Turnover of the Active Fraction of IRS1 Involves Raptor-mTOR- and S6K1-Dependent Serine Phosphorylation in Cell Culture Models of Tuberous Sclerosis (original) (raw)
Abstract
The TSC1-TSC2/Rheb/Raptor-mTOR/S6K1 cell growth cassette has recently been shown to regulate cell autonomous insulin and insulin-like growth factor I (IGF-I) sensitivity by transducing a negative feedback signal that targets insulin receptor substrates 1 and 2 (IRS1 and -2). Using two cell culture models of the familial hamartoma syndrome, tuberous sclerosis, we show here that Raptor-mTOR and S6K1 are required for phosphorylation of IRS1 at a subset of serine residues frequently associated with insulin resistance, including S307, S312, S527, S616, and S636 (of human IRS1). Using loss- and gain-of-function S6K1 constructs, we demonstrate a requirement for the catalytic activity of S6K1 in both direct and indirect regulation of IRS1 serine phosphorylation. S6K1 phosphorylates IRS1 in vitro on multiple residues showing strong preference for RXRXXS/T over S/T,P sites. IRS1 is preferentially depleted from the high-speed pellet fraction in TSC1/2-deficient mouse embryo fibroblasts or in HEK293/293T cells overexpressing Rheb. These studies suggest that, through serine phosphorylation, Raptor-mTOR and S6K1 cell autonomously promote the depletion of IRS1 from specific intracellular pools in pathological states of insulin and IGF-I resistance and thus potentially in lesions associated with tuberous sclerosis.
The insulin and insulin-like growth factor I (IGF-I) axes coordinate systemic nutrient utilization to meet the metabolic demands of higher organisms and, in so doing, perform essential roles in tissue growth and homeostasis and in the maintenance of euglycemia. In humans, systemic, non-cell-autonomous insulin resistance is a significant clinical feature of several pathologies, including obesity, dyslipidemia, inflammation, and infection, and is an ominous etiological prelude to overt type 2 diabetes. As such, the goal of diabetes research has been to therapeutically modulate insulin secretion and sensitivity as a means to correct the biochemical deficiencies that underlie aberrant glycemic control. In addition, the insulin/IGF-I axis serves a critical cell autonomous role to support cell and tissue growth, which perhaps has been most dramatically revealed in genetic studies of Drosophila melanogaster. Several insulin/IGF-I effectors, including phosphatase and tensin homolog (PTEN), class 1A phosphoinositide 3-kinase (PI3K), Ras, Akt, tuberous sclerosis gene products 1 and 2 (TSC1 and TSC2), Ras homology enriched in brain (Rheb), and the Peutz-Jehgers gene product LKB1, are tumor suppressors or proto-oncoproteins found mutated in human cancer or predispose to tumor development. Therefore, it is not surprising that dysregulation of this pathway has considerable oncogenic potential, underscoring the need for its meticulous regulation.
Genetic and biochemical approaches have independently confirmed novel roles for the Raptor-mTOR/S6K cassette in the regulation of cell autonomous insulin/IGF-I sensitivity by inhibiting requisite IRS1/2 function (for reviews, see references 8, 14, 28, and 37). This is remarkably overt in cell-based and animal models of the tumor predisposition syndrome, tuberous sclerosis, wherein loss-of-function of the TSC1-TSC2 complex gives rise to pathological hyperactivation of mTOR and S6K (7, 11, 19, 20, 36, 45, 47) and produces cell-autonomous insulin resistance (13, 20, 32, 39). In mouse embryo fibroblasts (MEFs) derived from TSC1- or TSC2-deficient animals, IRS1 and IRS2 are transcriptionally repressed (13, 39) through a mechanism requiring S6Ks (13). Cells deficient in either TSC1 or TSC2 display an accelerated rate of IRS1/2 degradation, which is rapamycin sensitive (39). Further regulation is achieved at the level of IRS1 phosphorylation as evidenced, in TSC2-null cells, by reduced electrophoretic mobility and S6K-regulated phosphorylation of IRS1 at S307 (13). Thus, TSC deficiency produces a robust state of insulin resistance by coordinating multiple negative regulatory inputs that converge on IRS1/2. Accordingly, little insulin or IGF-I-inducible PI3K activation is detected in TSC1−/− and TSC2−/− MEFs (13, 39), potentially explaining why Akt activation is repressed in tumors from the Eker rat model of tuberous sclerosis (20). It has been suggested that this may underlie the low frequency with which TSC lesions progress to malignancy (13) and may account for the hypersusceptibility of TSC1/2-deficient cells to apoptotic stimuli (13, 17, 39).
In the present study, we find that multiple components of the cell growth axis interact functionally with IRS1, affecting the phosphorylation of a subset of serine residues found frequently associated with insulin resistance. We corroborate the earlier finding that S6K1 directly phosphorylates IRS1 at S307 (13) and provide new evidence that S6K1 may phosphorylate IRS1 at S527 and potentially at additional residues in vivo. Despite the inability of S6K1 to phosphorylate IRS1 at the proline-directed sites—S312, S616, and S636/S639—in vitro, surprisingly, phosphorylation of these sites in vivo requires S6K1 activity. We present evidence that these phosphorylations promote the rapamycin-sensitive depletion of IRS1 from the high-speed pellet (HSP) fraction, providing a molecular explanation for the cell-autonomous insulin resistance observed in models of tuberous sclerosis.
MATERIALS AND METHODS
Antibodies and reagents.
The following antibodies were purchased from Cell Signaling Technology: pIRS1-[S616], pIRS1-[S636/S639], pRPS6-[S240/S244], and Akt substrate pSub-[RxRxxS/T]. The pIRS1-[S312], pS6K1-[T389], and p[Y] (clone 4G10) antibodies were purchased from Upstate Biotechnology. pIRS1-[S527] was purchased from Rockland Immunochemicals. IRS1 antibody was purchased from Santa Cruz Biotechnology. pIRS1-[S307] was generously provided by Jongsoon Lee (Harvard Medical School). Anti-Flag (clone M2) and anti-α-tubulin antibodies were purchased from Sigma. Anti-Myc (clone 9E10) and anti-HA (clone 12CA5) antibodies were prepared from ascites. Rapamycin and insulin were purchased from Sigma. _clasto_-lactacystin-β-lactone was purchased from Boston Biochem.
Plasmids and construction.
pcDNA3-IRS1 was kindly provided by Peter van der Geer (University of California, San Diego). Flag-IRS1 was generated by insertion of the 5′-Flag epitope by site-directed mutagenesis. Individual IRS1 point mutations and deletions were generated similarly using pcDNA3-Flag-IRS1 as the template. pRK5-Myc-mTOR(D2357E) was kindly provided by David Sabatini (Whitehead Institute, Massachusetts Institute of Technology). pRK7-HA-S6K1, pRK7-HA-S6K1(ED3E), pRK7-HA-S6K1(K100Q), pcDNA3-HA-S6K2, and pcDNA3-HA-S6K2(K112R) were generously supplied by John Blenis (Harvard Medical School). pRK5-Myc-Rheb was a gift from Paul Worley (Johns Hopkins University). pRK5-Myc-Rheb(I39K) and pRK5-Myc-Rheb(Q64L) were generated by site-directed mutagenesis and have been described elsewhere (39).
Cell culture and gene expression.
