A Fungal Family of Transcriptional Regulators: the Zinc Cluster Proteins (original) (raw)
Abstract
The trace element zinc is required for proper functioning of a large number of proteins, including various enzymes. However, most zinc-containing proteins are transcription factors capable of binding DNA and are named zinc finger proteins. They form one of the largest families of transcriptional regulators and are categorized into various classes according to zinc-binding motifs. This review focuses on one class of zinc finger proteins called zinc cluster (or binuclear) proteins. Members of this family are exclusively fungal and possess the well-conserved motif CysX2CysX6CysX5-12CysX2CysX6-8Cys. The cysteine residues bind to two zinc atoms, which coordinate folding of the domain involved in DNA recognition. The first- and best-studied zinc cluster protein is Gal4p, a transcriptional activator of genes involved in the catabolism of galactose in the budding yeast Saccharomyces cerevisiae. Since the discovery of Gal4p, many other zinc cluster proteins have been characterized; they function in a wide range of processes, including primary and secondary metabolism and meiosis. Other roles include regulation of genes involved in the stress response as well as pleiotropic drug resistance, as demonstrated in budding yeast and in human fungal pathogens. With the number of characterized zinc cluster proteins growing rapidly, it is becoming more and more apparent that they are important regulators of fungal physiology.
INTRODUCTION
The trace element zinc is required for proper function of a large number of proteins, including various enzymes. However, most zinc-containing proteins are transcription factors capable of binding DNA and are named zinc finger proteins. They are categorized into various families according to zinc-binding motifs. For example, the Cys2His2 family comprises hundreds of zinc finger proteins that are found in eukaryotes ranging from yeast to humans. In contrast, members of the zinc cluster protein family (or binuclear cluster) are exclusively fungal and possess the well-conserved motif CysX2CysX6CysX5-12CysX2CysX6-8Cys. The cysteine residues bind to two zinc atoms, which coordinate folding of the domain involved in DNA binding.
The family of zinc cluster proteins is best characterized for the budding yeast, Saccharomyces cerevisiae. The genome of this organism encodes over 50 known (or putative) zinc cluster proteins. The first- and best-studied zinc cluster protein is Gal4p, a transcriptional activator of genes involved in the catabolism of galactose. Zinc cluster proteins are also found in a variety of other fungal organisms, such as Kluyveromyces lactis, the fission yeast Schizosaccharomyces pombe, and the human pathogens Candida albicans and Aspergillus nidulans. This review is aimed at describing the structural and functional domains of zinc cluster proteins and summarizing their roles in fungal physiology as well as their modes of action in S. cerevisiae and other fungi.
ZINC FINGER PROTEINS: AN OVERVIEW
Zinc-binding proteins form one of the largest families of transcriptional regulators in eukaryotes, displaying variable secondary structures and enormous functional diversity. They are grouped together because they all harbor at least one common motif, the zinc finger. This motif was first identified in the Xenopus transcription factor TFIIIA 20 years ago (179), and the resolution of its three-dimensional solution structure a few years later revealed its protruding “finger-like” shape (145). The finger actually consists of one α helix and a pair of antiparallel β strands (287). In general, one or more zinc atoms are bound by cysteine or histidine residues. This stabilizes the domain and contributes to proper protein structure and function (135, 287). The majority of zinc finger proteins bind to DNA (and also to RNA in the case of TFIIIA), thereby playing important roles in transcriptional and translational processes (135). However, it should be noted that this superfamily of proteins is not solely restricted to binding nucleic acids. Newly identified zinc finger proteins are also involved in many other physiological roles, including mediating protein-protein interactions, chromatin remodeling, protein chaperoning, lipid binding, and zinc sensing (135). Of the DNA (or RNA)-binding variety, three major classes of zinc finger proteins have been established to date in eukaryotes, based on their unique and highly conserved consensus amino acid sequences. They are summarized in Table 1. Although they can be grouped together as zinc-binding transcription factors, each class has distinct structural properties.
TABLE 1.
Three major classes of eukaryotic zinc finger proteins
| Zinc finger class | Subclass(es) | Consensus amino acid sequence | Example |
|---|---|---|---|
| I (C2H2) | FOG (C2HC) | Cys-X2-4-Cys-X12-His-X3-5-His | Xenopus TFIIIA |
| II (C4) | GATA, nuclear receptors, LIM (C3H) | Cys-X2-Cys-X_n_-Cys-X2-Cys-X_n_-Cys-X2-Cys-X_n_-Cys-X2-Cys | Glucocorticoid receptor |
| III (C6) | Cys-X2-Cys-X6-Cys-X5-12-Cys-X2-Cys-X6-8-Cys | S. cerevisiae Gal4p |
Classes of Zinc Finger Proteins
Class I encompasses the Cys2His2 (C2H2) proteins and is often referred to as the classical zinc finger (reviewed in reference 287). It is one of the most common types of transcription factors found in eukaryotes, and these proteins contain two or more repeating zinc finger units. A well-known example in humans is the transcription factor Sp1 (152, 186). FOG proteins (_f_riend _o_f _G_ATA) are a subclass within this group because they contain standard zinc fingers (C2H2) along with a C2HC consensus sequence (265). Each repeating unit consists of a conserved amino acid sequence that interacts with one zinc atom. Moreover, members of this class binds to nucleic acids as monomers (135, 155).
Class II represents the Cys4 (C4) zinc fingers, which include the GATA, LIM, and nuclear receptor proteins. GATA transcription factors (GATA-1 to -6) bind to a DNA sequence called a GATA motif [(A/T)GATA(A/G)] in the regulatory regions of their target genes through two zinc finger domains (270). The mammalian glucocorticoid receptor represents an excellent example within this class (39); its structure has provided much information on the DNA-binding capabilities of this group. Unlike the first class, these proteins usually contain one zinc finger unit binding to DNA as homodimers or heterodimers (consisting of two C4 proteins). Usually, homodimers recognize inverted repeats within the target nucleic acid sequence, whereas heterodimers bind to direct repeats (135).
Class III (C6) zinc finger proteins contain a DNA-binding domain (DBD) that consists of six cysteine residues bound to two zinc atoms, and hence these have the names zinc cluster, zinc binuclear cluster, or Zn(II)2Cys6 (Zn2C6) proteins. This class of transcription factors is unique in that these proteins contain only one zinc finger unit that binds two zinc atoms. They may interact with DNA as monomers, homodimers, or heterodimers (156, 233, 260, 267). Furthermore, they are strictly fungal proteins. The Saccharomyces cerevisiae transcription factor Gal4p is arguably the most well-known and well-studied zinc cluster protein. Its classification as a zinc “cluster” protein and the resolving of its X-ray crystal structure over a decade ago (168, 199) became the driving force behind studies which further characterized it and other members within this fungal superfamily of transcription factors.
