST-p300-4B, resulting from the division of GST p300-4, were performed. In lanes 1, 8, and 12, 10% of the in vitro-translated proteins used for the interaction assays were loaded. The positions of 8E6 (upper part) and 8E2 (lower part) are indicated, as well as that of some marker proteins. In lanes 16 to 19, purified GST-p300-4 fusion protein or GST alone was incubated with bacterially expressed, purified His-tagged 8E6 protein. The presence of His-8E6 was analyzed by a Western blot developed with an antibody directed against the His tag. (B) Protein competition experiment. GST-p300-4 or GST alone was incubated with increasing amounts of His-8E6 for 1 h at 4°C. After several washes to remove unbound His-8E6, incubation either with 35S-labeled 8E2 (lanes 2 to 6) or 35S-labeled 8E6 (lanes 8 to 12) for another 2 h at 4°C followed. The binding of radiolabeled 8E2 or 8E6 was analyzed by autoradiography. After quantitating the signals with a PhosphorImager, the percentages bound by GST-p300-4 (lanes 2 to 4 and 8 to 10) or by GST (lanes 5, 6, 11, and 12) were calculated. Ten percent of the input of 35S-labeled 8E2 or 8E6 was loaded as a control (lanes 1 and 7).
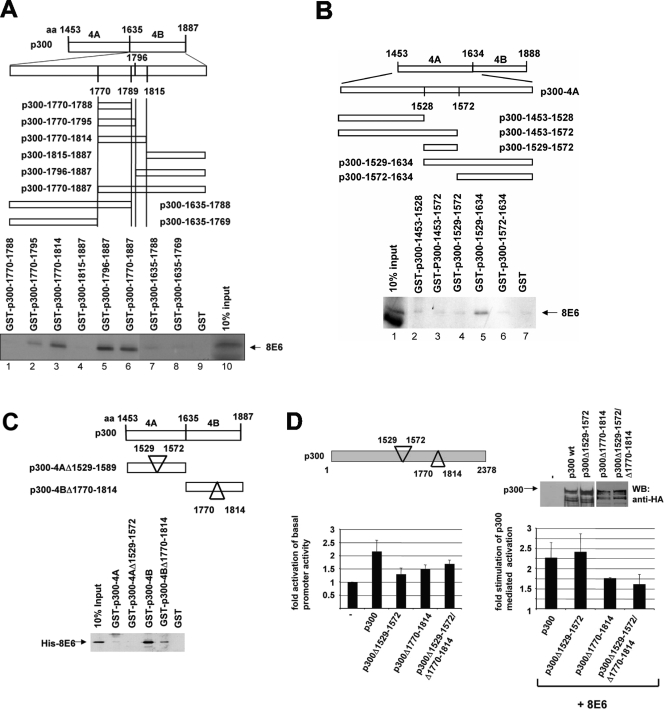
The segment of p300 from amino acids 1635 to 1887 bound by 8E6 was identified as a target sequence for a variety of transcription factors, such as p53, MyoD, YY1, c-fos, Notch, etc., indicating that this p300 sequence may serve as an important motif (35). In order to test the relevance of the binding of 8E6 to this domain for its activation of HPV8 transcription observed here, we initially mapped the 8E6 recognition motif by preparing additional GST-p300 fusion proteins representing eight partially overlapping fragments of p300-4B (Fig. 4A). Amino acids 1770 to 1814 of p300 are sufficient for binding of 8E6 (Fig. 4A, lane 3). Although GST-p300-1796-1887 precipitated 8E6 (lane 5) but not GST-p300-1815-1887 (lane 4), the residues from position 1796 to 1814 were not sufficient, since a GST fusion protein expressing only these amino acids was not able to bind to 8E6 (data not shown). A secondary structure mediated by surrounding p300 sequences may influence the folding allowing 8E6 to bind. Figure 3A suggests that an additional weak 8E6 binding site exists within p300-4A. The use of various GST deletion mutants covering p300-4A revealed that amino acids 1529 to 1572 are required, although not sufficient, for binding, since GST-p300-1529-1634 precipitated 8E6, in contrast to GST-p300-1572-1634 (Fig. 4B). The deletion of amino acids 1529 to 1572 within p300-4A abolished the weak binding of 8E6 to p300-4-A (Fig. 4C). Moreover, the deletion of amino acids 1770 to 1814 strongly reduced the interaction of His-tagged 8E6 with GST-p300-4B, although some residual binding was detectable (Fig. 4C). To directly test the involvement of the 8E6 motifs within p300-4 in the activation, we deleted these amino acids in the context of the entire p300. The transient transfections shown in Fig. 4D revealed that the deletion of the 8E6-interacting regions reduced the activation of 8NCR by p300, although the mutants are expressed at similar levels (Fig. 4D), indicating that specific functions of p300 on 8NCR-Luc are compromised. 8E6 enhanced the activations obtained by wild-type p300 as well as by p300Δ1529-1572 at 2.25-fold and 2.4-fold, respectively. In contrast, the 8E6-mediated activations of p300Δ1770-1814 or p300Δ1529-1572/1770-1814, lacking both 8E6-interacting regions, were significantly decreased to 1.75-fold (P = 0.040) or 1.6-fold (P = 0.026), respectively, after 8E6 has been coexpressed (Fig. 4D). Thus, the binding of 8E6 to amino acids 1770 to 1814 contributes to the stimulation of the coactivator function of p300.
