Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids (original) (raw)
Abstract
Oxidized phospholipids are thought to promote atherogenesis by stimulating endothelial cells (ECs) to produce inflammatory cytokines, such as IL-8. In studies with mouse models, we previously demonstrated that genetic variation in inflammatory responses of endothelial cells to oxidized lipids contributes importantly to atherosclerosis susceptibility. We now show that similar variations occur in cultured aortic ECs derived from multiple heart transplant donors. These variations were stably maintained between passages and, thus, reflect either genetic or epigenetic regulatory differences. Expression array analysis of aortic EC cultures derived from 12 individuals revealed that >1,000 genes were regulated by oxidized phospholipids. We have used the observed variations in the sampled population to construct a gene coexpression network comprised of 15 modules of highly connected genes. We show that several identified modules are significantly enriched in genes for known pathways and confirm a module enriched for unfolded protein response (UPR) genes using siRNA and the UPR inducer tunicamycin. On the basis of the constructed network, we predicted that a gene of unknown function (MGC4504) present in the UPR module is a target for UPR transcriptional activator ATF4. Our data also indicate that IL-8 is present in the UPR module and is regulated, in part, by the UPR. We validate these by using siRNA. In conclusion, we show that interindividual variability can be used to group genes into pathways and predict gene–gene regulatory relationships, thus identifying targets potentially involved in susceptibility to common diseases such as atherosclerosis.
Keywords: genetic, interleukin 8, atherosclerosis, unfolded protein response, network
Atherosclerosis, the major cause of heart disease, is characterized by the accumulation of cholesterol, inflammatory cells, smooth muscle cells, and fibrous elements beneath the endothelial cell (EC) monolayer that lines the artery wall (1). Although numerous risk factors for atherosclerosis, such as elevated blood pressure, hypercholesterolemia, and smoking, have been recognized, these factors do not alone account for the genetic contribution to risk (2). An important mechanism contributing to the recruitment of inflammatory cells in atherosclerosis is the induction of adhesion molecules, growth factors, and cytokines in vascular ECs by oxidized phospholipids, such as oxidized 1-palmitoyl-2-arachidonyl-_sn_-3-glycero-phosphorylcholine (oxPAPC) derived from lipoproteins trapped in the vessel wall (3).
We have previously demonstrated that ECs from different strains of mice show differences in the induction of inflammatory genes when treated with oxidized lipoproteins, and that these differences segregate with susceptibility to atherosclerosis (4, 5). Studies in human populations show significant variability in the plasma levels of inflammatory mediators associated with atherosclerosis, including IL-8 and C-reactive protein (6–8). The plasma levels of cytokines are influenced by genetic and environmental factors. In this study, we examined human aortic EC cultures derived from multiple heart transplant donors and showed the existence of striking differences in the level of inflammatory gene induction by oxPAPC. These differences were maintained between individual passages of the ECs and thus are likely to represent either genetic or epigenetic regulatory variations.
The mechanisms underlying the inflammatory effects of oxPAPC are not well understood, and here we describe a unique approach for understanding the global pathways involved. The approach is based on the fact that a natural population will exhibit multiple genetic or epigenetic variations that perturb the expression of many individual genes, as well as entire pathways. These variations can be quantitated by expression array profiling and analyzed by using systems biology approaches, which provide a way to bridge the gap between individual genes and complex biological systems (9–11). Here, we take advantage of the naturally occurring variability in human EC responses to oxidized phospholipids and show, using a gene coexpression network approach, that the unfolded protein response (UPR) pathway is one of the key regulators of IL-8 production.
Results
Variation in Inflammatory Responses of Primary Human Endothelial Cells to Oxidized Phospholipids.
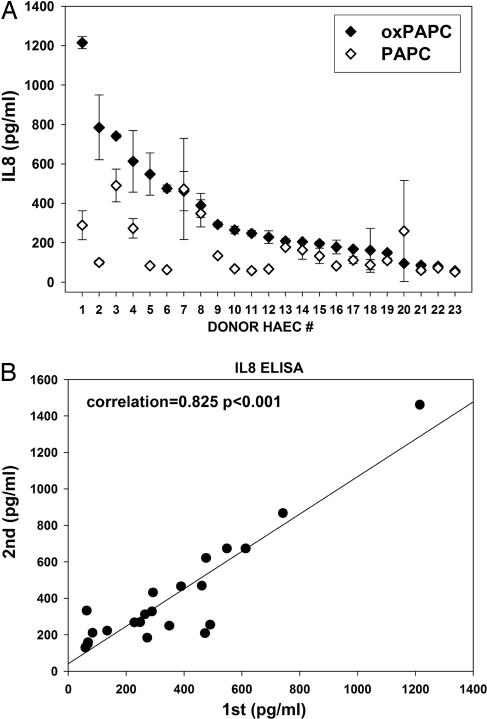
Based on our previous studies in mouse ECs (4, 5), we hypothesized that the inflammatory responses of human ECs to oxidized lipids may also differ among individuals in the population. We tested this possibility by performing an initial screen of IL-8 secretion in response to oxPAPC treatment in early passages of human aortic ECs (HAECs) derived from 23 separate heart transplant donors. As shown in Fig. 1A, we observed significant differences in the amount of IL-8 protein secreted into media after oxPAPC treatment. These differences between EC cultures were quite stable and preserved during multiple cell passages (Fig. 1B). We conclude that EC cultures derived from different individuals in the population exhibit either common genetic or epigenetic variations that perturb their responses to oxidized lipids.