ZINC CLUSTER PROTEINS
As stated above, the zinc binuclear cluster proteins (hereafter referred to as zinc cluster proteins) have been identified exclusively in fungi, although the other classes of zinc fingers are also present in this kingdom. For example, Msn2p, Msn4p, and Adr1p are all yeast transcription factors that contain a class I C2H2 motif (79). Zinc cluster proteins seem to belong predominantly to the ascomycete family, as only one (Lentinus edodes, PRIB protein) has been characterized in the basidiomycete family to date (61). Evolutionarily speaking, one hypothesis suggests that this unique Zn(II)2Cys6 motif appeared prior to the divergence of these two major fungal groups (260). Importantly, a multitude of recently identified zinc cluster proteins in Aspergillus, Candida, and Saccharomyces species, as well as Schizosaccharomyces pombe, are being studied (see Tables 3 to 5). The list of known zinc cluster proteins is growing rapidly, and the sequencing of other fungal genomes will allow for the identification of more transcription factors within this superfamily.
TABLE 3.
Classification of zinc cluster proteins in S. cerevisiaea
| Role | Gene name | Localization (reference[s])b | Function (reference[s]) |
|---|---|---|---|
| Sugar metabolism | GAL4 | C, N (32) | Activates genes involved in galactose metabolism (GAL1, GAL10) (24, 168) |
| RGT1 | C, N (235) | Activator/repressor of hexose transport genes (198) | |
| MAL13 | U | Part of MAL1 complex locus (35, 189) | |
| MAL33 | U | Activator of maltose genes in maltose metabolism, forms part of MAL3 complex locus (35, 189) | |
| MAL63 | U | Activator of maltose genes (35, 189) | |
| Amino acid, vitamin, and uracil metabolism | ARO80 | N | Activator of aromatic acid catabolic genes (108) |
| LEU3 | N (126) | Activator/repressor of leucine biosynthesis genes (76, 127, 297) | |
| LYS14 | C, N | Activator of lysine metabolic enzymes (68, 69) | |
| PUT3 | C, N | Induction of proline utilization genes (11, 242) | |
| THI2 (PHO6) | U | Activator of thiamine biosynthetic genes (194) | |
| ARG81 | C, N | Activator/repressor of arginine metabolism enzymes (210) | |
| CHA4 | C, N | Induction and basal expression of serine and threonine utilization, activates CHA1 (102) | |
| PPR1 | N (75) | Activates URA1 and URA3 (157) | |
| Miscellaneous | SEF1 | U | Compensates for the essential function of RPM2 in cell growth (87) |
| TEA1 | C, N | Activates transcription of Ty1 retrotransposon (86) | |
| STB4 | N | Interacts with Sin3p in yeast two-hybrid system (118) | |
| Chromatin remodeling | RSC3 | N | Essential component of the RSC chromatin-remodeling complex (8) |
| RSC30 | N | Subunit of the RSC chromatin-remodeling complex (8) | |
| Meiosis and mitosis | UME6 | N | Represses early meiotic genes (7, 202, 248) |
| CEP3 | M | Essential kinetochore component, chromosome segregation (143, 250) | |
| Nitrogen utilization | UGA3 | N | Activates GABA genes (277) |
| DAL81 | N | Activator of nitrogen catabolic genes, including allantoin and GABA genes (25, 40, 110, 277) | |
| PDR/stress response | PDR1 | N (52, 90) | Activator of PDR genes (15) |
| PDR3 | N (167) | Activator of PDR genes (53) | |
| PDR8 | C, N | Involved in PDR (100) | |
| YRM1 | C, N | Activator of PDR genes (158) | |
| YRR1 | C, N | Activator of PDR genes (46) | |
| HAL9 | C, N | Involved in salt tolerance (178) | |
| STB5 | C, N | Interacts with Sin3p, is an activator of PDR genes, and is involved in oxidative stress resistance (4, 118, 138) | |
| RDR1 | U | Repressor of PDR genes (97) | |
| RDS1 | U | Regulator of drug sensitivity (4) | |
| RDS2 | U | Regulator of drug sensitivity (4) | |
| WAR1 | N (131) | Activator of PDR12 in response to weak acid stress (131) | |
| ASG1 (YIL130W) | N | _A_ctivator of _s_tress response _g_enes (C. Wai and B. Turcotte, unpublished data) | |
| Peroxisome proliferation | OAF1 | C, N (116) | Activates genes involved in peroxisome proliferation (223) |
| PIP2 | C, N (116) | Activates genes involved in peroxisome proliferation (222) | |
| Ergosterol biosynthesis or uptake | UPC2 | C, N | Anaerobic sterol uptake, activator of ergosterol biosynthetic genes (44) |
| ECM22 | C, N | Activator of ergosterol biosynthetic genes (160, 274) | |
| SUT1 (YGL162W) | N (190) | Overexpression increases sterol uptake (190) | |
| SUT2 | U | Overexpression increases sterol uptake (190), multicopy suppressor of low activity of the cyclic AMP/proteinase kinase A pathway (226) | |
| Gluconeogenesis and | CAT8 | C, N | Activates genes needed for gluconeogenesis (96) |
| respiration | SIP4 | U | Snf1 kinase-dependent activator of gluconeogenesis genes (276) |
| HAP1 | U | Activates respiration genes (43, 205) | |
| Unknown | EDS1 (YBR033W) | U | Expression is dependent on Rpb2p (S. Vidan and M. Snyder, unpublished data) |
| TBS1 (YBR150C) | C, N | Δybr150c is sensitive to thiabendazole (62) | |
| YBR239C | C, N | Interacts with Rds2p in yeast two-hybrid system (75, 95, 109) | |
| YDR520C | C, N | Δ_ydr520c_ is slightly sensitive to caffeine (5) | |
| YER184C | U | ||
| YFL052W | U | Δ_yfl052w_ is hypersensitive to heat shock at 37°C (5) | |
| YJL103C | U | May be involved in oxidative phosphorylation (55) | |
| YJL206C | U | ||
| YKL222C | U | Δ_ykl222c_ is sensitive to caffeine (5) | |
| YKR064W | C, N | ||
| YLL054C | C | ||
| YLR278C | N | Δ_ylr278c_ is sensitive to caffeine (5) | |
| YNR063W | U |
TABLE 5.