FIG. 4.
Binding of 8E6 to a domain within C/H3 of p300 participates in stimulation of transcription by p300. (A) 8E6 requires amino acids 1770 to 1814 to bind to p300-4B. Eight partially overlapping GST-p300 fragments covering p300-4B (from amino acids 1635 to 1887), as indicated in the figure, were used in GST pulldown experiments with 35S-radiolabeled 8E6 to map the binding domain of 8E6 within p300-4B. (B) Amino acids 1529 to 1572 in p300-4A constitute a low-affinity site of 8E6. Five partially overlapping p300 fragments, covering p300-4A as indicated in the figure, were used in GST pulldown assays with 35S-labeled 8E6. (C) GST pulldown experiment with GST-p300-4A, GST-p300-4AΔ1529-1572 (lacking the low-affinity 8E6 binding site), GST-p300-4B, GST-p300-4BΔ1770-1814 (lacking the high affinity binding site), and His-8E6. (D) The high-affinity 8E6 binding site in p300-4 is required for full cooperation. RTS3b cells were transiently transfected with the 8NCR-Luc reporter construct (500 ng) and expression vectors for wild-type HA-p300, HA-p300Δ1529-1572 (lacking the low affinity 8E6 binding motif), HA-p300Δ1770-1815 (lacking the high affinity 8E6 binding motif), or HA-p300Δ1529-1572/1770-1814 (each 500 ng) either alone (graph on the left) or together with the expression vector for 8E6 (500 ng) as indicated in the figure. In the graph on the left, the fold activations of HPV8 promoter activity by wild-type p300 or the mutants from at least three independent experiments have been calculated, and the errors bars are given. In the graph on the right, the activation of the respective p300 proteins was set as 1 and the specific activations of 8E6 were calculated. A schematic drawing of p300 with the two deletions of the 8E6 binding motif is given above the graph. (The numbers refer to the corresponding amino acids of p300.) To verify the expression of the mutated p300 versions, extracts of C33A cells, transfected with the vector for HA-p300 or for the three deletion mutants, respectively, were analyzed by a Western blot developed with an antibody against the HA tag. The positions of the full-length p300 versions are indicated. The faster migrating bands may represent degradation products thereof. aa, amino acids; wt, wild type; WB, Western blot.
8E6 stimulates the early promoter P175 independent of 8E2 and p300.