Fig. 1.
Primary human aortic endothelial cells isolated from different donors exhibit differences in IL-8 induction in response to oxPAPC treatment. (A) HAEC lines derived from human individuals were examined for IL-8 induction in response to oxPAPC or control PAPC by ELISA. Individual HAEC lines from 23 donors were treated with oxPAPC (40 μg/ml) or PAPC (40 μg/ml) for 4 h, media were collected, and IL-8 levels were analyzed by ELISA. Absolute levels of secreted IL-8 protein (picograms per milliliter) after the treatment were plotted for each individual HAEC line. HAEC donors were numbered according to the level of IL-8 induction in response to oxPAPC. (B) Eleven individual HAEC cell lines were examined for differences in IL-8 protein secretion between successive cell passages. HAECs were treated with oxPAPC (40 μg/ml) or PAPC (40 μg/ml) for 4 h, and IL-8 protein was measured by ELISA (first passage). The same analysis was performed after freezing and replating cells to obtain the subsequent cell passage (second passage).
Identification of Genes Regulated by oxPAPC.
The observed differences in IL-8 secretion among individual cell lines indicated that we could expect similar variability at the level of gene transcription. Thus, we reasoned that we would be able to identify groups of functionally related, coexpressed genes (modules) that are responsive to oxPAPC by profiling endothelial cells derived from unrelated individuals in the population and testing for coexpression. Therefore, we profiled global mRNA expression levels in HAEC cultures derived from 12 individual donors, where each culture was treated with either unoxidized PAPC (40 μg/ml) as control or with oxPAPC (40 μg/ml) for 4 h. Each condition was profiled in duplicate, so that altogether 48 arrays were examined. Varying levels of secreted IL-8 protein among the 12 donors correlated significantly with the respective IL-8 mRNA levels (r = 0.824, P < 0.001), supporting the use of expression array data as a predictive measure of the inflammatory response to oxPAPC (Fig. 5, which is published as supporting information on the PNAS web site).
We restricted the analysis to genes differentially expressed between the control group and the group treated with oxPAPC. The selection criteria were as follows: (i) fold change of group means ≥1.5 or ≤0.667; (ii) t test P value ≤0.01; (iii) absolute difference of group means ≥500; and (iv) “QC” called present in at least one of the compared groups. For network analysis purposes, each individual array probe set was treated as a unique gene. This method yielded 1,043 differentially expressed genes (probe sets) with a corresponding mean false discovery rate (FDR) of 0.25% (see Materials and Methods). Some of these genes, including IL-8, HMOX1 and members of the sterol regulatory element-binding protein (SREBP) pathway, have previously been shown to be regulated by oxPAPC, but the vast majority represented novel oxPAPC targets (Table 1, which is published as supporting information on the PNAS web site; refs. 12 and 13).
Gene Coexpression Network of oxPAPC-Responsive Genes.
A gene coexpression network was constructed according to a recently described methodology (14). The connectivity (k) was determined for all network genes by taking the sum of their connection strengths (coexpression similarity) with all other genes in the network. To identify modules of highly coregulated genes, we used average linkage hierarchical clustering to group genes based on the topological overlap of their connectivity (see Materials and Methods for details).
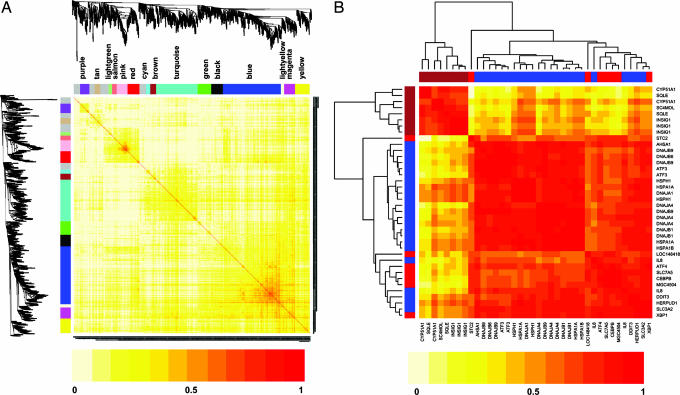
The resulting gene coexpression network was composed of 1043 oxPAPC-regulated genes separated into 15 modules of highly correlated genes. Each module was assigned a unique color identifier (Fig. 2A), with the remaining, poorly connected genes colored gray. In a topological overlap matrix (TOM) plot, the increasing color intensity indicates higher connectivity (coexpression similarity) among genes in the network (Fig. 2A). Genes with the greatest connectivity index (k) represent network “hubs” and are localized in the center of individual modules. Because the network is comprised of differentially expressed genes, the control group and the group treated with oxPAPC were distinguished by their gene expression profiles. Overall, the most highly induced/suppressed genes have the largest number of connections to other genes (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Gene coexpression network of the 1,043 genes regulated by oxPAPC. (A) Pearson correlations between expression profiles of all pairs of genes were transformed into network connection strengths (denoted by intensity of red color) by using a power function (see Materials and Methods). Average linkage hierarchical clustering with the topological overlap dissimilarity measure was used to identify gene coexpression modules, each of which was assigned a unique color. Rows and columns are symmetric and represent genes. (B) A color-coded version of the correlation matrix (Table 4) involving the SREBP genes from the brown module and the UPR genes from blue and red modules. The intensity of red color represents the absolute value of Pearson correlation coefficients. The rows and columns have been sorted by the gene clustering tree. Note that there are two broad clusters corresponding to SREBP and UPR genes.