Characterized zinc cluster proteins in other species
| Gene | Species | Function |
|---|---|---|
| ACEII | Trichoderma reesei | Activator involved in regulation of cellulase and xylanase genes (9) |
| AFLR | Aspergillus parasiticus, Aspergillus nidulans | Involved in regulation of the aflatoxin pathway (57, 58, 71, 289) |
| ALCR | Aspergillus nidulans | Activator of genes required for ethanol oxidation (133, 193, 200, 238) |
| AMYR | Aspergillus nidulans, Aspergillus oryzae, Aspergillus niger | Activator involved in amylolytic gene expression (83, 258) |
| ARCA | Aspergillus nidulans | Involved in the arginine catabolic pathway (60) |
| CLTA1 | Colletotrichum lindemuthianum | Involved in the switch between biotrophy and necrotrophy during infection (56) |
| CMR1 | Colletotrichum lagenarium | Involved in melanin biosynthesis (266) |
| CRG1 | Cercospora nicotianae | Involved in cellular resistance to cercosporin (37) |
| CTF1 α, CTF1 β | Nectria hematococca | Activator of cutinase genes (150, 151) |
| FACB | Aspergillus nidulans, Neurospora crassa | Activator of acetate regulatory genes (20, 261, 262) |
| FL | Neurospora crassa | Required for conidiophore morphogenesis (13, 14, 216) |
| LAC9 | Kluyveromyces lactis | Controls induction of the lactose-galactose regulon (147, 286, 290) |
| MLCR | Penicillium citrinum | Involved in ML-236B (compactin) biosynthesis (1) |
| MOC3 | Schizosaccharomyces pombe | Involved in sexual development, ascus formation, and stress response (80) |
| NIRA | Aspergillus nidulans | Regulator of nitrate assimilation (27, 188, 209, 247) |
| NIT4 | Neurospora crassa | Activator of the nitrate assimilatory pathway (70) |
| PDR1 | Candida glabrata | Activator of PDR genes (264) |
| PIG1 | Magnaporthe grisea | Involved in melanin biosynthesis (266) |
| PRNA | Aspergillus nidulans | Activator of genes involved in proline utilization (30, 82) |
| PRO1+ | Sordaria macrospora | Required for fruiting body development (175) |
| QUTA | Aspergillus nidulans | Regulates expression of genes involved in quinic acid utilization (149) |
| QUTH | Aspergillus nidulans | Possibly involved in the regulation of genes required for utilization of protocatechuic acid (136) |
| TAMA | Aspergillus nidulans | Involved in nitrogen regulation (49) |
| THI1 | Schizosaccharomyces pombe | Activator of several thiamine-repressible genes (65, 257) |
| UAY | Aspergillus nidulans | Activator involved in purine utilization and transport (31, 251) |
| XLNR | Aspergillus niger, Aspergillus oryzae | Controls expression of genes encoding xylanolytic enzymes (93, 94, 173, 174) |
| YNA2 | Hansenula polymorpha | Activator of the genes involved in nitrate assimilation (10) |
| ZFR1 | Fusarium verticillioides | Involved in regulation of fumonisin biosynthesis (73) |
Structural and Functional Domains
Like most transcription factors, zinc cluster proteins contain several functional domains apart from the cysteine-rich DBD, including the regulatory and activation domains. A model depicting functional domains is shown in Fig. 1.
FIG. 1.
Functional domains of zinc cluster proteins. Zinc cluster proteins can be separated into three functional domains: the DBD, the regulatory domain, and the acidic region. In addition, the DBD is compartmentalized into subregions: the zinc finger, the linker, and the dimerization domain. These regions contribute to DNA-binding specificity and to protein-DNA and protein-protein interactions (267). MHR, middle homology region.
The entire DBD is separated into three regions: the zinc finger, linker, and dimerization regions. Pioneer work done on Gal4p (activator of GAL genes) and Ppr1p (activator of URA genes) has elucidated much of the structural biology of these transcription factors. The metal-binding portion of the DBD is described as having two substructures; each is formed by three cysteines that are surrounded on both sides with basic amino acids and are separated by a loop (233). Together, these form a pair of short alpha helices, between which are nestled two zinc atoms bound and bridged by a total of six cysteine residues (78, 199). This cysteine-rich DBD is commonly located at the N terminus. However, at least two characterized C-terminal zinc cluster proteins also exist. They include S. cerevisiae Ume6p, as well as C. albicans Czf1p (283). Several mutagenic studies demonstrate the importance of the six cysteine residues in DNA binding and protein function (12, 50, 79, 111, 204, 205, 248, 260, 293). Other residues found within the metal-binding motif are equally important. For example, a conserved proline located in the loop between the two substructures provides flexibility (168), while a highly conserved lysine residue (sometimes replaced by arginine, histidine, or glutamine) is positioned between the second and third cysteines (168, 169, 233). X-ray crystallography of the S. cerevisiae Gal4p and Ppr1p DBDs performed by Marmorstein et al. (168) confirmed that these proteins bind as homodimers (Fig. 2). In fact, the cysteine-rich regions of these two proteins are remarkably similar. The zinc clusters of the homodimer complexes recognize a pair of CGG nucleotide triplets, interacting via major-groove contacts. This not only reflects the high degree of homology among members of this protein class but also suggests that other domains/factors must influence DNA targeting by these transcriptional regulators (see below).
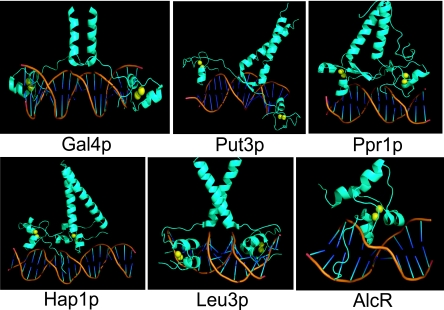
FIG. 2.
Crystal structures of the DBDs of some Zn(II)2Cys6 regulators. AlcR binds as a monomer (28), while Gal4p, Put3p, Ppr1p, Leu3p, and Hap1p bind as homodimers. Gal4p, Put3p, and Ppr1p recognize inverted DNA repeats (168, 169, 253, 278). Leu3p and Hap1p bind to everted and direct repeats, respectively (72, 89, 98, 124, 294). Yellow spheres correspond to zinc atoms.
At least two known zinc cluster proteins do not require the cysteine-rich DBD. Both S. cerevisiae Dal81p and Aspergillus nidulans TamAp proteins appear fully functional when their zinc clusters are deleted or disrupted (25, 49). Three other members of this superfamily in S. cerevisiae do not bind to DNA directly. The RSC3 and RSC30 genes encode proteins which make up part of the chromatin-remodeling complex RSC (_r_emodel the _s_tructure of _c_hromatin) (8), while Cep3p is an important component of the kinetochore complex (143, 250).
With a few exceptions, the requirement for zinc in stabilizing protein folding and function in this transcription factor class is obvious. However, several key experiments performed over a decade ago illustrate that zinc can be replaced by other metal ions, while still allowing for proper protein function. In determining the X-ray crystal structure of Gal4p, Marmorstein et al. showed that Cd2+-containing crystals were of better quality than those containing Zn2+ ions (168). In addition, the nuclear magnetic resonance (NMR) solution structure of Ume6p was solved by demonstrating that zinc could also be replaced with cadmium (7). Importantly, both groups showed that these proteins bind to DNA in a metal ion-dependent manner.
The linker region is located C-terminally to the zinc cluster motif. It can take on very different forms, and sequence alignments show no similarities between linkers in various zinc cluster proteins. For example, the linker region for Gal4p extends along one DNA strand, contacting the phosphodiester backbone (168). In Ppr1p, the linker region is made up of antiparallel β sheets (169). Moreover, the Hap1p DBD also targets two CGG triplets, but in a direct-repeat orientation, as opposed to the case for Gal4p, Put3p, and Ppr1p, where the CGG triplets are inverted (Fig. 2). The crystal structure of Hap1p shows that the protein dimer is asymmetrical and that the linker regions of both monomers interact exclusively with different residues, occupying two very different environments (124). Various linker regions between zinc cluster proteins that recognize similar nucleotides can explain this region's role in contributing to DNA-binding specificity. Replacing the zinc cluster motif of one protein with another does not affect DNA targeting, although switching linker regions does (166, 214). Moreover, mutations in the Gal4p and Ppr1p linker regions also affect DNA binding and proper protein function (111). It is, therefore, proposed that the linker region provides a rigid scaffold, mediating DNA binding to a preferred sequence and preventing binding to any alternate sites (166).