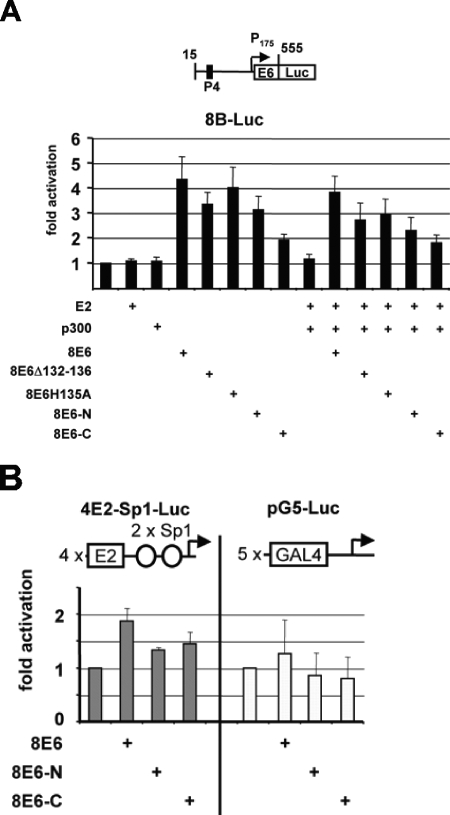
Our data presented thus far suggest that the E6 oncoprotein of HPV8 contributes to the activation of the late promoter by binding to a motif within the C/H3 region of the cellular coactivator p300 via amino acids 132 to 136. When using the 8NCR-Luc reporter construct containing the early and the late promoter, an effect of 8E6 on the early promoter would have been overridden by the strong late P7535. To investigate the role of 8E6 on P175 in transient transfection experiments, we used a reporter construct named 8B-Luc, comprised of HPV8 sequences from positions 15 to 555, thus P175 and its promoter proximal region. Luciferase activity from 8B-Luc was not affected by overexpression of 8E2 and/or p300, indicating that 8E2 and p300 do not regulate this promoter in this construct. However, 8E6, 8E6Δ132-136, and 8E6H135A activated transcription by 4.3-, 3.5-, and 4.0-fold, respectively (Fig. 5A). The coexpression of 8E2 and p300 with 8E6, E6Δ132-136, or 8E6H135A did not further stimulate activations mediated by the various 8E6 proteins. The notion that 8E6 uses a mechanism independent of 8E2 and p300 to stimulate the early promoter, at least within this construct, is also supported by the observation that 8E6-N, inactive on P7535, activated P175 3.8-fold, while 8E6-C, in combination with 8E2 and p300, an efficient activator of the late promoter (see Fig. 1C and 2A), only had a small effect when the 8B-Luc construct was used (Fig. 5A). The activation of HPV8 early promoter activity by 8E6 is not due to a nonspecific effect on transcription, since the coexpression of 8E6, 8E6-N, or 8E6-C did not activate two other control promoters (Fig. 5B).
FIG. 5.
8E6 can activate the early HPV8 promoter independently of E2 and p300. (A) Transient transfections of RTS3b cells with the reporter construct 8B-Luc, containing the 3′ region of the HPV8 NCR from nucleotides 15 to 555. In this part of the NCR, only the E2 binding site P4 and the early promoter P175 are present, as indicated in the figure. The reporter construct has been cotransfected with 500 ng expression vector for 8E6, 8E6Δ132-136, 8E6H135A, 8E6-N, and 8E6-C, respectively, either alone or together with 10 ng E2 and 500 ng p300 expression vector, as indicated beneath the graphs. The fold activations of three independent experiments have been calculated, and the error bars are given. (B) Transient transfection of RTS3b cells with an expression vector for 8E6, 8E6-N, or 8E6-C (each 500 ng) and two synthetic reporter constructs. The structure of both is shown above the graphs. The results represent the means of three independent experiments.
DISCUSSION
Our data presented here show for the first time that an E6 oncoprotein contributes to the regulation of HPV transcription. Transient transfections and RPA revealed that 8E6 activates both the early and the late promoter of HPV8. The use of various 8E6 mutants provides evidence that each of the two promoters is stimulated via a specific, distinct mechanism. While early promoter activation by 8E6 does not involve p300, it may be a major determinant for the late promoter's activity in HPV8, since P7535 activity increased after overexpression of p300. p300 may represent a central platform allowing the HPV8-encoded proteins E2 and E6 to mediate their effects on P7535. Our data suggest that 8E2 is one of the DNA-bound transcription factors which recruits p300 to the promoter via protein-protein interaction (33). We provide several lines of evidence that the induction of HPV8 late gene expression by 8E6 requires direct contact with p300. The deletion of a small motif from amino acids 132 to 136 within 8E6 abolished the interaction with p300 and significantly reduced the 8E6-mediated activation of P7535 in combination with 8E2 and p300, as revealed by coimmunoprecipitation and transient transfections. In addition, an 8E6 with a mutation of H135A retained weak binding to p300 and some activation of p300-mediated transcription, indicating that this amino acid may be critical. The deletion or the point mutation did not lead to defective 8E6 proteins, since 8E6Δ132-136 and 8E6H135A were also able to activate the early promoter, further supporting the specific role these amino acids play in cooperation with p300. The increase of basal P7535 transcription after coexpression of p300 in the absence of E2 may reflect the involvement of p300 in the transcription of the HPV8 late promoter activated by cellular factors. Increasing the concentrations of p300, which has been shown to be limiting under physiological conditions (19), may enable cellular factors such as AP1 and RUNX1, which have been shown to regulate the HPV8 late promoter (16, 45, 48) and to use p300 as coactivator (3, 20), to recruit more p300, resulting in higher transcriptional activity. Obviously, the 8E6-mediated stimulation of this transcription strictly depends on the interaction with p300, since 8E6Δ132-136 failed to stimulate here.