TSC1−/−/p53+/+, TSC2−/−/p53−/−, and p53−/− MEFs were graciously provided by David Kwiatkowski (Harvard Medical School) and were propagated in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) fetal calf serum and ciprofloxacin. The TSC2−/− (R2) and TSC2−/− (R4) cell lines have been described elsewhere (39). HEK293 and HEK293T cells were acquired from the American Type Culture Collection and were maintained and passaged in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (vol/vol) and ciprofloxacin. HEK293 and HEK293T cells were transiently transfected by lipofection using Effectene transfection reagent (QIAGEN) according to the manufacturer's protocol. Cells were processed and analyzed at 48 to 72 h posttransfection.
Cell lysis and immunoprecipitation.
Cells were harvested by scraping in standard lysis buffer (40 mM HEPES [pH 7.5], 120 mM NaCl, 1% NP-40 [vol/vol] 1 mM EDTA, 10 mM pyrophosphate, 10 mM β-glycerophosphate, 50 mM NaF, 1 mM dithiothreitol [DTT], 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, and 1 μg of aprotinin/ml). Lysates were rotated end-over-end at 4°C for 20 min and clarified by centrifugation at 15,000 × g for 15 min. Protein concentrations were determined spectrophotometrically using a DC protein assay kit (Bio-Rad). Lysates were either mixed with sample buffer and heated to 100°C for 5 min or subjected to immunoprecipitation.
For immunoprecipitation, precleared lysates normalized for cell protein were incubated with the appropriate antibody and mixed end-over-end for 2 h or overnight at 4°C. Protein A-agarose or protein G-agarose beads were added for an additional hour, and immune complexes were isolated by centrifugation. Immunoprecipitates were washed three times with lysis buffer and heated for 5 min to 100°C in sample buffer or subjected to in vitro phosphorylation.
Subcellular fractionation.
An HSP comprised of crude cell membranes and cytoskeletal components was separated from cytosol by differential centrifugation of postnuclear hypotonic lysates. Briefly, cells were rinsed in ice-cold phosphate-buffered saline and scraped in hypotonic lysis buffer (10 mM HEPES pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 100 μM EGTA, 20 mM NaF, 200 μM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, and 1 μg of aprotinin/ml) and then extracted by Dounce homogenization with 100 strokes using a type-B pestle. Nuclei and unbroken cells were cleared from the extract by centrifugation at 3,000 × g for 5 min. The postnuclear supernatant was fractionated into the HSP and cytosol by ultracentrifugation at 175,000 × g for 1.25 h. When necessary, the cytosolic fraction was concentrated by using a Centricon-5 (5-kDa molecular mass cutoff) centrifugal filter (Millipore). The HSP was solubilized in standard lysis buffer on a vortex shaker for 20 min. Insoluble material was then removed from the HSP fraction by centrifugation at 15,000 × g for 15 min. Protein concentrations were determined spectrophotometrically using a DC protein assay kit (Bio-Rad). Lysates were mixed with sample buffer and heated to 100°C for 5 min.
In vitro phosphorylation of IRS1 by S6K1.
To prepare the substrate, Flag epitope-tagged wild-type or mutant IRS1 was expressed in HEK293T cells and immunoprecipitated overnight from precleared lysates using anti-Flag (M2) antibody. The immune complexes were washed three times with lysis buffer and three times with phosphatase reaction buffer (50 mM Tris-HCl [pH 7.5], 100 μM disodium EDTA, 5 mM DTT, 0.01% [vol/vol] Brij 35). Dephosphorylation was carried out overnight at 25°C in phosphatase reaction buffer using recombinant λ protein phosphatase (400 U/reaction). Immune complexes were then washed three times with lysis buffer and three times with kinase assay buffer (20 mM morpholinepropanesulfonic acid [pH 7.2], 25 mM β-glycerol phosphate, 5 mM EGTA, 5 mM Na3VO4, 5 mM DTT).
HA-S6K1 was expressed in HEK293T cells, and the cells were either left untreated or cultured in the presence of 100 nM rapamycin for 1 h. Cell lysates were prepared, and HA-S6K1 was immunoprecipitated from precleared lysates overnight at 4°C with anti-HA (12CA5) antibody. Immune complexes were washed three times with lysis buffer and three times with kinase assay buffer. Anti-Flag and anti-HA immunoprecipitates were then combined and assayed for in vitro phosphorylation. Kinase assays were performed for 30 min at 30°C in the presence of 18.75 mM MgCl2, 125 μM unlabeled ATP, and 10 μCi of [γ-32P]ATP (3,000 Ci/mmol). Alternatively, S6K1 was assayed in anti-HA immunoprecipitates using a synthetic RXRXXS/T substrate peptide (AKRRRLSSLRA; 50 μM). The incorporation of 32P into the peptide was measured by liquid scintillation counting as previously described (38).
Phosphopeptide mapping and phosphoamino acid analysis.
Phosphopeptide mapping and phosphoamino analysis was carried out essentially as detailed elsewhere (2). Briefly, Flag-IRS1 phosphorylated in vitro by S6K1 was separated from other contaminating proteins by immunoprecipitation with anti-Flag antibody, followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The resulting gel was dried and subjected to autoradiography. Radiolabeled Flag-IRS1 was then excised and twice serially extracted from the gel in elution buffer (50 mM NH4HCO3 [pH 7.5], 1% [vol/vol] β-mercaptoethanol, and 0.2% [vol/vol] SDS) overnight. Flag-IRS1 was precipitated from the eluate with trichloroacetic acid in the presence of 20 μg of RNase A as the carrier protein for 1 h on ice. Precipitates were washed with 100% ice-cold ethanol and then allowed to air dry. Methionine and cysteine residues were oxidized in performic acid for 1 h on ice. This preparation was subjected to two rounds of digestion with TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin, and the residual NH4HCO3 was removed by four successive rounds of lyophilization. The phosphopeptides were dissolved in pH 1.9 buffer (2.2% [vol/vol] formic acid and 7.8% glacial acetic acid [pH 1.9]) and spotted onto cellulose thin-layer chromatography plates. The peptides were then separated in the electrophoretic dimension by high-voltage electrophoresis (1,000 V) for 45 min in pH 1.9 buffer. The plates were dried for 1 h, and peptides were separated in the chromatographic dimension in phosphochromotography buffer (37.5% _n_-butanol [vol/vol], 25% pyridine [vol/vol], 7.5% glacial acetic acid) overnight. Phosphopeptide maps were visualized by autoradiography.
For phosphoamino acid analysis, approximately 300 cpm of labeled and oxidized IRS1 was lyophilized, dissolved in 6 N HCl, and heated to 110°C for 1 h. The resulting phosphoamino acids, together with exogenously added phosphoserine, phosphothreonine, and phosphotyrosine standards, were separated by two-dimensional high-voltage electrophoresis. Phosphoamino acids were visualized by ninhydrin staining and autoradiography.
RESULTS
S6K1 acts genetically downstream of Rheb and Raptor-mTOR to induce IRS1 serine phosphorylation.