The dimerization region is the last element within the DBD and is typically positioned C terminal to the linker. The majority of zinc proteins contain this region, which is made up of heptad repeats similar to those found in leucine zippers (233). These heptad repeats form a highly conserved coiled-coiled structure which is most likely responsible for dimerization and protein-protein interactions. Importantly, this coiled-coil element is absent in S. cerevisiae Ume6p, which is one of the two characterized zinc cluster proteins containing a C-terminal DBD. This evidence suggests that Ume6p most likely acts as a monomer (202, 260).
The regulatory domain contains an important region displaying lesser homology among most members within this protein class, and it is termed the middle homology region. This region is what separates the DBD from the C-terminal acidic region. Although not always present in all zinc cluster proteins, this region, which spans about 80 amino acids, is thought to play a role in regulating the transcriptional activity of these factors (233, 267). This model is based on several studies in which the deletion of this region often renders zinc cluster proteins constitutively active. For example, removal of a region encompassing the middle homology region in S. cerevisiae Hap1p results in transcriptional activation even in the absence of the inducing molecule, heme; this suggests an additional role of oxygen sensing for this region in Hap1p as well (205). When a similar region is deleted in S. cerevisiae Leu3p, the protein is permanently activated (76, 297). Other examples include the Pdr1p and Pdr3p mutants that contain gain-of-function mutations within this region, implying that it possesses an inhibitory role (53, 128).
Most often C-terminally located, the acidic domain acts as an activation domain (233, 267). This is not a conserved domain, and its function/structure within this superfamily of transcriptional regulators is varied and not well defined. In both Pdr1p and Pdr3p, gain-of-function mutations have also been found in this motif (29, 196). Interestingly, several predicted transmembrane motifs are located in the activation domain of the C. albicans Upc2p (163, 243). This supports the hypothesis that this transcription factor may be membrane anchored in the cytoplasm prior to cleavage and translocation to the nucleus (243). Strangely, the deletion of only the last 10 C-terminal amino acids from Uga3p results in a totally inactive form of the transcription factor (M.-A. Sylvain and B. Turcotte, unpublished data). Clearly, this domain plays an important yet individualized role in each zinc cluster protein.
Binding Elements and DNA-Binding Specificity
Many studies show that zinc cluster proteins recognize highly related elements containing trinucleotide sequences in single or repeat forms, in either a symmetrical or an asymmetrical format. CGG triplets are common, although variations within these binding elements have also been reported (Table 2). Because these highly conserved transcriptional regulators all target very similar sequences, several strategies are needed to generate a vast repertoire of binding sites. This ensures that the required protein is able to carry out its own specific regulatory task (156, 233, 260, 267). As stated above, many factors influence DNA targeting and binding by zinc cluster proteins. In terms of protein structure, many components of the DBD contribute substantially in binding to target DNA. Moreover, nucleotides surrounding the CGG triplets also determine DNA-binding affinities to some extent (195). However, two very important determinants of DNA-binding specificity are the orientation of the CGG triplets and the spacing between these triplets.
TABLE 2.
DNA motifs recognized by zinc cluster proteins in S. cerevisiae
| Zinc cluster | Motif(s) (reference[s])a | Cross-regulation/autofeedback loop/ ChIP-chip binding (reference[s])b |
|---|---|---|
| Arg81p | TGACTCY (162) | Arg81p (92) |
| Aro80p | CCGNgRNTWRCCGMSAKTTGCCG (162) | Aro80p (92) |
| Cat8p | YCCNYTNRKCCG (221) | Ume6p (92) |
| Cep3p | Binds to the CDEIII element of centromeric DNA (63) | |
| Cha4p | tGCGAtgaR (162) | |
| Dal81p | GAAAATTGCGTTT (271), AAAAGCCGCGGGCGGGATT (162) | Uga3p (92) |
| Ecm22p | TCGTATA (274) | |
| Gal4p | CGG-N11-CCG (272) | |
| Hap1p | CGG-N6-CGG (294), CGG-N3-TANCGG-N3-TA (89) | Hap1p (92, 104) |
| Leu3p | CCGG-N2-CCGG (98), CCGGTMCCGG (162) | |
| Lys14p | TCCRNYGGA (16) | |
| Mal63p | MGC-N9-MGS (244) | |
| Oaf1p | CGG-N3-TNRN8-12CCG (223) | |
| Pdr1p | TCCGCGGA (120, 165) | |
| Pdr3p | TCCGCGGA (51, 98, 120, 165) | Pdr1p, Pdr3p (51, 92, 138) |
| Pdr8p | TCCG(A/T/C)GGA (100) | |
| Pip2p | CGG-N3-TNRN8-12CCG (223) | Oaf1p, Pip2p (223) |
| Ppr1p | TTCGG-N6-CCGAA (153) | |
| Put3p | CGG-N10-CCG (242), CGGGAAGCCM-N3-c (162) | Stb4p (92) |
| Rds1p | KCGGCCGa (92) | |
| Rgt1p | CGGANNA (123), SYCGGAAAAA (162) | |
| Sip4p | TCCATTSRTCCGR (221), CCRTYCRTCCG (276), CGGNYNAATGGRR (92) | Cat8p (276), Ume6p (92) |
| Stb4p | TCGg-N2-CGA (92) | Hal9p (92) |
| Stb5p | CGGNStTAta (92), CGGNSNTA (138) | Stb5p (92, 138) |
| Sut1p | CGCG (215), GCSGSG-N2-SG (92), gCSGgg (162) | Sut1p (92) |
| Tbs1p | Oaf1p, Pip2p (92) | |
| Tea1p | CGG-N10-CCG (86) | |
| Thi2p | GMAAcYNTWAgA (92), GMAACYSWWAGARCY (162) | |
| Uga3p | AAAARCCGCSGGCGGSAWT (255), CCGCSSGCGG (195), SGCGGNWttt (107) | |
| Ume6p | TCGGCGGCT (285), taGCCGCCSa (92) | Oaf1p, Pip2p, Cat8p, Sip4p (92) |
| Upc2p | TCGTATA (274) | Upc2p (2) |
| War1p | CGG-N23-CCG (131) | |
| Ydr520Cp | tCtCCGGCGga (162) | |
| Yjl103Cp | Ume6p (92) | |
| Yrr1p | WCCGYKKWW (144), TttTGTTACSCR (162) | Pdr1p, Pdr3p, Yrm1p, Yrr1p (158, 296) |
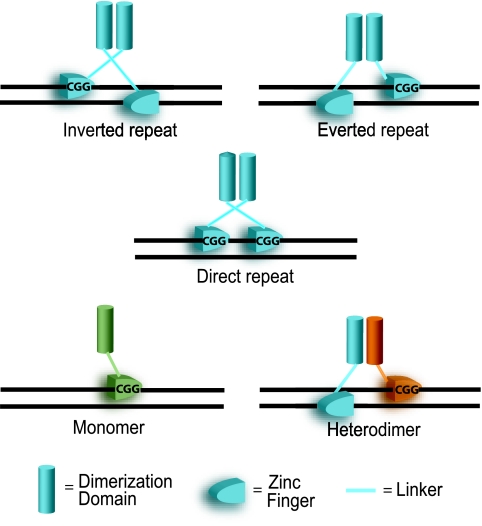
Zinc cluster proteins can bind as homodimers to CGG triplets that are oriented in everted, inverted, or direct repeats. Gal4p, Put3p, and Ppr1p bind to inverted repeats whereby the zinc clusters of each monomer face each other (Fig. 2). Leu3p and Pdr3p exemplify how a homodimer binding to an everted repeat consists of two zinc clusters facing away from one another, while a Hap1p homodimer contains two zinc clusters facing the same direction in order to bind to a direct repeat (Fig. 2). Figure 3 represents a model illustrating this differential binding. Spacing between trinucleotide sequences is critical for zinc cluster proteins that bind to CGG triplets in the same conformation. For example, Gal4p binds to inverted CGG triplets spaced by 11 bp (CGG-N11-CCG), whereas Put3p binds to CGGs separated by 10 bp (CGG-N10-CCG) (11, 242, 272).