Genetic or Epigenetic Variations Perturb Multiple Genes and Pathways Involved in Responses to Oxidized Phospholipids.
Because of the indicated importance of hubs in the network (15), we ranked genes within each of the 15 modules based on their intramodular connectivity (k.in) to identify module hubs (Table 1). Interestingly, INSIG1, a key regulator of the SREBP pathway activation (16), had the highest connectivity in the brown module and represented the main hub. The brown module was also the most highly enriched in SREBP-regulated genes (Fisher’s exact test, P < 10−9; Table 2, which is published as supporting information on the PNAS web site). Altogether, eight of the 26 total genes in the brown module were SREBP targets (17).
Next, we focused on examining modules containing the IL-8 gene. There are two probes on the array chip measuring IL-8, both of which were present within the blue module. Examination of the blue module revealed the presence of a large number of molecular chaperones (DNAJA1, DNAJA4, DNAJB1, DNAJB6, and DNAJB9) and heat shock proteins (HSPA1A, HSPA1B, and HSPH1), many of which are known to be induced by ER stress as part of the UPR pathway (Table 1) (18). The blue module also contained several other UPR targets including DDIT3, ATF3, and HERPUD1 and ranked first in terms of enrichment for UPR target genes (P < 10−12).
Additional searches for UPR genes revealed that the red module contained several additional genes known to be regulated by the UPR (ATF4, XBP1, CEBPB, SLC7A5, and STC2) (19). Although only moderately enriched in UPR genes, four of the top 10 most connected genes (hubs) in the red module were UPR members, including two key UPR transcription factors ATF4 and XBP1 (Tables 1 and 3, which are published as supporting information on the PNAS web site) (20).
Induction of UPR-target genes was a strong indication that oxPAPC induces the UPR pathway in HAECs. The UPR has three branches, each acting via separate transcription factor represented by ATF4, ATF6, and XBP1 (20). The blue module contained large number of ER chaperones, which are known to be regulated primarily by XBP1, whereas many of the red module UPR genes (SLC7A5, ATF4, and STC2) are known targets of ATF4 (18, 19, 21). This finding suggests that differences in transcriptional regulatory mechanisms may underlie separation of genes into modules. This was visualized by further clustering analysis (Fig. 2B and Table 4, which is published as supporting information on the PNAS web site), which showed that the blue and red UPR subgroups of genes were very similar to each other but strikingly different from the SREBP genes present in the brown module.
The other modules also contained genes that could be functionally related, although many of the identified transcripts have not been functionally characterized. For example, the yellow module contained HMOX1 and other genes involved in the response to oxidative stress such as GCLM and GCLC (Table 1) (13).
In an effort to further understand the functional significance of the network modules, we carried out a gene-enrichment analysis with respect to GO ontology and KEGG pathway databases. This proved to be problematic, given that <25% of the network genes were annotated in the ontology databases and only 10% were present in the KEGG pathway database. Nevertheless, the blue module was enriched for genes with heat shock protein and chaperone activity, and the brown module for genes involved in sterol biosynthesis (Table 5, which is published as supporting information on the PNAS web site).
Network Application: The UPR Pathway Contributes to Expression of the Inflammatory Cytokine IL-8 and the Gene MGC4504.
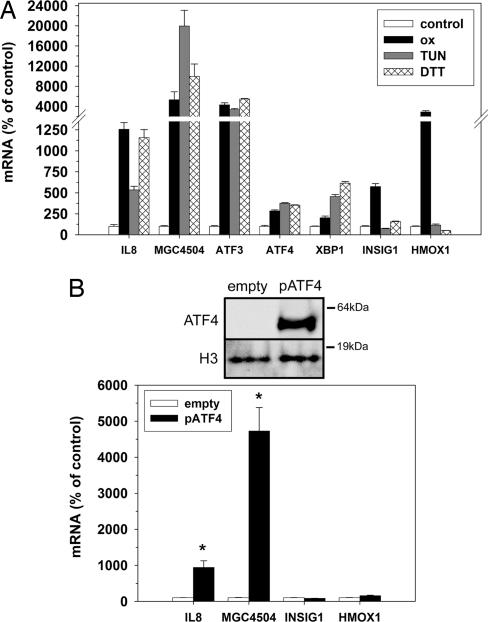
As illustrated above, the constructed network contains modules with genes functionally linked on the basis of their mRNA regulation. This approach can therefore be applied to identification of biologically important regulatory mechanisms, in this case the oxPAPC-mediated IL-8 induction. Alternatively, it can help in the functional annotation of uncharacterized genes. To address this, we focused on IL-8 (blue module) and the MGC4504, a gene of unknown function in the red module. MGC4504 was chosen based on its highest connectivity value among unannotated genes in the red (UPR) module (Table 1). To examine whether IL-8 and MGC4504 expression is regulated by the UPR, we incubated HAECs with two well established inducers of ER stress and UPR, tunicamycin and DTT (22). Treatment with either tunicamycin or DTT resulted in induction of IL-8 (5- and 11-fold, respectively) and MGC4504 (200- and 100-fold, respectively). UPR genes ATF3, ATF4, and XBP1 were also induced, with only marginal changes observed in non-UPR genes, INSIG1 (brown module), and HMOX1 (yellow module) (Fig. 3A).