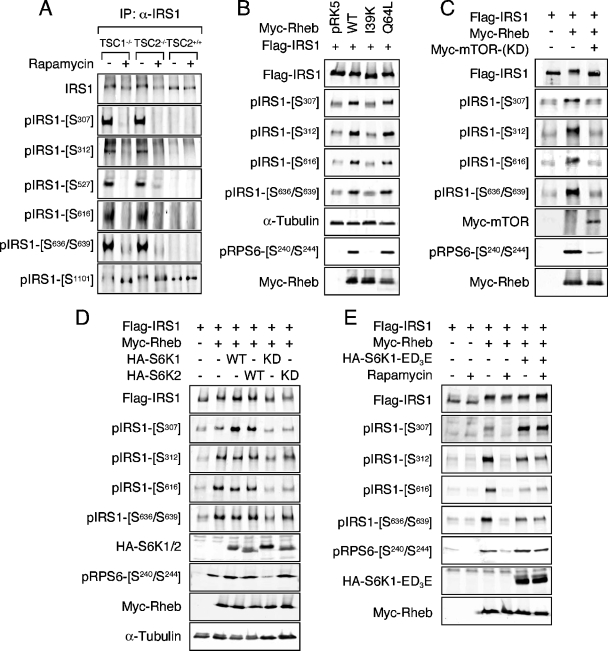
Because of the retarded migration of IRS proteins from TSC2-deficient MEFs during SDS-polyacrylamide gel electrophoresis (13), we reasoned that, owing to the size of IRS proteins (∼180 kDa), phosphorylation at multiple residues is likely to account for the shift in mobility. Due to transcriptional repression and enhanced protein turnover, the endogenous level of IRS1 in these cells is greatly diminished (13, 39). Therefore, in order to obtain a sufficient amount of IRS1 to assay for phosphorylation, IRS1 was immunoprecipitated prior to analysis. Using an antibody panel directed against multiple serine phosphorylation sites known to be linked to various states of insulin resistance and highly conserved between IRS1 and IRS2, the phosphorylation of a number of residues (S307, S312, S527, S616, and S636/S639) was found to be significantly induced in TSC-deficient cells compared to wild-type controls (Fig. 1A). Moreover, the phosphorylation of each of these sites was inhibited in cells exposed to rapamycin for 1 h, indicating acute regulation by Raptor-mTOR downstream of the TSC1-TSC2 complex. The phosphorylated residues assayed fall into two general substrate contexts: RXRXXS/T (S307, S527, and S1101), often regarded as an Akt phosphorylation consensus, and S/T,P (S312, S616, and S636), a motif preferred by proline-directed kinases (boldface residues are phosphorylated). Multiple RXRXXS/T sites are present in IRS1, not all of which exhibited elevated phosphorylation, however, in TSC1/2-null cells (e.g., S1101 [Fig. 2A ], a site reportedly phosphorylated by PKCθ [24]).
FIG. 1.
The TSC/Rheb/Raptor-mTOR/S6K regulates IRS1 serine phosphorylation. (A) TSC1-null, TSC2-null, or wild-type (+/+) MEFs were cultured in serum-free medium overnight, followed by the addition of 100 nM rapamycin for 1 h. IRS1 was immunoprecipitated and immunoblotted with the indicated antibodies. (B) HEK293 cells were cotransfected with Flag-IRS1 and either empty vector (pRK5) or individual Myc-Rheb constructs as indicated. Lysates were prepared from serum-starved cells, and immunoblot analysis of exogenous and endogenous proteins was carried out. Under these conditions, endogenous IRS1 phosphorylation is insignificant. (C) A dominant-interfering allele of Myc-mTOR (KD) was included in transfections of HEK293 with Flag-IRS1 and Myc-Rheb. (D) Flag-IRS1 was coexpressed with Myc-Rheb in HEK293 cells in the presence or absence of wild-type or dominant-interfering alleles of HA-S6K1 or HA-S6K2. Cells were deprived of serum overnight prior to harvest. (E) Flag-IRS1 was coexpressed with Myc-Rheb and a rapamycin-resistant allele of HA-S6K1 (ED3E). Serum-starved cells were treated or not treated with 100 nM rapamycin for 1 h prior to lysis.
FIG. 2.
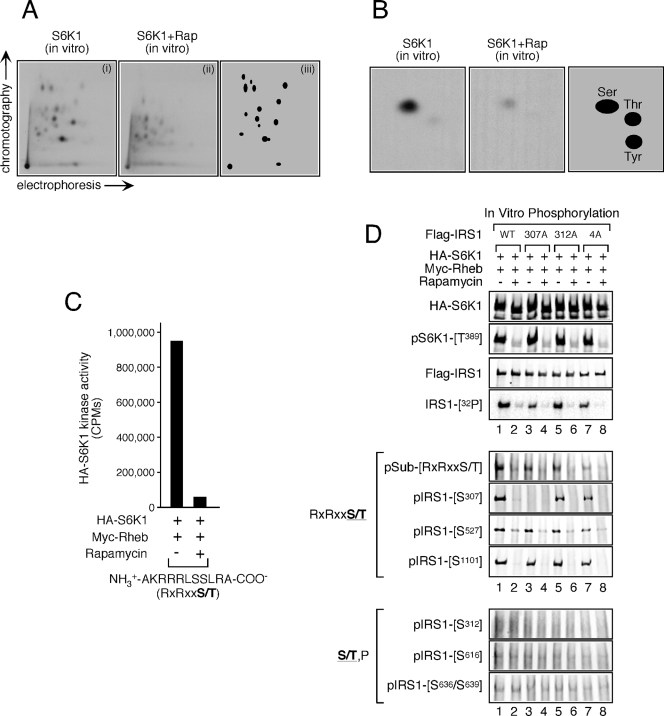
In vitro phosphorylation of IRS1 by S6K1. (A) Wild-type Flag-IRS1 was expressed in HEK293T cells, purified, and dephosphorylated as indicated in Materials and Methods. HA-S6K1 was separately coexpressed with Myc-Rheb in HEK293T cells, which were treated or not for 1 h with 100 nM rapamycin. HA-S6K1 was subsequently purified and combined with dephosphorylated Flag-IRS1. Phosphopeptide mapping of the in vitro phosphorylation product was then carried out. Panels show S6K1 phosphorylation of IRS1 before (i) and after (ii) rapamycin treatment. Panel iii schematically represents the distribution of rapamycin-sensitive phosphopeptides. (B) Phosphoamino acid analysis of in vitro S6K1-phosphorylated Flag-IRS1 before (left panel) and after (middle panel) rapamycin treatment. The right panel shows schematically the distribution of rapamycin-sensitive phosphoamino acids. (C) HA-S6K1 was prepared as described in panel A and used to phosphorylate an RXRXXS/T peptide substrate. The relative kinase activity toward this peptide is presented. (D) Wild-type, S307A, S312A, and 4A (S616A/S636A/S666A/S736A) Flag-IRS1 constructs were expressed in HEK293T cells, purified, and dephosphorylated as in panel A. These substrates were mixed with HA-S6K1 expressed in and purified from HEK293T cells coexpressing Myc-Rheb before and after treatment with 100 nM rapamycin for 1 h. In vitro phosphorylation was then carried out, and 32P incorporation was monitored by autoradiography. Aliquots from the same reaction were subjected to immunoblot analysis to determine expression level and site-specific phosphorylation.