FIG. 3.
A model for zinc cluster protein DNA recognition. Zinc cluster proteins preferentially bind to CGG triplets that can be oriented in three different configurations: the inverted, everted, and direct repeats. The orientation of CGG triplets and the nucleotide spacing between the triplets are the two major determinants of DNA-binding specificity (166). Zinc cluster proteins can also bind as monomers (in green) as well as homodimers (two molecules in blue) and heterodimers (one molecule in blue and one in orange).
As more zinc cluster proteins are characterized, the presence of monomers and heterodimers predicts that realistically, many variations and combinations of this paradigm most likely occur. One possibility is that heterodimers comprised of members within this family bind preferentially to slightly different combinatorial elements, increasing the number of potential binding sites for this protein class. The physiological presence of Pdr1p and Pdr3p homodimers and Pdr1p/Pdr3p heterodimers in yeast provides evidence supporting this theory (167).
It remains to be seen whether or not other structures/motifs/binding elements may also be important in DNA recognition but have yet to be characterized. A recent large-scale study employing a powerful strategy for genome-wide location analysis demonstrates that many zinc cluster proteins can regulate other target genes through elements different from those initially identified (92). The technique combined the chromatin immunoprecipitation (ChIP) of approximately 200 tagged transcriptional regulators in S. cerevisiae (including many zinc cluster proteins) with DNA microarrays consisting of all the intergenic sequences within the yeast genome. This approach is useful when identifying additional targets for one transcriptional regulator, as well as novel DNA recognition sites (Table 2). However, it also demonstrates how some binding elements do not fit the standard model for zinc cluster proteins, implying that other factors determining DNA-binding specificity can go undetected and have not yet been elucidated.
Mechanisms of Action
The Gal4p superfamily encompasses a wide variety of pivotal, albeit individualized, roles within the cell, and they employ a range of mechanisms in order to do so. Like for many transcriptional regulators, a multitude of strategies exists in order to control their transcriptional activity. These can include nuclear-cytoplasmic shuffling, DNA binding, phosphorylation, and unmasking of the activation domain (236, 249). This section describes some of the known mechanisms in which zinc cluster proteins are transported, activated, aided, or coordinated in order to perform their specific tasks.
Although zinc cluster protein homodimers were once perceived as the “norm” in regulating target genes, recent work demonstrates that many proteins within this class are found predominantly as monomers or heterodimers under physiological conditions. A classic example of a monomer is the Aspergillus AlcR protein. NMR spectroscopy clearly shows that one monomeric zinc cluster binds to an element in the alcA promoter, which contains the sequence CGTGCGGATC (28). Monomer or dimer status can sometimes be inferred based on the sequence of its target regulatory element. It is proposed that Rgt1p most likely acts as a monomer because its target sequence contains a single trinucleotide, CGGANNA (123). Using these two examples, other zinc cluster proteins that could also potentially regulate target genes as monomers include the S. cerevisiae proteins Upc2p and Ecm22p, as well as their homologue Upc2p in Candida albicans. They activate transcription of ERG genes, which encode enzymes needed for ergosterol biosynthesis, acting through DNA response elements that contain the consensus sequence CGTATA (163, 274). A peculiar exception is Ume6p. Its zinc cluster is localized at the C terminus, and no coiled-coil dimerization region is predicted (233, 260). It was postulated that Ume6p acted as a monomer (248), and this was confirmed when its NMR structure was resolved (7). However, a close examination of its preferred binding sites shows that they actually include two perfect CGG triplets in inverted or direct-repeat orientations. Clearly, NMR spectroscopy and crystallography are currently some of the only methods that can determine for certain the dimerization status of members within this protein class.
Several zinc cluster proteins within the pleiotropic drug resistance (PDR) network are able to heterodimerize (see below) (3, 167), although how these heterodimers differentially regulate genes is still unknown. It has long been known that zinc cluster proteins Oaf1p and Pip2p differentially regulate genes by forming heterodimers (115, 117, 223). They regulate genes involved in peroxisome proliferation by acting through oleate response elements (OREs) in the promoters of their target genes FOX1, FOX3, and CTA1 (101, 115, 117). In vitro binding assays performed by Rottensteiner et al. (223) show that the Oaf1p/Pip2p heterodimer better binds to the FOX1 ORE than to the FOX3 ORE. They concluded that specific sequence differences within the ORE, as well as homodimeric or heterodimeric complexes, must influence promoter recognition (223).
While an Oaf1p homodimer maintains basal levels of target genes, an Oaf1p/Pip2p heterodimer complex is preferred in the upregulation of genes when cells are grown using oleate as a carbon source (115, 117, 223). Zinc cluster proteins can also form heterodimers with members of other transcription factor families, such as members of the MADS box family. Arg81p (ArgRIIp) dimerizes with MADS box proteins ArgRIp and Mcm1p in order to regulate genes that encode enzymes implicated in arginine metabolism (6). These three proteins all possess DBDs, but they must form the three-component complex in order to bind to DNA in vitro in an arginine-dependent manner (6). Amar et al. also suggest that Arg81p acts as the arginine sensor in this complex, because two regions directly N terminal to the DBD share sequence homologies with an arginine-binding pocket in the Escherichia coli ArgR repressor (6).