Fig. 3.
UPR regulates expression of IL-8 and MGC4504. (A) Effect of UPR inducers on gene expression. HAECs were treated for 4 h in medium alone (control), medium containing oxPAPC (50 μg/ml), or UPR-inducing agents tunicamycin (10 μg/ml) or DTT (1 mM). Expression levels of IL-8 (blue module), MGC4504 (red module), INSIG1 (brown module), HMOX1 (yellow module), and UPR genes from blue and red module (ATF3, ATF4, and XBP1) were measured by quantitative PCR (Q-PCR) as described in Materials and Methods. Q-PCR results are expressed as the mean differences for each treatment group and control group (set at 100%) ± 1 SD. (B) Effect of ATF4 overexpression on IL-8 and MGC4504. HeLa cells were transfected with an expression plasmid for human ATF4 (pATF4) or a control plasmid (empty) as described in Materials and Methods. Forty-eight hours after transfection, expression levels of IL-8, MGC4504, INSIG1, and HMOX1 mRNA were analyzed by Q-PCR. ATF4 protein levels in isolated nuclear extracts were measured by immunoblotting. Histone H3 (H3) antibody was used as a protein-loading control. Asterisks indicate significantly different mean expression value of pATF4 group from control group (P < 0.005).
Next, we searched the promoters (the 2-kb 5′ flanking region) of all of the network genes for regulatory elements known to confer responsiveness to the UPR (18, 23). Sixty-nine of the 746 network genes with unique promoters contained at least one of the four searched UPR-binding sites. These included many known UPR genes (Table 6, which is published as supporting information on the PNAS web site). This analysis revealed that the MGC4504 promoter contains a highly conserved (human, mouse, rat) C/EBP-ATF composite element 5′-TTGCATCA-3′ at −323 to −315 bp, which is known to bind ATF4 and lead to induction of UPR genes DDIT3 and HERPUD1 (23). None of the searched sites were present in the proximal promoter of IL-8.
To examine whether ATF4 expression can lead to IL-8 and MGC4504 induction, we transfected HeLa cells with a plasmid constitutively expressing ATF4. HeLa cells were used due to the extremely low plasmid transfectability of primary human EC. These cells were previously shown to respond to oxPAPC treatment much like primary endothelial cells, up-regulating IL-8, activating SREBP (12), and inducing the UPR (P.S.G., unpublished data). Expression of ATF4 resulted in a significant induction of IL-8 and MGC4504 mRNA (9- and 47-fold, respectively) (Fig. 3B). No significant changes were observed in expression of INSIG1 and HMOX1.
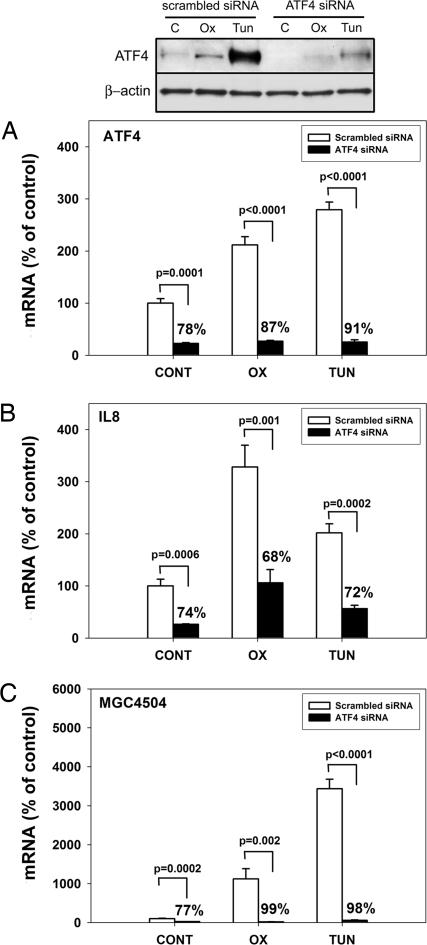
To determine whether ATF4 is required for MGC4504 and IL-8 induction by oxPAPC, we used an siRNA approach (see Materials and Methods). Oligo-based siRNA targeting effectively reduced the endogenous ATF4 mRNA (control, 78%; oxPAPC, 87%; tunicamycin, 91%) and protein levels in primary HAECs (Fig. 4A). ATF4 down-regulation resulted in a significant decrease in IL-8 mRNA levels (control, 74%; oxPAPC, 68%; tunicamycin, 72%) and secreted protein levels (Fig. 4B and Fig. 7, which is published as supporting information on the PNAS web site). Inhibition of ATF4 had a striking impact on MGC4504 expression, which decreased on average by >90% relative to control siRNA (Fig. 4C). The siRNA targeting was selective, without significant decrease in INSIG1 and HMOX1 in any of the treatment conditions (Fig. 7). Altogether, these data confirm the network-based predictions and demonstrate that ATF4 is a necessary mediator of MGC4504 expression in HAECs. These data also indicate that ATF4 is a significant regulator of IL-8 expression.