Natural mutations in the TSC2 GAP domain cause disease and disrupt the in vitro Rheb GAP activity of TSC2 (9, 42, 48), suggesting that GTP loading of Rheb is an important aspect of the molecular pathogenesis of tumor formation in tuberous sclerosis. Since the cellular Rheb GAP activity provided by TSC2 is limiting, when overexpressed, Rheb accumulates in the active form (16, 23, 25). In HEK293 cells, ectopic expression of wild-type Rheb or an activated variant that is heavily GTP-charged (Q64L [23]) induced phosphorylation of the RxRxxS/T site S307 and the S/T,P sites S312, S616, and S636/S639 (Fig. 1B), representing the same subset of IRS1 phosphorylation sites detected in TSC1/2-null MEFs (Fig. 2A). In contrast, an inactive Rheb mutant in which the switch I domain has been disabled (I39K [25]) failed to stimulate the phosphorylation of any of these sites (Fig. 1B). Phosphorylation of IRS1 at S1101 was not affected in HEK293 cells expressing Rheb (data not shown), similar to observations of TSC1/2-deficient MEFs (Fig. 1A). The basal phosphorylation of S527 in ectopic IRS1 was substantially elevated when overexpressed, confounding our attempts to determine whether Rheb genuinely induced phosphorylation of this site. IRS1 serine phosphorylation closely correlated with endogenous S6 phosphorylation, indicating that the activation of endogenous S6K1 was also induced by Rheb (Fig. 1B).
To establish that Raptor-mTOR is necessary for Rheb-induced IRS1 phosphorylation at this collection of residues, two criteria were satisfied: a sensitivity to rapamycin (Fig. 1E) and genetic interference by dominant-negative, kinase-deficient mTOR (Fig. 1C). Once again, endogenous S6 phosphorylation tightly followed IRS1 serine phosphorylation under these conditions (Fig. 1C and E).
In TSC1/2-null cells and in cells ectopically expressing Rheb, S6K1 is highly active and yet remains sensitive to rapamycin (11, 19, 21, 30, 36, 47). In fact, by virtue of its TOS motif and C-terminal tail, S6K1 is a direct substrate of Raptor-mTOR, which phosphorylates T389 within the S6K1 hydrophobic motif (1). The tight correlation between endogenous S6 phosphorylation and IRS1 phosphorylation in cells expressing Rheb suggested that S6K1 might signal to IRS1 distal to the TSC1-TSC2 complex, Rheb, and Raptor-mTOR. We therefore coexpressed IRS1 and Rheb with S6K1 or S6K2 and sought to determine whether expression of wild-type S6K could potentiate IRS1 phosphorylation and, conversely, whether kinase-dead S6K could inhibit IRS1 phosphorylation at the subset of Rheb-inducible sites. Whereas the expression of Rheb enhanced IRS1 serine phosphorylation of all sites assayed, the addition of wild-type S6K1 or S6K2 only potentiated the phosphorylation of S307 (Fig. 1D). This finding is consistent with S6K1 directly phosphorylating IRS1 at S307 (reference 13 and see below). Rheb induced maximal phosphorylation of endogenous S6, as ectopic S6K1 or S6K2 did not further augment this effect. Surprisingly, coexpression of kinase-dead S6K alleles, particularly S6K1, inhibited not only the phosphorylation of S307 but of other sites as well, including S312, S616, and S636/S639, indicating that S6K1 function is necessary for IRS1 phosphorylation at the entire Rheb-regulated subset of sites. These data are in accord with those of Um et al. (46), who demonstrated that siRNA-mediated knockdown of S6K1 reduces phosphorylation of human IRS1 at S312 and S636/S639 in insulin-treated HeLa cells (46). We further validated the involvement of S6K1 in IRS1 phosphorylation utilizing a rapamycin-resistant, gain-of-function S6K1 mutant (ED3E). Whereas treatment of cells with rapamycin inhibited IRS1 serine phosphorylation at all Rheb-regulated residues, coexpression of S6K1-ED3E, completely rescued phosphorylation of S307 and partially rescued S312, S616, and S636/S639 after rapamycin treatment (Fig. 1E).
S6K1 phosphorylates IRS1 in vitro.
The observation that S6K1 modulates the phosphorylation status of several residues raises the possibility that S6K1 may directly phosphorylate IRS1 at these sites. IRS1 was an excellent substrate for S6K1 since tryptic phosphopeptide maps of in vitro phosphorylated IRS1 revealed at least 15 phosphopeptides, which could correspond to as many as 15 sites of phosphorylation (Fig. 2A). Furthermore, phosphoamino acid analysis indicated that serine phosphorylation accounted for the bulk of phosphate incorporation (Fig. 2B). To begin to ascertain which residues were phosphorylated by S6K1 in vitro, wild-type IRS1 and a panel of IRS1 constructs bearing alanine substitutions at serine phosphorylation sites detected in vivo [S307A, S312A, and S616A/S636A/S666A/S736A (4A)] were assayed for 32P incorporation and site-specific phosphorylation. Generally, 32P incorporation was similar between wild-type and S→A IRS1 mutants (lane 1 versus lanes 3, 5, and 7), although a slight reduction was noted for the S307A mutant (lane 1 versus lane 3), suggesting that S307 represents a major site of phosphorylation (Fig. 2D). S6K1 showed a strong preference for the phosphorylation of sites in an RXRXXS/T over S/T,P context since S307, S527, and S1101 were readily phosphorylated in a rapamycin-sensitive manner (Fig. 2D), as was an RXRXXS/T peptide substrate (Fig. 2C). Furthermore, an antibody raised against a generic RXRXXS/T phosphorylation consensus recognized IRS1 when phosphorylated by S6K1 (Fig. 2D). On the other hand, no phosphorylation of S/T,P sites was detected in this assay, suggesting that S312, S616, and S636/S639 are not likely to be phosphorylated by S6K1 in vivo. Given that phosphorylation of S1101 was not upregulated in TSC1/2-deficient MEFs (Fig. 1A), the in vitro phosphorylation of this residue likely represents nonphysiological phosphorylation. Although some of the other phosphopeptides in the map might also be due to phosphorylation of nonphysiological sites, it seems likely that S6K1 can phosphorylate IRS1 at residues in addition to S307 and S527 in vivo.
IRS1 depletion from the HSP fraction is regulated through serine phosphorylation.
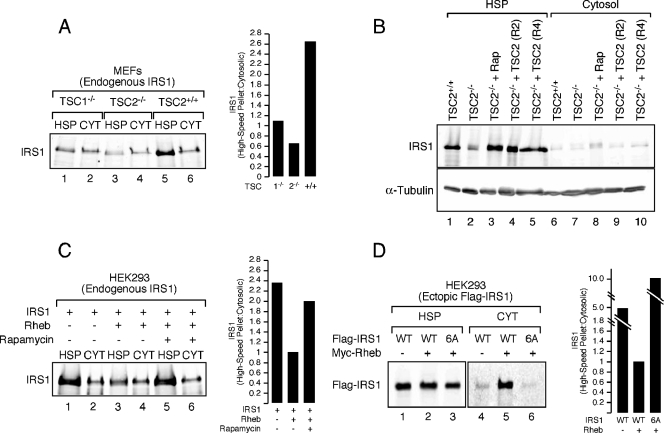
Most IRS1 in the cell appears to exist in two subcellular fractions: the cytosol and the HSP (also called the low-density membrane fraction). Because of the appearance in the HSP of tyrosine-phosphorylated IRS1 and IRS1-associated p85 and PI3K activity during acute and sustained insulin stimulation, the HSP likely represents the active signaling fraction of IRS1 (4, 5, 18, 41). We previously observed increased IRS1/2 degradation rates in TSC1/2-deficient cells (39) and therefore sought to determine whether Raptor-mTOR and S6K1 could promote IRS1 depletion from a particular intracellular fraction in models of tuberous sclerosis. IRS1 was surveyed in HSP and cytosolic fractions of postnuclear, hypotonic lysates prepared from TSC1−/−, TSC2−/−, and wild-type MEFs. In wild-type cells at steady-state, most cellular IRS1 was retained in the HSP fraction, with <30% present in cytosol, comparing these fractions normalized for total protein (Fig. 3A). As previously shown, total IRS1 was reduced in TSC1/2-deficient cells (13, 39), but, critically, the proportion of total IRS1 in the HSP was substantially decreased. Treatment of TSC2−/− MEFs with rapamycin for 24 h restored IRS1 in the HSP to wild-type levels (Fig. 3B, lane 3 versus lane 1), as did retroviral vector reconstitution of TSC2-null cells with wild-type TSC2 (Fig. 3B, lanes 4 and 5 versus lane 1). Similar to the effects of TSC deficiency, we noted that ectopic expression of Rheb in HEK293 cells reduced the abundance of endogenous IRS1 (39) (Fig. 3C). Despite this, the ratio of HSP to cytosolic IRS1 recapitulated that observed in TSC-deficient cells (see Fig. 3A) and was reversed by a 24-h rapamycin treatment (Fig. 3C), indicating that the Raptor-mTOR complex regulates the abundance of IRS1 in the HSP and thus the active fraction of IRS1.