Zinc cluster proteins can further coordinate the transcriptional control of target genes alone or in coordinated networks with other members of this class. They can do so by acting through one or more DNA recognition sites. For example, at least three zinc cluster proteins (Pdr1p, Pdr3p, and Rdr1p) regulate the PDR5 gene (encoding an ATP-binding cassette [ABC] transporter involved in PDR) by acting through the same pleiotropic drug response elements (PDREs). Conversely, Rgt1p acts alone but requires multiple sites within the promoters of hexose transport genes (123). Another scenario depicts one gene's promoter being regulated by at least two different zinc cluster proteins, acting through two different and exclusive recognition sites. Such is the case for several β-oxidation genes. Ume6p represses the transcription of the genes CTA1, POX1, FOX2, and FOX3 by acting through a URS1 element, while the Oaf1p/Pip2p heterodimer positively regulates the same genes through OREs in the same promoters (234).
Self-Regulation and Positive Feedback Loops
Several members of this class regulate the expression of other zinc cluster proteins (Table 2). Others are self-regulated, forming a positive feedback loop. In response to oleate, the Oaf1p/Pip2p heterodimer has an additional role in self-activating the PIP2 gene through another ORE in its own promoter (223). Yrr1p not only is regulated by another zinc cluster protein, Yrm1p (158), but also forms part of an autoregulatory feedback loop. It has been detected at its own promoter in ChIP assays (296). Similarly, Pdr3p is positively autoregulated, as well as being regulated by the Pdr1p zinc cluster protein. Pdr3p controls its own transcription through two PDREs in its promoter (51) and is described in greater detail below. Two other zinc cluster proteins involved in gluconeogenesis, Cat8p and Sip4p, are in this category. Evidence suggests that they regulate themselves by using a complex autoregulatory pathway involving cross talk between the two activators (99, 276). Hap1p is another zinc cluster protein that falls under this umbrella. It regulates genes involved in respiration (134, 295), but its own activity is in part autoregulated (104). ChIP-chip experiments also demonstrate that Stb5p is bound to is own promoter (92, 138). Lastly, studies show that the activator of ERG genes, Upc2p, and its Candida albicans homologue appear to be involved in positive autoregulation loops (2, 163, 243).
Nuclear Import of Zinc Cluster Proteins and Localization
In order to carry out their functions as transcriptional regulators, members within the Gal4p superfamily must first be localized to the nucleus. Thus, zinc cluster proteins can be categorized based on their initial location within the cell, prior to activating or repressing transcription of their target genes. The first group consists of those that are permanently present in the nucleus, while the second group resides in the cytoplasm.
Many zinc cluster proteins that make up the first group are constitutively localized within the nucleus on a permanent basis. These include Lys14p, Oaf1p, War1p, Put3p, and Leu3p (11, 59, 126, 127, 131, 234). It has been demonstrated that Oaf1p, War1p, Put3p, and Leu3p are constitutively bound to their target promoters. Put3p is an activator of PUT (proline utilization) genes that encode enzymes required for proline metabolism. Although it is always bound to a promoter, its activity is controlled by direct interaction with proline (236, 237). Similarly, Leu3p is bound to the promoters of leucine biosynthesis genes (LEU4, ILV2, and ILV5) but is activated only when a leucine precursor, α-isopropylmalate, is present (126, 254). In addition, it is proposed that Ppr1p, an activator of genes in the pyrimidine biosynthetic pathway, is bound to its target promoters in an inactive state until it is activated by a metabolic intermediate or effector molecule (75). It is postulated that many constitutively active zinc cluster proteins, or “condition-invariant” regulators such as the examples listed above, although not yet identified, must be controlled in this manner (92).
Zinc cluster proteins taking part in transcriptional regulation but initially localized in the cytoplasm must somehow be imported into the nucleus. However, the mechanisms by which they do so are just starting to be clarified. Nuclear import of transcription factors in eukaryotes includes many exclusive pathways. In general, transport of proteins across the nuclear membrane is mediated through nuclear pores wherein soluble transport receptors bind to nuclear localization signals (NLSs) on their target molecules (84, 85, 187). NLSs usually consist of one or two short stretches of basic amino acid residues (reviewed in reference 187). In general, proteins to be shuffled into the nucleus are bound by the α/β importin heterodimer. The α subunit acts as the bridge between the NLS-containing cargo protein and the β subunit, which carries the cargo through the nuclear pore.
Most nuclear import is orchestrated by the importin β receptor family (84), but many nonclassical NLSs on target molecules requiring less conventional nuclear import pathways are also reported (192). Many different importin β or importin-β-like proteins have been characterized in mammalian and yeast cells. Although no general strategy for the import of zinc cluster proteins has been deciphered, a few NLSs have been identified in Aspergillus PrnA and AlcR, as well as in S. cerevisiae Gal4p and Pdr1p. Gal4p interacts directly (without the help of the α subunit) with the importin β receptor yeast homologue Rsl1p/Kap95p complex, as well as with another importin β called Nmd5p (32, 33). Pdr1p uses the Pse1p/Kap121p complex, which is another member of the yeast importin β-related family (52). AlcR requires three importin β-related proteins: Kap104p, Sxm1p, and Nmd5p. In addition, the NLSs of these aforementioned proteins are located in the N terminus, within or very close to the DBD (192). Thus, differences in nuclear import for a few zinc cluster proteins reflect the many different mechanisms required to fulfill this task, as in higher eukaryotes.
A large-scale protein localization project performed by Huh et al. has provided much insight into zinc cluster proteins and others with respect to their location within the cellular environment (106). Their findings, as well as those of other studies based on the localization of zinc cluster proteins, are summarized in Table 3. The locations of other characterized proteins are assumed based on their functions; Cep3p forms part of the kinetochore complex and should therefore be localized to microtubules and the centromere (106, 143, 250), whereas Rsc3p and Rsc30p share a chromatin-remodeling function and most likely also carry out their roles solely in the nucleus (8).
Activation by Phosphorylation
Several zinc cluster proteins are activated by a phosphorylation or dephosphorylation event. For instance, Gal4p is activated upon phosphorylation. In the absence of galactose, Gal80p represses Gal4p activity by covering its activation domain (161; reviewed in references 236 and 263). Gal4p is phosphorylated at multiple sites (184, 185, 228, 229). Under noninducing conditions, an unphosphorylated form and a phosphorylated form of Gal4p are observed. The presence of the inducer galactose results in the appearance of a second phosphorylated form associated with transcriptionally active Gal4p. Phosphorylation at only a single serine residue (Ser699) in the C-terminal activation domain appears to be necessary for activation (228). However, phosphorylation of Ser699 is not absolutely required for Gal4p activity, since a Ser699-Ala Gal4p mutant shows transcriptional activity in cells lacking Gal80p or in the presence of high galactose levels (219). From these observations, Rohde et al. (219) suggested a model in which phosphorylation of Gal4p is required for an acute response to galactose.
Two other zinc cluster proteins, Pdr1p and Pdr3p, have also been identified as phosphoproteins, although the distinct roles carried out by the phosphorylated isoforms have not yet been elucidated (167). Mamnun et al. eliminated several possibilities for the C-terminally phosphorylated form of Pdr3p, including nuclear localization, dimer formation, and proteolytic turnover (167). In addition, Cat8p and Sip4p are two other members of this family that are characterized phosphoproteins and are activators of genes involved in gluconeogenesis. Both become phosphorylated during the derepression of target genes (34, 212, 276). Likewise, Rgt1p's DNA-binding ability is also regulated by phosphorylation. When cells are grown in glucose, Rgt1p is phosphorylated. This inhibits its binding to its target promoters, thereby preventing its transcriptional repression of hexose transporter genes (74, 123, 182).