Since the deletion of the five amino acids within 8E6 did not completely abolish activation of 8E6 in the presence of coexpressed 8E2 and p300, we cannot exclude that an interaction between 8E2 and 8E6 may contribute. In the case of genital HPVs, it has been demonstrated that E2 and E6 proteins directly bind to each other, regulating their activity (13). We tested for a direct interaction of both HPV8 proteins. We did not succeed in coprecipitating coexpressed 8E6 with 8E2, although bacterially expressed, purified His-tagged 8E6 was specifically retained by purified GST-8E2 (data not shown). Such an interaction may contribute to the stabilization of a ternary complex between p300, 8E6, and 8E2. The existence of a ternary complex is based on results obtained by the protein competition experiment (Fig. 3B) indicating that 8E2 and 8E6 do not compete for binding to p300-4 and on the observation that the high-affinity sites of 8E2 and 8E6 in p300 are not overlapping (Fig. 3A).
Our data suggest that the contact of 8E6 with amino acids 1770 to 1814 within the C/H3 region is required for 8E6-mediated activation, since the deletion of this 45-amino-acid segment, encoding the here-identified high-affinity 8E6 binding site, significantly impaired the stimulation of p300 activity by 8E6. Interestingly, the virally encoded inhibitors of CBP/p300, E1A and 16E6, bind to this domain as well (3, 5, 15, 25, 35, 38, 52). The deletion of amino acids 1770 to 1814 within p300 led to a reduction of basal HPV8 late promoter activation by p300. This part of p300 and the homologous segment in CBP are recognized by a number of important transcriptional regulators, including p53 (2, 14, 26), c-fos (3, 36), YY1, TFIIB, E2F, RNA helicase A, and p/CAF (6). A decreased recruitment of p300Δ1770-1814 due to the lack of the interaction domain for cellular DNA binding factors binding to the HPV 8NCR, such as the members of the AP1 complex, p53 and YY1 (1, 16, 37), or for components of the preinitiation complex may be the basis for this reduced activity of p300Δ1770-1814. Although the deletion of amino acids 1529 to 1572, located within the HAT domain and containing a second, here-identified low-affinity 8E6 binding site, affected p300 activity as well, it did not impair the coactivation by 8E6. Thus, this motif may not be relevant. We cannot exclude an involvement of the 8E6 binding sites within the amino-terminal or carboxyl-terminal part of p300 for the stimulating effect of 8E6 under physiological conditions as well. By the overexpression of the factors as occurred here, we may reduce the requirement for multiple contacts, since the high intracellular concentrations favor a complex formation. The binding of 8E6 to multiple sites within p300 may be required for the formation of a stable complex under limiting endogenous concentrations of p300 (19, 51).
The residues of 16E6 homologous to amino acids 132 to 136 of 8E6 have been described to be involved in p300 or CBP binding and correlated with the inhibition of the coactivator function of CBP/p300 (38, 52). Thus, we conclude that E6 proteins are targeted via this motif to p300. Why does the targeting of 8E6 and 16E6 to p300 lead to complete opposite effects on the coactivator function of p300? One major feature of 16E6 is its binding to E6 associated protein E6AP, a cellular ubiquitin ligase, which determines a variety of target proteins, such as p53, E6TP1, and the PDZ domain, containing partner proteins for degradation (28). Although the contact of 16E6 with E6AP seems to be dispensable for repression of transcription by p53, which was shown to be achieved by 16E6-mediated inhibition of the p300 HAT activity (50), it may be relevant for other targets. 16E6 has been demonstrated to bind to and induce the degradation of a variety of cellular factors which function as coactivators of transcription, among which are ADA3 and AMF1/Gps2 (7, 10, 23). It seems feasible that the interaction of 8E6 with such factors may not lead to their degradation, resulting in a more stable complex efficient in transcription. For example, the published interaction between 8E6 and AMF1/Gps2 (7), which may act as a bridging factor between E2 and p300 (39), may stabilize a multiprotein complex, resulting in an increase of the availability of AMF1/Gps2 for E2. Further experiments are necessary to decipher the mechanism of how 8E6 protein modulates p300 activity.