FIG. 3.
Depletion of IRS1 from the HSP fraction in TSC1/2-null or Rheb-overexpressing cells. (A) TSC1−/−, TSC2−/−, and wild-type MEFs were deprived of serum for 4 h and then lysed under hypotonic conditions and fractionated into HSP and cytosolic (CYT) components. Equivalent amounts of cell protein were loaded, and the ratio of HSP to cytosolic IRS1 was calculated (as shown in a histogram). (B) IRS1 was detected in the HSP fraction from TSC2+/+ MEFs, TSC2−/− MEFs, TSC2−/− MEFs treated with 100 nM rapamycin for 24 h, and two clonal lines of TSC2−/− MEFs reconstituted with retroviral TSC2 (R2 and R4). (C) HEK293 cells were transfected with empty vector or Myc-Rheb and serum deprived overnight in the presence or absence of 100 nM rapamycin. Quantitation of the HSP/cytosolic ratio of endogenous IRS1 was calculated and is presented in the accompanying histogram. (D) HEK293 cells were cotransfected with either empty vector or Myc-Rheb and wild-type or 6A (S307A/S312A/S616A/S636A/S666A/S736A) Flag-IRS1. Equivalent amounts of protein were loaded for each panel. However, the abundance of detectable Flag-IRS1 is lower in the cytosolic fraction, presumably because it is not as tightly regulated when overexpressed. The relative ratios of HSP to cytosolic exogenous Flag-IRS1 were then calculated and are presented in the histogram to the right.
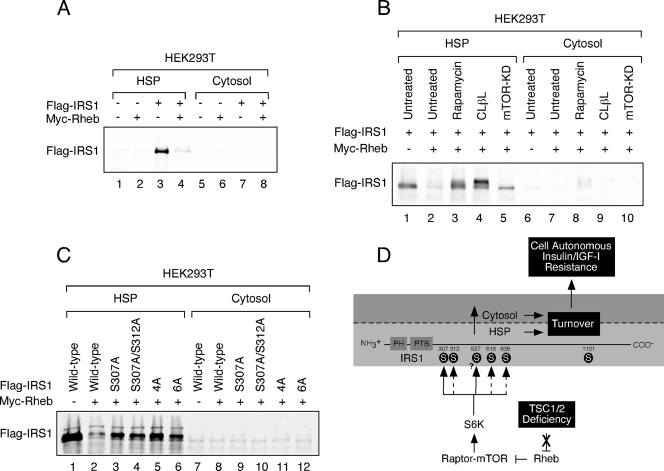
To determine whether the Raptor-mTOR- and S6K1-regulated phosphorylation sites identified in the present study are required for retention of IRS1 in the HSP, we collectively mutated these residues to alanine [S307A/S312A/S616A/S636A/S666A/S736A (6A)] and monitored the subcellular distribution of IRS1. Although a portion of exogenous IRS1 partitioned to the cytosolic fraction when coexpressed with Rheb, the 6A mutant failed to do so (Fig. 3D), suggesting that site-specific serine phosphorylation of IRS1 regulates subcellular distribution in this cell culture model. We have consistently failed to observe, in contrast to findings with endogenous IRS1, a decrease in the total abundance of ectopic IRS1 when coexpressed with Rheb in HEK293 cells. Presumably, this is due to the limiting ability of the endogenous destruction machinery to turn over an expanded pool of IRS1 in HEK293 cells. In contrast, exogenous Flag-IRS1 was specifically depleted from the HSP when coexpressed with Rheb in HEK293T cells (Fig. 4A), an effect that was reversed by rapamycin or expression of kinase-dead mTOR (Fig. 4B). Although the proteasome inhibitor, _clasto_-lactacystin-β-lactone, stabilized Flag-IRS1 in drug-treated cells, we were surprised to find that this occurred in the HSP (Fig. 4B), suggesting that IRS1 turnover may take place in different cellular compartments depending on cell type. Analysis of a set of Flag-IRS1 S→A mutants coexpressed with Rheb in HEK293T cells highlights the important roles played by individual phosphorylations (Fig. 4C). S307A and S307A/S312A mutants both partially resisted depletion from the HSP in Rheb-expressing cells, as did the quadruple alanine mutant (4A), where all four PI3K-flanking SP sites were substituted. Combining the S307A/S312A and 4A mutations (6A) did not further enhance IRS1 retention in the HSP.
FIG. 4.
IRS1 depletion from the HSP in HEK293T cells. (A) HEK293T cells expressing Flag-IRS1 and/or Myc-Rheb were serum deprived for 4 h and then fractionated and immunoblotted with anti-Flag antibody. (B) HEK293T cells transfected with Flag-IRS1 were cotransfected with empty vector or Myc-Rheb and Myc-mTOR (KD) as indicated. Cells were either left untreated or cultured in the presence 100 nM rapamycin or 5 μM _clasto_-lactacystin-β-lactone (CLβL) for 48 h. (C) Myc-Rheb was coexpressed with a panel of Flag-IRS1 mutants (defined in the legend of Fig. 2D and 3D) as indicated. The HSP fraction was prepared and immunoblotted with anti-Flag antibodies. The expressions of these mutants are similar at steady state (data not shown). (D) Proposed model for IRS1 regulation in the context of TSC deficiency. When TSC1 or TSC2 is inactivated, signaling through the Rheb/Raptor-mTOR/S6K axis is constitutive. S6K directly phosphorylates IRS1 on the RXRXXS/T site, S307 and potentially S527 (solid arrows) but not S1101. Raptor-mTOR or other kinases phosphorylate IRS1 on the S/T,P sites, S312, S616, and S636/9, which is somehow indirectly regulated by S6K1 (hatched arrows. The phosphorylation of these sites collectively contributes to the depletion of IRS1 from the HSP fraction. This either involves phosphorylation-mediated redistribution of IRS1 from the HSP to the cytosol, where it is subsequently degraded, or degradation directly in the HSP. The mode of IRS1 turnover is likely to be cell type dependent. IRS1 turnover produces cell-autonomous insensitivity to insulin or IGF-I.