Fig. 4.
Effect of the ATF4 siRNA on IL-8 and MGC4504 expression. HAECs were transfected with siRNA directed against human ATF4 or with control scrambled oligonucleotide (see Materials and Methods). Transfected cells were treated for 4 h with medium only (control), oxPAPC (50 μg/ml), or tunicamycin (10 μg/ml). mRNA expression levels of ATF4 (A), IL-8 (B), and MGC4504 (C) were measured by Q-PCR. The levels of ATF4 protein were measured in isolated nuclear extracts by immunoblotting (A). Q-PCR data were set at 100% for untreated cells (control) incubated with scrambled siRNA. Percentage values above bars indicate the mean expression decrease in the ATF4 siRNA group versus control siRNA group for each treatment ± 1 SD.
Discussion
Aortic endothelial cells are a crucial source of inflammatory cytokines contributing to atherosclerosis. We previously showed that certain inbred strains of mice differing in susceptibility to atherosclerosis exhibit striking differences in the response of aortic ECs to oxidized lipoproteins and that, in genetic crosses, the susceptibility to atherosclerosis segregated with the responsiveness of ECs to oxidized lipoproteins (4, 5). In the present study we examined the responses of human aortic EC cultures isolated from different heart transplant donors to oxPAPC. Striking differences were observed in the expression of many genes important in atherosclerosis, including IL-8 cytokine (Fig. 1A). These differences are unlikely to be caused by experimental variability or variation in culturing conditions, as indicated by high preservation of inflammatory responsiveness among successive cell passages (Fig. 1B). Although we hypothesize that the genetic makeup of each individual contributes to observed variation in IL-8 expression, we cannot exclude possible role of epigenetic changes taking place before and after the collection of cell explants. Epigenetic changes are caused by heritable changes in gene expression that are independent of nucleotide sequence. They include methylation of DNA, biochemical modifications of core histones and modulation of the higher-order chromatin structure (24). For most of the cytokines studied, plasma level heritability is ≈50% with environmental factors likely playing a significant role (25).
Expression array analyses of a subset of the EC cultures revealed regulation of surprisingly large number of genes (>1,000) by oxPAPC. These changes were unlikely due to toxic effects of oxPAPC, because oxPAPC at concentrations up to 50 μg/ml was not toxic to ECs and had no effect on caspase-3 activity (Fig. 8, which is published as supporting information on the PNAS web site).
We hypothesized that we could identify groups of genes that are biologically related with respect to regulatory mechanisms. This was based on the assumption that variation in the expression level of a key transcription factor(s) or the degree of receptor activation would perturb a whole series of downstream target genes. Our approach explores either genetic or epigenetic variability to focus on mechanisms underlying clinically highly relevant interactions of oxidized phospholipids and HAECs. Our results demonstrate that this approach can be used to subdivide genes into pathways on the basis of the regulation of their expression. For example, genes involved in the SREBP pathway were highly induced in some EC cultures but not others, whereas the UPR pathway genes also exhibited very similar expression profiles among the EC cultures but the overall responses were quite distinct from genes of the SREBP pathway (Fig. 2B). Thus, there appeared to be multiple genetic or epigenetic variations that are responsible for independent regulation of the individual pathways in response to oxPAPC.
In addition to segregating genes into pathways, we showed that the intramodular network connectivity is a very useful measure for identifying biologically important genes within pathways. A case in point is the red and brown modules, where hub genes with high connectivity value included some of the key regulators of the UPR (ATF4 and XBP1) and SREBP (INSIG1) pathways.
One of the exciting features of gene networks is their ability to identify novel target genes. Using MGC4504 as an example, we showed that this previously uncharacterized gene represents a UPR-target, induction of which requires ATF4. MGC4504 and ATF4 were both among the top six hub genes in the red (UPR) module. To illustrate the advantage of the network approach, we used simple correlation and analyzed all network genes on the basis of their correlation with ATF4 in oxPAPC-treated cells. MGC4504 ranked as 99th most correlated gene with ATF4. Therefore, using simple correlation, this gene would likely have been missed. Based on its protein sequence, MGC4504 appears to be a highly conserved human homologue of a putative protein mediating cation transport in bacteria, which could play an important role in ER homeostasis.