In contrast to the late promoter, the activation of the early promoter does not require 8E2 and/or p300, since its activity remained unaffected after the coexpression of these factors. Consequently, activation of the early promoter by 8E6 and corresponding mutants was not enhanced by 8E2 and p300. The notion that a distinct mechanism is responsible for early promoter activation is supported by observations that 8E6-N, which had no effect on P7535, significantly activated P175, and in contrast 8E6-C only slightly stimulated the early promoter. The mechanism of activation of the early promoter by 8E6 is unknown. We speculate that a direct interaction with cellular DNA binding transcription factors binding to the P175 promoter proximal region, which includes YY1 (37), may recruit 8E6 to the promoter region, allowing superactivation. Moreover, the binding of 8E6 to components of the TFIID complex such as TAF28, TAF135, and TBP, which has been shown in vitro (10), may contribute.
Our data further support an essential role for p300 in regulation of the HPV8 life cycle. In normal skin, p300 is hardly detected in the basal cells, while its expression increases in the more differentiated layers (33). Thus, in the differentiated cells, large amounts of p300 are available which allow the viral 8E2 and 8E6 proteins to activate the late promoter by targeting p300. Other differentiation dependently expressed transcription factors may contribute to the activation as well. As a consequence of the small amounts of p300 in the basal layers, the early promoter's transcription seems to be independent of p300 and E2, which has been show to be expressed from the late promoter (48). Thus, both factors cannot be used by 8E6. Early promoter activation by 8E6 should result in an upregulation of its own expression. High levels of 8E6 may modulate the cellular environment in favor of the viral life cycle. For example, via stimulation of the coactivator function of p300, which is required for keratinocyte differentiation (32), the differentiation state of the infected keratinocytes may be affected by 8E6. In summary, the results presented here suggest an unexpected novel function of an HPV E6 protein as a coactivator of HPV gene expression. 8E6 enhances the activity of both the early and the late promoter and thus displays essential functions to efficiently complete the viral life cycle.
Acknowledgments
We thank Manuela Rehtanz, Sharon S. Willer, and Herbert Pfister for critical reading of the manuscript and helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (STE 604/3-3), the Wilhelm Sander-Stiftung (2003.098.1), and the Köln Fortune Program, Faculty of Medicine, University of Cologne.
REFERENCES
- 1.Akgül, B., P. Karle, M. Adam, P. G. Fuchs, and H. Pfister. 2003. Dual role of tumor suppressor p53 in regulation of DNA replication and oncogene E6 promoter activity of epidermodysplasia verruciformis-associated human papillomavirus type 8. Virology 308**:**279-290. [DOI] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., V. V. Ogryzko, K. Gardner, A. Giordano, S. A. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53 dependent signal pathways. Cell 89**:**1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14**:**4758-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouallaga, I., S. Teissier, M. Yaniv, and F. Thierry. 2003. HMG-I(Y) and the CBP/p300 coactivator are essential for human papillomavirus type 18 enhanceosome transcriptional activity. Mol. Cell. Biol. 23**:**2329-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravarti, D., V. V. Ogryzko, H.-V. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and P/CAF acetyltransferase activity. Cell 96**:**393-403. [DOI] [PubMed] [Google Scholar]
- 6.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Science 114**:**2363-2373. [DOI] [PubMed] [Google Scholar]
- 7.Degenhardt, Y. Y., and S. J. Silverstein. 2001. Gps2, a protein partner for human papillomavirus E6 proteins. J. Virol. 75**:**151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11**:**1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbel, M., S. Carl, S. Spaderna, and T. Iftner. 1997. A comparative analysis of the interactions of the E6 proteins from cutaneous and genital papillomaviruses with p53 and E6AP in correlation to their transforming potential. Virology 239**:**132-149. [DOI] [PubMed] [Google Scholar]