At least two zinc cluster proteins are phosphorylated in response to an external stress. War1p is responsible for the upregulation of the gene encoding the ABC transporter Pdr12p in response to weak acid stress. Data suggest that War1p is rapidly phosphorylated in the presence of sorbate, benzoate, and propionate, most likely in order to activate transcription (131). Similarly, Put3p is differentially phosphorylated in the cells' response to different nitrogen sources (105).
Promoter Occupancy
The promoter occupancy of some zinc cluster proteins is influenced by external factors leading to variation in binding at target sites. For example, binding of Upc2p and Ecm22p, two regulators of ergosterol biosynthesis genes, is influenced by treatment with lovastatin, an inhibitor of the ergosterol pathway. While the level of Upc2p is increased at ERG3, the level of the closely related zinc cluster protein Ecm22p is reduced at this same promoter (48). Similarly, genome-wide location analysis (ChIP-chip) revealed that binding of Stb5p is enhanced at some target genes when cells are treated with the oxidative agent diamide. This treatment also leads to Stb5p binding at additional target genes unoccupied by Stb5p in untreated cells (138).
Recruitment of Chromatin Remodelers, Histone Modifications, and Cofactors
Eukaryotic DNA is tightly packaged into chromatin, hampering transcription by limiting DNA accessibility to transcriptional activators and other factors making up the transcriptional machinery. Zinc cluster proteins sometimes require the aid of chromatin-remodeling complexes, histone-modifying enzymes, and/or transcriptional cofactors in order to surmount the repressive nature of chromatin and facilitate gene transcription. This section describes some of the known relationships between zinc cluster proteins and the chromatin remodelers/cofactors that they recruit at their target promoters.
A well-characterized histone acetyltransferase in S. cerevisiae is the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex. It is highly conserved throughout evolution (P/CAF complex in humans) (125). Spt and Ada proteins have separate functions apart from the acetyltransferase activity of the Gcn5p subunit (125). A number of yeast genes, including _GAL1_-10 and PDR5, are SAGA dependent (19, 77, 139, 170). The Gal4p activation domain is responsible for recruiting the SAGA complex to the _GAL1_-10 promoter, although transcription is not dependent on the Gcn5p subunit (139).
As described previously, at least three zinc cluster proteins (Pdr1p, Pdr3p, and Rdr1p) regulate the transcription of PDR5 via PDREs dispersed throughout the promoter (3, 4, 97, 119, 120, 125). An interaction between Pdr1p and the SAGA complex was detected via a two-hybrid assay several years ago, and evidence suggested that perhaps this interaction actually caused an inhibitory effect on PDR5 expression (170). It has since been clarified that SAGA is needed to actually activate transcription of PDR5 (77). Spt3p and Spt20p/Ada5p subunits, but not Gcn5p, are needed to activate transcription (77). The PDR5 promoter is also occupied by other coactivators, including the mediator complex and the chromatin-remodeling SWI/SNF complex (77).
Many yeast genes are negatively regulated by histone deacetylases (HDACs). At least six HDACs exist in yeast. The RPD3, HDA1, HOS1, HOS2, HOS3, and SIR2 yeast genes encode HDACs (18). A well-characterized HDAC complex in yeast is the Rpd3p/Sin3p complex. The Rpd3p component exerts the histone deacetylase activity (259), while Sin3p is characterized as a corepressor that depends on Rpd3p to coordinate its repressive effect (114, 279). The Rpd3/Sin3p HDAC complex negatively regulates a variety of genes implicated in numerous cellular processes. Kadosh and Struhl (114) emonstrated that the Ume6p zinc cluster protein relies on this histone-modifying complex in order to repress its target genes. In addition, they showed that only a small region within the Ume6p protein is necessary to recruit this complex to a specific promoter (114). Interestingly, two additional zinc cluster proteins, encoded by the STB4 and STB5 genes, interact with the Sin3p corepressor in a two-hybrid assay (118), although a relationship between these interactions and inhibitory transcriptional activity has not been established. Rgt1p is yet another zinc cluster protein that depends on a corepressor in order to exert its repressive effect. It interacts physically with the corepressor Ssn6p in order to negatively regulate HXT genes when glucose sources are depleted (197, 208). The Ssn6p-Tup1p complex was first characterized as a general repressor of transcription in yeast (121). Since then, its functional association with multiple HDACs has been elucidated (47, 281, 291).
As their names imply, the ATP-dependent chromatin-remodeling complexes require ATP hydrolysis in order to carry out their chromatin-disrupting function. They are typically composed of several protein subunits. A genome-wide study has revealed that approximately 5% of all yeast genes are SWI/SNF dependent (103). At least two zinc cluster proteins need the SWI/SNF complex at target promoters. Côté et al. showed that Gal4p binding is facilitated and stimulated by the SWI/SNF complex (42). Targeting of SWI/SNF to GAL1 following galactose induction required the presence of Gal4p (146). Hap1p (an activator of respiration genes, including CYC1 and CYC7) also relies on a functional SWI/SNF chromatin remodeler for transcriptional activity (88).
Other ATP-dependent chromatin remodelers in yeast include the ISWI (_i_mitation _swi_tch)-based family and the RSC (_r_emodels the _s_tructure of _c_hromatin) complex. ISWI complexes are known to organize or displace nucleosomes by sliding them along a stretch of DNA, and this can lead to either repression or activation of target genes (125). The RSC complex is similar to the SWI/SNF complex in that it also contains a large number of subunits. Ume6p is yet another example of a zinc cluster protein that recruits the Isw2p subunit to carry out repression of its target genes, while cooperating with the HDAC complex mentioned earlier (66, 81). It has also been demonstrated that transcriptional activation by Gal4p fusion proteins requires members of the ISW-based family (148, 180). As stated above, the RSC3 and RSC30 genes encode zinc cluster proteins that form part of the RSC megacomplex. The zinc clusters of both proteins are needed for proper protein complex function (8). Whether or not they interact directly with DNA by binding to a consensus sequence or whether their DBD motif helps target the complex to RSC-dependent promoters has yet to be elucidated.
ZINC CLUSTER PROTEINS IN SACCHAROMYCES CEREVISIAE
The study of zinc cluster proteins in budding yeast has provided much of the framework for understanding fungal transcriptional regulators and their functions within the cell. Sequencing of the Saccharomyces cerevisiae genome has allowed for the identification of 55 members within this family (5, 233, 260, 267), based on the well-conserved consensus amino acid sequence of the Zn(II)2Cys6 motif. This makes it one of the largest families of transcription factors in yeast. Moreover, they can act as repressors, as activators, or as both activators and repressors for certain genes (267). For instance, Rgt1p and Ume6p are both activators and repressors of glucose transport and early meiotic genes, respectively (110, 198). It has also been recently demonstrated that Stb5p acts as an activator and a repressor in the presence of oxidative stress (138).