The power of this network approach is further highlighted by the fact that we used a relatively small sample of individuals. Even though successful in predicting gene–gene regulatory relationships, our study using heart transplant donor ECs does not allow for follow up comparison of the cell culture results with plasma cytokine levels of individuals. However, similar network approaches could be applied to large human populations, using easily obtainable cells such as blood monocytes, and further combined with SNP analysis. This approach would permit comparison of the cell culture and in vivo data as well as examination of any genetic variations controlling the expression of genes contributing to atherosclerosis and other complex traits.
A particularly important finding of the present study is that HAEC exposure to oxidized phospholipids results in the induction of the UPR, which directly participates in modulating the inflammatory response. Interestingly, homocysteine, a common risk factor for cardiovascular disease, is also known to induce UPR in ECs (26) as well as the synthesis of MCP1 and IL-8 (27). Our study indicates that UPR, and more specifically, ATF4, is an important mediator of IL-8 expression in HAECs. Previous studies have shown that induction of IL-8 by oxPAPC is complex and likely mediated by multiple pathways, including c-src and SREBP (12, 28). Our findings are consistent with this, and we show that UPR appears to be required, but its induction by tunicamycin is not sufficient, to maximally induce IL-8 expression levels in HAEC. This finding is in contrast to the MGC4504 gene, which behaved as a sole UPR-target. Tunicamycin interferes with protein processing and secretion in the ER, which is likely the reason for the observed decrease in IL-8 protein secreted into medium after tunicamycin treatment, despite increased IL-8 mRNA levels.
Aside from the SREBP and UPR, many as yet unknown pathways are likely to be mediating endothelial responses to oxPAPC. KEGG and GO enrichment analysis of the network was relatively uninformative, because only a limited number of genes on the array were present in these databases. Future studies using targeted disruption of hub genes, particularly those coding for transcription factors, may provide greater insight into underlying gene–gene regulatory relationships.
Materials and Methods
Reagents and Antibodies.
OxPAPC was prepared and analyzed by mass spectrometry to confirm the lipid profile described (29). All reagents were determined to have <2 pg/ml LPS as determined by a kit from MA Biowhittaker (Walkersville, MD; catalog no. 5064U). Antibodies used were from the following sources: ATF4 (catalog no. sc-200; Santa Cruz Biotechnology, Santa Cruz, CA), beta-actin antibody (catalog no. A2066; Sigma, St. Louis, MO), Histone H3 antibody (catalog no. 06-599; Upstate Cell Signaling, Charlottesville, VA). Western blots were developed by using ECL-Plus reagent (Amersham Pharmacia, Pittsburgh, PA). siRNA duplexes were from Qiagen (Valencia, CA), and PCR primers were from Invitrogen (Carlsbad, CA) (see Table 7, which is published as supporting information on the PNAS web site). A plasmid expressing human ATF4 was kindly provided by Pierre Fafournoux (from INRA, Theix, France).
Cell Culture and siRNA Knock-Down Experiments.
Human aortic endothelial cells were isolated from aortic samples retrieved at the time of organ harvest for cardiac transplantation and used at passages 4–7 (see Supporting Text, which is published as supporting information on the PNAS web site). For ELISA screening assays, cells were cultured in 96-well dishes, and for gene expression array analysis, cells were cultured in 35-mm dishes. For siRNA experiments, cells were transfected for 16 h with 5 nM siRNA in medium containing 10% FBS using HiPerFect reagent (Qiagen) according to the manufacturer’s protocol and treated 40 h after transfection. Cells were treated with PAPC or oxPAPC (10–50 μg/ml), tunicamycin (10 μg/ml), or DTT (1 mM) in medium containing 1% FBS. HeLa cells were transfected by using FuGene 6 (Roche, Basel, Switzerland) at a ratio of 1:3 of μg DNA to FuGene and harvested 24 or 48 h after transfection.
Nuclear Protein Extraction and Immunoblotting.
Nuclear protein extraction was performed by using standard procedures (see Supporting Text). Isolated protein (20 μg) was separated on 4–12% PAGE and transferred to PVDF membrane. After blocking using 5% dry milk, membranes were incubated in primary antibodies (1:1,000 dilution) followed by appropriate secondary antibodies for 1 h at room temperature. ECL-Plus (Amersham Pharmacia, Uppsala, Sweden) reagent was used to detect bound antibodies.
ELISA.
IL-8 levels in HAEC supernatants were measured by using an ELISA kit (Quantikine Immunoassay R & D Systems, Minneapolis, MN) as described (30).
Expression Array Analysis.
Twelve individual HAEC lines were treated in duplicate (in 35-mm dishes) for 4 h in media containing PAPC (40 μg/ml) or oxidized PAPC (40 μg/ml). Cytoplasmic RNA was isolated by using an RNAeasy kit (Qiagen) and analyzed on an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) to assess RNA integrity. Double-stranded cDNAs were synthesized from total RNA by using the cDNA Synthesis System (Invitrogen, Carlsbad, CA). Biotin-labeled cRNA was generated and used to probe Affymetrix (Santa Clara, CA) Human Genome U133A and B arrays according to the manufacturer’s recommendations (Affymetrix). The Microarray Suite 5.0 software (Affymetrix) was used to analyze image data and make the absolute call for each measurement. The array data were normalized with the median invariant method (31). To filter out differentially expressed genes, we adopted the standard t test in a pairwise comparison involving PAPC and oxPAPC. All genes in the network followed a normal distribution except one (238755_at) based on the Kolmogorov–Smirnov test (cutoff P value = 0.05). The normality test for each gene was based on 24 measurements (absolute gene expression values) from 12 PAPC cell lines and 12 oxPAPC cell lines. The 1,043 array probe sets that passed the selection criteria were used for the gene coexpression network analysis. The computed mean false discovery rate (FDR) was 0.25% (see Supporting Text). In the subsequent analyses, we also used the absolute value of the t test P value as a measure of gene significance. Thus, the gene significance measures the responsiveness to oxPAPC treatment.