- 10.Enzenauer, C., G. Mengus, A.-C. Lavigne, I. Davidson, H. Pfister, and M. May. 1998. Interaction of human papillomavirus 8 regulatory proteins E2, E6 and E7 with components of the TFIID complex. Intervirology 41**:**80-90. [DOI] [PubMed] [Google Scholar]
- 11.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94**:**2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation and development. Genes Dev. 14**:**1553-1577. [PubMed] [Google Scholar]
- 13.Grm, H. S., P. Massimi, N. Gammoh, and L. Banks. 2005. Crosstalk between the human papillomavirus E2 transcriptional activator and the E6 oncoprotein. Oncogene 24**:**5149-5164. [DOI] [PubMed] [Google Scholar]
- 14.Gu, W., X.-L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387**:**819-823. [DOI] [PubMed] [Google Scholar]
- 15.Hamamori, Y., V. Sartorelli, V. Ogryzko, P. L. Puri, H. Y. Wu, J. Y. Wang, Y. Nakatani, and L. Kedes. 1999. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96**:**405-413. [DOI] [PubMed] [Google Scholar]
- 16.Horn, S., H. Pfister, and P. G. Fuchs. 1993. Constitutive transcriptional activator of epidermodysplasia verruciformis-associated human papillomavirus 8. Virology 196**:**674-681. [DOI] [PubMed] [Google Scholar]
- 17.Huang, S.-M., and D. J. McCance. 2002. Downregulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J. Virol. 76**:**8710-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iftner, T., S. Bierfelder, Z. Csapo, and H. Pfister. 1988. Involvement of human papillomavirus type 8 genes E6 and E7 in transformation and replication. J. Virol. 62**:**3655-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S.-C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85**:**403-414. [DOI] [PubMed] [Google Scholar]
- 20.Kitabayashi, I., A. Yokoyama, K. Shimizu, and M. Ohki. 1998. Interaction and functional cooperation of the leukemia-associated factors AML-1 and p300 in myeloid cell differentiation. EMBO J. 17**:**2994-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyono, T., A. Hiraiwa, and M. Ishibashi. 1992. Differences in transforming activity and coded amino acid sequences among E6 genes of several papillomaviruses associated with epidermodysplasia verruciformis. Virology 186**:**628-639. [DOI] [PubMed] [Google Scholar]
- 22.Kiyono, T., A. Hiraiwa, S. Ishii, T. Takahashi, and M. Ishibashi. 1994. Inhibition of p53-mediated transactivation by E6 of type 1, but not type 5, 8, or 47, human papillomavirus of cutaneous origin. J. Virol. 68**:**4656-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, A., Y. Zhao, G. Meng, M. Zeng, S. Srinivasan, L. M. Delmolino, Q. Gao, G. Dimri, G. F. Weber, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 22**:**5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, D., B. Lee, J. Kim, D. W. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 275**:**7045-7051. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. S., R. H. See, T. Deng, and Y. Shi. 1996. Adenovirus E1A downregulates cJun- and JunB-mediated transcription by targeting their coactivator p300. Mol. Cell. Biol. 16**:**4312-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lill, N. L., S. R. Grossman, D. Ginsberg, J. A. DeCaprio, and D. Livingston. 1997. Binding and modulation of p53 by p300/CBP co-activators. Nature 387**:**823-827. [DOI] [PubMed] [Google Scholar]
- 27.Lipari, F., G. A. McGibbon, E. Wardrop, and M. G. Cordingley. 2001. Purification and biophysical characterization of a minimal functional domain and of an N-terminal Zn2+-binding fragment from the human papillomavirus Type 16 E6 protein. Biochemistry 40**:**1196-1204. [DOI] [PubMed] [Google Scholar]
- 28.Longworth, M. S., and L. A. Laimins. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68**:**362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN-β enhanceosome is required for synergistic activation of transcription. Mol. Cell 1**:**277-287. [DOI] [PubMed] [Google Scholar]
- 30.Meyers, C., M. G. Frattini, and L. A. Laimins. 1994. Tissue culture techniques for the study of human papillomaviruses in stratified epithelia. Academic Press, Orlando, Fla.