DISCUSSION
Regulation of IRS1 serine phosphorylation by S6Ks.
The TSC1-TSC2 complex performs an integral function in insulin and IGF-I signal transduction and therefore presumably in retrograde regulation as well. Functionally, TSC deficiency or ectopic Rheb expression resembles a state of hyperinsulinemia or chronic insulin action insofar as constitutive feedback regulation is enforced. Indeed, several studies have found that insulin-induced serine phosphorylation of IRS1/2 is highly sensitive to rapamycin (3, 10, 12, 43), particularly during the period where S6Ks are most active (∼30 min poststimulation). S6K1 is an essential effector for retrograde regulation of IRS1, as demonstrated by the finding that siRNA-directed S6K1 knockdown precludes insulin-induced phosphorylation of S307, S312, and S636/S639 (13, 46). Our data further indicate that the kinase activity of S6K1 is required for both direct and indirect regulation of IRS1 through serine phosphorylation.
We have confirmed the conclusion that IRS1 S307 is direct S6K target site in cell culture models of tuberous sclerosis (13) and expanded this original observation to show that S6K1 can phosphorylate several additional residues in IRS1, with S527 being a second potential in vivo site in this model. The phosphorylation of S527 by S6K1 may be linked and, perhaps unique to, model systems in which the Raptor-mTOR/S6K axis is pathologically active. Under such conditions, S6Ks may “promiscuously” phosphorylate substrates that are not otherwise recognized when their activity is held in check. In support of this hypothesis, rapamycin fails to inhibit insulin-induced S527 phosphorylation in C2C12 myotubes (data not shown) and yet reproducibly does so in TSC1/2-deficient MEFs (Fig. 1A). In HEK293 cells, we have consistently observed elevated basal phosphorylation of all IRS1 sites evaluated in the present study when IRS1 is ectopically expressed. This is especially true for S527, for which we were unable to observe strong induction with coexpressed Rheb, and thus our further in vivo characterization of this site was limited.
Our finding that the expression of wild-type, but not kinase-deficient, S6K1 not only induced phosphorylation of S307, a direct RxRxxS/T target site, but also the phosphorylation of S312, S616, and S636/S639, which are SP sites, is unexpected and suggests that S6Ks facilitate phosphorylation of IRS1 through the activation of a S312/616/636 proline-directed kinase or inhibition of a phosphatase that dephosphorylates these sites. Alternatively, phosphorylation of IRS1 by S6Ks could induce an IRS1 conformation permissive for phosphorylation by proline-directed kinases, such as mitogen-activated protein kinases (12, 22). Tzatsos and Kandror (44) have recently shown that Raptor-mTOR can phosphorylate IRS1 S636/S639 in vitro and further demonstrated that Raptor siRNA inhibits basal and Rheb-induced phosphorylation of this site. It was also shown by this group that kinase-dead S6K1 suppressed the serum-induced phosphorylation of this site and S307, suggesting that S6K1 is indeed required for this phosphorylation (44). We have further substantiated a role for S6K1 in controlling S307, S312, S616, and S636/S639 by showing that a highly active, rapamycin-resistant, gain-of-function S6K1 allele (S6K1-ED3E) not only induced the phosphorylation of these residues but did so in the presence of rapamycin, a condition where Raptor-mTOR is inhibited. These data argue that despite the ability of Raptor-mTOR to phosphorylate S636/S639 in vitro, other kinases whose phosphorylation of IRS1 at S312, S616, and S636/S639 is facilitated by S6K1 are predominant in vivo. The data presented here reveal that S6Ks play a greater role in IRS1 serine phosphorylation than previously thought.
Posttranslational regulation of IRS1 in cell-based models of tuberous sclerosis.
Several lines of evidence suggest that the HSP fraction harbors the population of IRS1 that actively participates in signal transduction. Tyrosine phosphorylation of IRS1 and IRS2 is detected within 1 min after insulin stimulation in this compartment, and most acute (<10 min poststimulation with insulin) IRS1-associated PI3K activity is detected here (18). As early as 10 min after insulin stimulation, total IRS1 begins to disappear from the HSP and accumulates in the cytosolic fraction (5, 18, 41). It is notable that IRS1 is not detected in purified plasma membrane preparations (4, 5, 18), indicating that the HSP fraction of IRS1 that is engaged by the active insulin or IGF-I receptor must be juxtaposed, but not intrinsic to, the plasma membrane. During insulin stimulation, IRS1 appears in the cytosol complexed with the p85 regulatory subunit of PI3K (41), although this may represent a sequestration complex comprised of tyrosine phosphorylated IRS1 and monomeric p85 (26). This complex between IRS1 and monomeric p85 form cytosolic foci detectable by immunofluorescence approximately 10 min after IGF-I stimulation and are devoid of the p110 catalytic subunit of PI3K, indicating that the complex is nonfunctional (26). Pretreatment of 3T3-L1 adipocytes with the proteasome inhibitor, lactacystin, preserves IRS1 levels in the cytosolic fraction rather than the HSP (41). Therefore, the likely site of IRS1 turnover, at least in this cell culture system, is within the cytosol. Inhibition of PI3K and Raptor-mTOR, respectively, with wortmannin and rapamycin protects against the depletion of IRS1 from the HSP, and to a lesser extent, from the cytosol (5, 41). In the present study, IRS1 was significantly depleted from the HSP in both TSC1- and TSC2-null MEFs and in cells that overexpress Rheb, whereas the cytosolic fraction of IRS1, albeit reduced, was essentially unaltered. This likely reflects the chronic nature of constitutive absence of TSC genes or constitutive presence of Rheb, both of which simulate chronic insulin action as far as Raptor-mTOR activation is concerned.
It is clear from previous work in 3T3-L1 adipocytes that the during the course of insulin-induced depletion of IRS1, the cytosolic pool of IRS1 is detected only transiently (within 4 h [5, 41]) prior to the destruction of all cellular IRS1. Because the Raptor-mTOR axis is constitutively active in cell culture models of tuberous sclerosis, this cycle of expression of IRS1 in the HSP, followed by destruction, is also constitutive, resulting in the net depletion of IRS1 from the HSP, as well as total cellular IRS1. The data presented in Fig. 4B indicate that IRS1 depletion may occur directly from the HSP in some cell types, since pretreatment of HEK293T cells with the proteasome inhibitor, _clasto_-lactacystin-β-lactone, resulted in accumulation of IRS1 in the HSP rather than the cytosol. In contrast, wild-type IRS1 was detected in the cytosolic compartment in HEK293 cells expressing Rheb, suggesting that ectopic IRS1 turnover in this cell culture model likely occurs in the cytosol (Fig. 3D). Cumulatively, the data presented here and in our earlier study (39) support a model of IRS regulation wherein TSC deficiency links HSP depletion of IRS1 to turnover (Fig. 4D). We found that residual IRS1 was heavily serine phosphorylated in TSC1/2-deficient MEFs or in cells that expressed Rheb ectopically. In these cell models, a subset of serine phosphorylations was required for IRS1 depletion from the HSP fraction and, furthermore, IRS1/2 proteins displayed an accelerated rate of destruction. Critically, each of these processes, i.e., IRS1 serine phosphorylation, HSP depletion, and protein degradation, were all exquisitely sensitive to rapamycin.
Implications for cell-autonomous insulin resistance in tumorigenesis.