Gene Coexpression Network Construction and Analysis.
A weighted gene coexpression network was constructed to identify groups of genes (modules) involved in various activated pathways following a previously described algorithm (14). Briefly, we first computed the Pearson correlation between each pair of 1,043 selected genes yielding a similarity (correlation) matrix (_s_ij). Then, a power function, _a_ij = Power(_s_ij, β) ≡ |_s_ij|β, was used to transform the similarity matrix into an adjacency matrix A = [_a_ij], where _a_ij is the strength of a connection between two nodes (genes) i and j in the network. The connectivity (k) was determined for all differentially expressed genes by taking the sum of their connection strengths with all other genes in the network. A characteristic of many biological networks is that they are approximately scale-free (32). The parameter β was chosen by using the scale-free topology criterion (14), such that the resulting network connectivity distribution approximated scale-free topology (Fig. 9, which is published as supporting information on the PNAS web site). A major advantage of weighted networks is that they are highly robust with respect to the choice of the parameter β. R software code, a tutorial, and a technical report for generating weighted gene coexpression networks are available on request. The adjacency matrix was then used to define a measure of node dissimilarity, based on the topological overlap matrix. To identify gene modules, we performed hierarchical clustering on the topological overlap matrix. The details of the algorithm are available on request. Fisher’s exact test was used to determine whether enrichment in pathway genes within the module is random (see Supporting Text).
Real-Time QPCR Analysis.
In a typical experiment each treatment was done in triplicate. RNA was isolated from cells by using RNeasy isolation kit (Qiagen). One microgram of total RNA was reverse transcribed by using random hexamers and Superscript-III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed by using an ABI Prism 7700 (Applied Biosystems, Foster City, CA) and SYBR Green detection (SYBR Green Taq ready mix; Sigma). Primers were designed by using PrimerQuest software (IDT Technologies) and verified by using BLAST search. Sequences of primers can be found in Table 7. The PCR consisted of a 2-min step at 94°C and 40 cycles of 94°C for 15 s, 60°C for 1 min, and 72°C for 1 min, and ending with a slow heating step from 55°C to 95°C to generate the melting curve data. The correct sizes of the PCR products were verified by agarose gel electrophoresis. Serial dilutions of the pooled samples were used to construct the standard curve and determine the real-time PCR efficiency for each primer pair by using the ABI Prism 7700 software. Each individual sample cDNA was analyzed separately and corrected for the β2M expression. The final data are expressed as an average relative expression of the treatment group versus the control group (set as 100%).
Supplementary Material
Supporting Information
Acknowledgments
This work has been supported by National Institutes of Health Grants HL30568 (to A.J.L. and J.A.B.), HL64731 (to J.A.B.), and HL72367 (to S.F.N.); an unrestricted research award from Bristol–Myers Squibb (to A.J.L.); an American Heart Association (AHA) Postdoctoral Fellowship award (to P.S.G.); an AHA Predoctoral Fellowship award (to N.M.G.); and the Laubisch Fund [University of California, Los Angeles (UCLA)]. Microarray work was performed in the UCLA DNA Microarray Facility and supported by the UCLA/National Heart, Lung, and Blood Institute Shared Microarray Resource.
Abbreviations
EC
endothelial cell
oxPAPC
oxidized 1-palmitoyl-2-arachidonyl-_sn_-3-glycero-phosphorylcholine
UPR
unfolded protein response
HAEC
human aortic EC
SREBP
sterol regulatory element-binding protein
Q-PCR
quantitative PCR.
Footnotes
Conflict of interest statement: W.-P.Y., A.H., A.T., and T.G.K. are employees of Bristol–Myers Squibb Pharmaceutical Research Institute.