- 31.Mink, S., B. Haenig, and K.-H. Klempnauer. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell. Biol. 17**:**6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Missero, C., E. Calautti, R. Eckner, J. Chin, L. H. Tsai, D. M. Livingston, and G. P. Dotto. 1995. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc. Natl. Acad. Sci. USA 92**:**5451-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76**:**11042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munoz, N. 2000. Human papillomavirus and cancer: the epidemiological evidence. J. Clin. Virol. 19**:**1-5. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor, M. J., H. Zimmerman, S. J. Nielsen, H.-U. Bernard, and T. Kouzarides. 1999. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J. Virol. 73**:**3574-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oelgeschlager, M., R. Janknecht, J. Krieg, S. Schreek, and B. Luscher. 1996. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 15**:**2771-2780. [PMC free article] [PubMed] [Google Scholar]
- 37.Pajunk, H. S., C. May, H. Pfister, and P. G. Fuchs. 1997. Regulatory interactions of transcription factor YY1 with control sequences of the E6 promoter of human papillomavirus type 8. J. Gen. Virol. 78**:**3287-3295. [DOI] [PubMed] [Google Scholar]
- 38.Patel, D., S.-M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18**:**5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, Y.-C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74**:**5872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfister, H., P. G. Fuchs, S. Majewski, S. Jablonska, S. Pniewska, and M. Malejczyk. 2003. High prevalence of epidermodysplasia verruciformis-associated human papillomavirus DNA in actinic keratoses of the immunocompetent population. Arch. Dermatol. Res. 295**:**273-279. [DOI] [PubMed] [Google Scholar]
- 41.Pfister, H., and J. ter Schegget. 1997. Role of HPV in cutaneous premalignant and malignant tumors. Clin. Dermatol. 15**:**335-347. [DOI] [PubMed] [Google Scholar]
- 42.Purdie, K. J., C. J. Sexton, C. M. Proby, M. T. Glover, A. T. Williams, J. N. Stables, and I. M. Leigh. 1993. Malignant transformation of cutaneous lesions in renal allograft patients: a role for human papillomavirus. Cancer Res. 53**:**5328-5333. [PubMed] [Google Scholar]
- 43.Schaper, I. D., G. P. Marcuzzi, S. J. Weissenborn, H. U. Kasper, V. Dries, N. Smyth, P. G. Fuchs, and H. Pfister. 2005. Development of skin tumors in mice transgenic for early genes of human papillomavirus type 8. Cancer Res. 65**:**1394-1400. [DOI] [PubMed] [Google Scholar]
- 44.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63**:**1129-1136. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, H.-M., G. Steger, and H. Pfister. 1997. Competitive binding of viral E2 protein and mammalian core-binding factor to transcriptional control sequences of human papillomavirus type 8 and bovine papillomavirus type 1. J. Virol. 71**:**8029-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steger, G., and H. Pfister. 1992. In vitro expressed HPV 8 E6 protein does not bind p53. Arch. Virol. 125**:**355-360. [DOI] [PubMed] [Google Scholar]
- 47.Stubenrauch, F., I. M. Leigh, and H. Pfister. 1996. E2 represses the late gene promoter of human papillomavirus type 8 at high concentrations by interfering with cellular factors. J. Virol. 70**:**119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stubenrauch, F., J. Malejczyk, P. G. Fuchs, and H. Pfister. 1992. Late promoter of human papillomavirus type 8 and its regulation. J. Virol. 66**:**3485-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stubenrauch, F., and H. Pfister. 1994. Low-affinity E2-binding site mediates downmodulation of E2 transactivation of the human papillomavirus type 8 late promoter. J. Virol. 68**:**6959-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas, M. C., and C.-M. Chiang. 2005. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independent of inducing p53 degradation. Mol. Cell 17**:**251-264. [DOI] [PubMed] [Google Scholar]
- 51.Yao, T.-P., S. P. Oh, M. Fuchs, N.-D. Zhou, L.-E. Chang, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93**:**361-372. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann, H., R. Degenkolbe, H.-U. Bernard, and M. J. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73**:**6209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zimmermann, H., C.-H. Koh, R. Degenkolbe, M. J. O'Connor, A. Müller, G. Steger, J. J. Chen, Y. Lui, E. J. Androphy, and H.-U. Bernard. 2000. Interaction with CBP/p300 enables the bovine papillomavirus type 1 E6 oncoprotein to downregulate CBP/p300 mediated transactivation by p53. J. Gen. Virol. 81**:**2617-2623. [DOI] [PubMed] [Google Scholar]