We and others (13, 39) have found the retrograde insulin signal to be aberrantly transduced in models of tuberous sclerosis, not conventionally considered an insulin-resistant disorder. The data in the present study significantly expand these observations. We describe here new sites of IRS1 serine phosphorylation not previously linked to tuberous sclerosis. We further demonstrate that IRS1 serine phosphorylation is functionally linked to HSP depletion, collectively providing a molecular basis for the cell-autonomous insulin resistance intrinsic to cells with TSC deficiency. The cell autonomous insulin resistance that accompanies TSC deficiency is likely to produce a unique tumor biology. Although such tumors retain dysplastic and hypertrophic characteristics, survival deficiencies may also exist. In fact, similar to MEFs from TSC1- and TSC2-deficient animals, tumors from the Eker rat model of tuberous sclerosis show little PI3K/Akt activation and low malignant potential (20). Two genetic studies of PTEN and TSC2 single knockout and compound heterozygous knockout mice have revealed that PTEN haploinsufficiency leads to Akt activation and a synergistic activation of the Raptor-mTOR/S6K axis in the TSC2 heterozygous background (27, 29), which one group found to correlate with a more aggressive cancer phenotype (29). It was argued, therefore, that the limited activation of Akt observed in TSC2 deficiency may be causally linked to reduced progression to malignancy. The Peutz-Jehgers tumor predisposing syndrome shares striking cell biological similarities with tuberous sclerosis, namely, similar tumor histology, inappropriately active Raptor-mTOR signaling, reduced Akt activation, and sensitivity to cellular stresses (6, 40). We have found that IRS1 function is repressed in MEFs null for the Peutz-Jehgers tumor suppressor product, LKB1 (data not shown), indicating that mTOR-mediated feedback regulation may be relevant in this tumor syndrome as well. Furthermore, in cell-based models of the PTEN-associated hamartoma syndromes, IRS proteins are greatly depleted and are heavily phosphorylated when ectopically expressed (33, 34). Indeed, loss of PTEN correlates positively with IRS1 depletion in human prostate-derived, metastatic tumors (15).
Pharmacologically, inhibition of the Raptor-mTOR/S6K axis with rapamycin has been shown to induce Akt activation in a subset of human cancer cells (31, 35), although this response does not appear to be universally conserved (35). Although this does provide a rationale for combined therapeutic inhibition of Raptor-mTOR/S6K and IGF-IR pathways, it could be imagined that rapamycin alone may prove as efficacious as the combination in many settings, since prolonged rapamycin treatment inhibits resynthesized mTOR and thereby limits the assembly and function of the Rictor-mTOR complex (35). A greater understanding of the oncogenic settings that give rise to robust feedback inhibition through the Raptor-mTOR/S6K cassette and identification of the oncogenic phenotypes that rely on this feedback should facilitate the rationale design of new cancer therapeutics that exploit this phenomenon.
Acknowledgments
We thank David Kwiatkowski for providing wild-type and TSC1- and TSC2-null MEFs; Peter van der Geer (University of California, San Diego), John Blenis (Harvard Medical School), Paul Worley (Johns Hopkins University), and David Sabatini (Whitehead Institute) for cDNAs; Jongsoon Lee (Harvard Medical School) for the pIRS1-[S307] antibody used in this study; and Jill Meisenhelder for help with peptide mapping.
O.J.S. was supported by National Institutes of Health Grant T32-CA09523, by an NRS Award (GM67407), and a Pioneer Fellowship. This study was supported by USPHS grants CA14195 and CA82683 to T.H. from the National Cancer Institute. T.H. is a Frank and Else Schilling American Cancer Research Professor.
REFERENCES
- 1.Ali, S. M., and D. M. Sabatini. 2005. Structure of S6K1 determines if Raptor-mTOR or Rictor-mTOR phosphorylates its hydrophobic motif site. J. Biol. Chem. 280**:**19445-19448. [DOI] [PubMed] [Google Scholar]
- 2.Boyle, W. J., and H. T. P. van der Geer. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201**:**110-149. [DOI] [PubMed] [Google Scholar]
- 3.Carlson, C. J., M. F. White, and C. M. Rondinone. 2004. Mammalian target of rapamycin regulates IRS-1 serine 307 phosphorylation. Biochem. Biophys. Res. Commun. 316**:**533-539. [DOI] [PubMed] [Google Scholar]
- 4.Clark, S. F., S. Martin, A. J. Carozzi, M. M. Hill, and D. E. James. 1998. Intracellular localization of phosphoinositide 3-kinase and insulin receptor substrate-1 in adipocytes: potential involvement of a membrane skeleton. J. Cell Biol. 140**:**1211-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, S. F., J.-C. Molero, and D. E. James. 2000. Release of insulin receptor substrate proteins from an intracellular complex coincides with the development of insulin resistance. J. Biol. Chem. 275**:**3819-3826. [DOI] [PubMed] [Google Scholar]
- 6.Corradetti, M. N., K. Inoki, N. Bardessy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18**:**1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el-Hashemite, N., H. Zhang, E. P. Henske, and D. J. Kwiatkowski. 2003. Mutation in TSC2 and activation of mammalian target of rapamycin signaling pathway in renal angiomyolipoma. Lancet 361**:**1348-1349. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, T. L., and M. F. White. 2004. Signaling pathways: the benefits of good communication. Curr. Biol. 14**:**R1005-R1007. [DOI] [PubMed] [Google Scholar]
- 9.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11**:**1457-1466. [DOI] [PubMed] [Google Scholar]
- 10.Giraud, J., R. Leshan, Y.-H. Lee, and M. F. White. 2004. Nutrient-dependent and insulin-stimulated phosphorylation of insulin receptor substrate-1 on serine 302 correlates with increased insulin signaling. J. Biol. Chem. 279**:**3447-3454. [DOI] [PubMed] [Google Scholar]
- 11.Goncharova, E. A., D. A. Goncharova, A. Eszterhas, D. S. Hunter, M. K. Glassberg, R. S. Yeung, C. L. Walker, D. Noonan, D. J. Kwiatkowski, M. M. Chou, R. A. J. Panettieri, and V. P. Krymskaya. 2002. Tuberin regulates p70 S6 kinase activation and ribosomal protein s6 phosphorylation. J. Biol. Chem. 277**:**30958-30967. [DOI] [PubMed] [Google Scholar]
- 12.Gual, P., T. Gremeaux, T. Gonzalez, Y. Le Marchand-Brustel, and J. F. Tanti. 2003. MAP kinases and mTOR mediate insulin-induced phosphorylation of insulin receptor substrate-1 on serine residues 307, 612 and 632. Diabetologia 46**:**1532-1542. [DOI] [PubMed] [Google Scholar]
- 13.Harrington, L. S., G. M. Findlay, A. Gray, T. Tolkacheva, S. Wigfield, H. Rebholz, J. Barnett, N. R. Leslie, S. Cheng, P. R. Shepherd, I. Gout, C. P. Downes, and R. F. Lamb. 2004. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166**:**213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington, L. S., G. M. Findlay, and R. F. Lamb. 2005. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem. Sci. 30**:**35-42. [DOI] [PubMed] [Google Scholar]