References
- 1.Libby P. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 2.Lusis A. J., Mar R., Pajukanta P. Annu. Rev. Genomics Hum. Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 3.Berliner J. A., Watson A. D. N. Engl. J. Med. 2005;353:9–11. doi: 10.1056/NEJMp058118. [DOI] [PubMed] [Google Scholar]
- 4.Shi W., Haberland M. E., Jien M. L., Shih D. M., Lusis A. J. Circulation. 2000;102:75–81. doi: 10.1161/01.cir.102.1.75. [DOI] [PubMed] [Google Scholar]
- 5.Shi W., Wang N. J., Shih D. M., Sun V. Z., Wang X., Lusis A. J. Circ. Res. 2000;86:1078–1084. doi: 10.1161/01.res.86.10.1078. [DOI] [PubMed] [Google Scholar]
- 6.Boekholdt S. M., Peters R. J., Hack C. E., Day N. E., Luben R., Bingham S. A., Wareham N. J., Reitsma P. H., Khaw K. T. Arterioscler. Thromb. Vasc. Biol. 2004;24:1503–1508. doi: 10.1161/01.ATV.0000134294.54422.2e. [DOI] [PubMed] [Google Scholar]
- 7.Ridker P. M., Hennekens C. H., Buring J. E., Rifai N. N. Engl. J. Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 8.Boisvert W. A., Curtiss L. K., Terkeltaub R. A. Immuol. Res. 2000;21:129–137. doi: 10.1385/ir:21:2-3:129. [DOI] [PubMed] [Google Scholar]
- 9.Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N., Colinayo V., Ruff T. G., Milligan S. B., Lamb J. R., Cavet G., et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 10.Cheung V. G., Jen K. Y., Weber T., Morley M., Devlin J. L., Ewens K. G., Spielman R. S. Cold Spring Harbor Symp. Quant. Biol. 2003;68:403–407. doi: 10.1101/sqb.2003.68.403. [DOI] [PubMed] [Google Scholar]
- 11.Barabasi A. L., Oltvai Z. N. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 12.Yeh M., Cole A. L., Choi J., Liu Y., Tulchinsky D., Qiao J. H., Fishbein M. C., Dooley A. N., Hovnanian T., Mouilleseaux K., et al. Circ. Res. 2004;95:780–788. doi: 10.1161/01.RES.0000146030.53089.18. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa K., Navab M., Leitinger N., Fogelman A. M., Lusis A. J. J. Clin. Invest. 1997;100:1209–1216. doi: 10.1172/JCI119634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B., Horvath S. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. article 17. [DOI] [PubMed] [Google Scholar]
- 15.Han J. D., Bertin N., Hao T., Goldberg D. S., Berriz G. F., Zhang L. V., Dupuy D., Walhout A. J., Cusick M. E., Roth F. P., Vidal M. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 16.Engelking L. J., Liang G., Hammer R. E., Takaishi K., Kuriyama H., Evers B. M., Li W. P., Horton J. D., Goldstein J. L., Brown M. S. J. Clin. Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton J. D., Goldstein J. L., Brown M. S. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee A. H., Iwakoshi N. N., Glimcher L. H. Mol. Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K., Kaufman R. J. J. Biol. Chem. 2004;279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- 21.Ito D., Walker J. R., Thompson C. S., Moroz I., Lin W., Veselits M. L., Hakim A. M., Fienberg A. A., Thinakaran G. Mol. Cell. Biol. 2004;24:9456–9469. doi: 10.1128/MCB.24.21.9456-9469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Back S. H., Schroder M., Lee K., Zhang K., Kaufman R. J. Materials and Methods. 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y., Hendershot L. M. J. Biol. Chem. 2004;279:13792–13799. doi: 10.1074/jbc.M313724200. [DOI] [PubMed] [Google Scholar]
- 24.Mager J., Bartolomei M. S. Nat. Genet. 2005;37:1194–1200. doi: 10.1038/ng1664. [DOI] [PubMed] [Google Scholar]
- 25.Berrahmoune H., Lamont J., Fitzgerald P., Visvikis-Siest S. Clin. Chem. Lab. Med. 2005;43:671–684. doi: 10.1515/CCLM.2005.116. [DOI] [PubMed] [Google Scholar]
- 26.Austin R. C., Lentz S. R., Werstuck G. H. Cell Death Differ. 2004;11(Suppl. 1):S56–S64. doi: 10.1038/sj.cdd.4401451. [DOI] [PubMed] [Google Scholar]
- 27.Poddar R., Sivasubramanian N., DiBello P. M., Robinson K., Jacobsen D. W. Circulation. 2001;103:2717–2723. doi: 10.1161/01.cir.103.22.2717. [DOI] [PubMed] [Google Scholar]
- 28.Yeh M., Gharavi N. M., Choi J., Hsieh X., Reed E., Mouillesseaux K. P., Cole A. L., Reddy S. T., Berliner J. A. J. Biol. Chem. 2004;279:30175–30181. doi: 10.1074/jbc.M312198200. [DOI] [PubMed] [Google Scholar]
- 29.Watson A. D., Leitinger N., Navab M., Faull K. F., Horkko S., Witztum J. L., Palinski W., Schwenke D., Salomon R. G., Sha W., et al. J. Biol. Chem. 1997;272:13597–13607. doi: 10.1074/jbc.272.21.13597. [DOI] [PubMed] [Google Scholar]
- 30.Yeh M., Leitinger N., de Martin R., Onai N., Matsushima K., Vora D. K., Berliner J. A., Reddy S. T. Arterioscler. Thromb. Vasc. Biol. 2001;21:1585–1591. doi: 10.1161/hq1001.097027. [DOI] [PubMed] [Google Scholar]
- 31.Li C., Wong W. H. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravasz E., Somera A. L., Mongru D. A., Oltvai Z. N., Barabasi A. L. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information