- 15.Hellawell, G. O., G. D. Turner, D. R. Davies, R. Poulsom, S. F. Brewster, and V. M. Macaulay. 2002. Expression of the type 1 insulin-like growth factor receptor is up-regulated in primary prostate cancer and commonly persists in metastatic disease. Cancer Res. 62**:**2942-2950. [PubMed] [Google Scholar]
- 16.Im, E., F. C. von Lintig, J. Chen, S. Zhuang, W. Qui, S. Chowdhury, P. F. Worley, G. R. Boss, and R. B. Pilz. 2002. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene 21**:**6356-6365. [DOI] [PubMed] [Google Scholar]
- 17.Inoki, K., T. Zhu, and K. L. Guan. 2003. Tsc2 mediates cellular energy response to control cell growth and survival. Cell 115**:**577-590. [DOI] [PubMed] [Google Scholar]
- 18.Inoue, G., B. Cheatham, R. Emkey, and C. R. Kahn. 1998. Dynamics of insulin signaling in 3T3-L1 adipocytes. Differential compartmentalization and trafficking of insulin receptor substrate (IRS)-1 and IRS-2. J. Biol. Chem. 273**:**11548-11555. [DOI] [PubMed] [Google Scholar]
- 19.Jaeschke, A., J. Hartkamp, M. Saitoh, W. Roworth, T. Nobukuni, A. Hodges, J. Sampson, G. Thomas, and R. Lamb. 2002. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 159**:**217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenerson, H. L., L. D. Aicher, L. D. True, and R. S. Yeung. 2002. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 62**:**5645-5650. [PubMed] [Google Scholar]
- 21.Kwiatkowski, D. J., H. Zhang, J. L. Bandura, K. M. Heiberger, M. Glogauer, N. el-Hashemite, and H. Onda. 2002. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11**:**525-534. [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. H., J. Giraud, R. J. Davis, and M. F. White. 2003. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J. Biol. Chem. 278**:**2896-2902. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., K. Inoki, and K. L. Guan. 2004. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24**:**7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y., T. J. Soos, X. Li, J. Wu, M. DeGennaro, X. Sun, D. R. Littman, M. J. Birnbaum, and R. D. Polakiewicz. 2004. Protein Kinase C θ inhibits insulin signaling by phosphorylating IRS1 at Ser1101. J. Biol. Chem. 279**:**45304-45307. [DOI] [PubMed] [Google Scholar]
- 25.Long, X., Y. Lin, S. Ortiz-Vega, K. Yonezawa, and J. Avruch. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15**:**702-713. [DOI] [PubMed] [Google Scholar]
- 26.Luo, J., S. J. Field, J. Y. Lee, J. A. Engelman, and L. C. Cantley. 2005. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. J. Cell Biol. 170**:**455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, L., J. Teruya-Feldstein, N. Behrendt, Z. Chen, T. Noda, O. Hino, C. Cordon-Cardo, and P. P. Pandolfi. 2005. Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression. Cell 19**:**1779-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning, B. D. 2004. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167**:**399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning, B. D., M. N. Logsdon, A. I. Lipovsky, D. Abbott, D. J. Kwiatkowski, and L. C. Cantley. 2005. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 19**:**1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onda, H., P. B. Crino, H. Zhang, R. D. Murphey, L. Rastelli, B. E. Gould-Rothberg, and D. J. Kwiatkowski. 2002. Tsc2 null murine neuroepithelial cells are a model for human tuber giant cells, and show activation of an mTOR pathway. Mol. Cell Neurosci. 21**:**561-574. [DOI] [PubMed] [Google Scholar]
- 31.O'Reilly, K. E., F. Rojo, Q. B. She, D. Solit, G. B. Mills, D. Smith, H. Lane, F. Hofmann, D. J. Hicklin, D. L. Ludwig, J. Baselga, and N. Rosen. 2006. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66**:**1500-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radimerski, T., J. Montagne, M. Hemmings-Mieszczak, and G. Thomas. 2002. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 16**:**2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiss, K., J. Y. Wang, G. Romano, F. B. Furnari, W. K. Cavenee, A. Morrione, X. Tu, and R. Baserga. 2000. IGF-I receptor signaling in a prostatic cancer cell line with a PTEN mutation. Oncogene 19**:**2687-2694. [DOI] [PubMed] [Google Scholar]
- 34.Reiss, K., J. Y. Wang, G. Romano, X. Tu, F. Peruzzi, and R. Baserga. 2001. Mechanisms of regulation of cell adhesion and motility by insulin receptor substrate-1 in prostate cancer cells. Oncogene 20**:**490-500. [DOI] [PubMed] [Google Scholar]
- 35.Sarbassov, D. D., S. M. Ali, S. Sengupta, J. H. Sheen, P. P. Hsu, A. F. Bagley, A. L. Markhard, and D. M. Sabatini. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22**:**159-168. [DOI] [PubMed] [Google Scholar]
- 36.Shah, O. J., and T. Hunter. 2004. Critical role of T-loop and H-motif phosphorylation in the regulation of S6 kinase 1 by the tuberous sclerosis complex. J. Biol. Chem. 279**:**20816-20823. [DOI] [PubMed] [Google Scholar]
- 37.Shah, O. J., and T. Hunter. 2005. Tuberous sclerosis and insulin resistance: unlikely bedfellows reveal a TORrid affair. Cell Cycle 4**:**122-127. [DOI] [PubMed] [Google Scholar]
- 38.Shah, O. J., S. R. Kimball, and L. S. Jefferson. 2000. Among translational effectors, p70S6k is uniquely sensitive to inhibition by glucocorticoids. Biochem. J. 347**:**389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah, O. J., Z. Wang, and T. Hunter. 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14**:**1650-1656. [DOI] [PubMed] [Google Scholar]
- 40.Shaw, R. J., N. Bardessy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6**:**1-9. [DOI] [PubMed] [Google Scholar]
- 41.Takano, A., I. Usui, T. Haruta, J. Kawahara, T. Uno, M. Iwata, and M. Kobayashi. 2001. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol. Cell. Biol. 21**:**5050-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13**:**1259-1268. [DOI] [PubMed] [Google Scholar]
- 43.Tremblay, F., A. M. Gagnon, A. Veilleux, A. Sorisky, and A. Marette. 2005. Activation of the mammalian target of rapamycin pathway acutely inhibits insulin signaling to Akt and glucose transport in 3T3-L1 and human adipocytes. Endocrinology 146**:**1328-1337. [DOI] [PubMed] [Google Scholar]
- 44.Tzatsos, A., and K. V. Kandror. 2006. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via Raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol. Cell. Biol. 26**:**63-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlmann, E. J., W. Li, D. K. Scheidenhelm, C.-L. Gau, F. Tamanoi, and D. H. Gutmann. 2004. Loss of tuberous sclerosis complex 1 (Tsc1) expression results in increased Rheb/S6K pathway signaling important for astrocyte cell size regulation. Glia 47**:**180-188. [DOI] [PubMed] [Google Scholar]
- 46.Um, S. H., F. Frigerio, M. Watanabe, F. Picard, M. Jaoquin, M. Sticker, S. Fumagalli, P. R. Allegrini, S. C. Kozma, J. Auwerx, and G. Thomas. 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431**:**200-205. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, H., G. Cicchetti, H. Onda, H. B. Koon, K. Asrican, N. Bajraszewski, F. Vazquez, C. L. Carpenter, and D. J. Kwiatkowski. 2003. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Investig. 112**:**1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5**:**578-581. [DOI] [PubMed] [Google Scholar]