Roles
A plethora of cellular processes is orchestrated by members of the Gal4p superfamily. These processes include sugar metabolism, gluconeogenesis and respiration, amino acid metabolism and vitamin synthesis, mitosis, meiosis, chromatin remodeling, nitrogen utilization, and peroxisome proliferation, as well as the stress response and PDR (see below). Table 3 classifies their initially characterized functions into several broad categories and is a compilation of several works (106, 156, 194, 260, 267). Many of these transcriptional regulators not only have more than one distinct role but can also have overlapping functions. They often coordinate gene regulation of different subsets of genes together or at different times. For instance, Ume6p plays a role in nonfermentative metabolism (234), but its primary roles seem to be regulation of early meiotic genes as well as repressing expression of arginine biosynthesis enzymes (7, 22, 110, 125, 202, 224, 248). Another example is Upc2p, whose primary function is in activating ergosterol biosynthesis genes but which also plays secondary roles in anaerobic sterol uptake and expression of DAN/TIR mannoprotein genes (2, 38, 284). Similarly, zinc cluster proteins Pdr1p and Pdr3p are known for their primary roles in regulating PDR genes, but they also regulate hexose transport genes HXT9 and HXT11, as well as recently being implicated in the transcriptional control of sphingolipid biosynthesis genes (91, 129). Moreover, Pdr3p has other functions that do not include Pdr1p, such as retrograde signaling, as well as a novel role in controlling DNA damage-inducible genes MAG1 and DDI1 (298).
Two zinc cluster proteins are essential. Cep3p is part of the kinetochore complex needed during mitosis (143, 250), and Rsc3p is a subunit within the SWI/SNF-like chromatin remodeler RSC. RSC is fairly abundant in cells, and is required for the activation of a number of genes (125). Lastly, Stb5p is also essential but only in certain genetic backgrounds (4, 191, 197).
Amino Acid Metabolism
A number of zinc cluster proteins are involved in controlling expression of genes required for amino acid metabolism. For example, Leu3p is involved in regulating synthesis of branched amino acids (for a detailed review, see reference 127). Cha4p controls expression of genes for catabolism of serine and threonine, while the activator Lys14p is specific for lysine synthesis. Aro80p, another zinc cluster protein, controls expression of genes involved in catabolism of aromatic amino acids (tryptophan, phenylalanine, and tyrosine). These amino acids can be metabolized to alcohols (e.g., tryptophol) for use as a nitrogen source. ARO9 encodes an aromatic aminotransferase involved in the first catabolic step of aromatic amino acids. ARO9 expression is increased in the presence of aromatic acids and repressed in the presence of a rich nitrogen source such as ammonia (108). Aro80p positively regulates expression of ARO9 through a DNA element called UASaro found in its promoter region (108). Diploid yeast cells can switch to an invasive filamentous form when starved for nitrogen (164). Interestingly, some aromatic alcohols, such as tryptophol, promote morphogenesis (36). Expression of ARO9 and ARO10 (another key gene for production of aromatic alcohols) is dependent on cell density or low ammonia concentration and is subject to autoregulation by tryptophol, a process that requires Aro80p (36). Thus, the zinc cluster protein Aro80p is part of a quorum-sensing system bridging environmental conditions to morphogenesis (245).
Like many other zinc cluster proteins, Cha4p activates transcription of target genes in a classical way by binding to specific sequences found in their promoter regions. For example, the Cha4p-dependent expression of CHA1 (encoding an l-serine/l-threonine deaminase) is induced in the presence of serine or threonine for utilization as nitrogen sources. The CHA1 promoter contains two UASCHAs that confer serine or threonine induction when placed in front of a heterologous promoter (21, 102). Interestingly, Cha4p also controls expression of the serine biosynthetic gene SER3 indirectly via SRG1 (171, 172). The SRG1 gene, which does not encode a protein, is located just upstream of the SER3 gene. Expression of SRG1 causes transcriptional interference resulting in repression of SER3. Cha4p binds to the SRG1 promoter and is activated in the presence of serine, resulting in SRG1 transcription and, indirectly, in SER3 repression (172).
Multidrug Resistance
A large proportion of zinc cluster proteins (at least 12 in S. cerevisiae) have been implicated in the cell's response to stress and multidrug resistance. Clearly, the fungal cell must rely on this group of regulators to communicate external or internal environmental pressures. Multidrug resistance, or PDR, is a widespread phenomenon that is highly conserved. It is found throughout evolution in organisms ranging from bacteria to humans and is defined as the cell's ability to become resistant to a multitude of structurally and functionally different cytotoxic compounds (113, 183, 230). PDR is caused by the overexpression of membrane-associated protein pumps and, consequently, expulsion of a wide range of molecules, including antimicrobial drugs (246). In bacteria and other microorganisms such as fungi, multidrug resistance is an evolved and evasive mechanism that presents a major obstacle in the prevention of infectious disease. It also poses many problems in food preparation and agricultural industries. In human beings, acquired multidrug resistance in tumor cells hampers effective chemotherapy. Although most drugs are used against human diseases (cancer) or pathogenic microorganisms (bacteria, protozoans, or fungi), many of the underlying mechanisms in acquired drug tolerance appear to be highly conserved, even among very distantly related organisms (227). Therefore, Saccharomyces cerevisiae is an excellent eukaryotic model for providing insight into the phenomenon of pleiotropic resistance.
Many distinct strategies in yeast have been characterized, in relation to how cells respond to different stresses or harmful molecules that can make up an ever-changing cellular environment. They range from very specific regulatory pathways to widespread reactions and are broadly characterized into two interconnected networks: the stress response and the PDR network. Pathways induced in response to stress can buffer external factors such as heat shock, low pH, weak acids, and high osmolarity (288). As mentioned above, PDR is most often mediated by the upregulation of multidrug efflux pumps or protein transporters, of which there are two types: the ABC transporters and members of the major facilitator superfamily. Two prominent families of transcriptional regulators are equally significant contributing factors in either or both of these networks. They are the bZip protein family (reviewed in reference 183) and the zinc cluster proteins.
Implicated Transcriptional Regulators
Zinc cluster proteins are implicated in PDR because many of them positively regulate the genes that encode drug efflux pumps, thereby conferring drug resistance (Fig. 4). Moreover, drug tolerance and acquired drug resistance in Saccharomyces cerevisiae are often traced back to hyperactive or gain-of-function mutations harbored within some of these transcriptional regulators. Pdr1p and Pdr3p are two zinc cluster proteins that have been named the master regulators of drug resistance in budding yeast (reviewed in reference 183). Pdr1p was first characterized by a number of dominant multidrug-resistant alleles that were mapped to its gene's location (213, 232). Pdr3p was initially identified as a gene that conferred resistance to the mitochondrial inhibitor mucidin (252). Together, Pdr1p and Pdr3p are responsible for the regulation, both positive and negative, of multiple genes related to PDR. They act on target genes by binding to PDREs in the promoters of target genes (119, 120, 240). Target genes linked directly to the PDR phenomenon include the ABC transporters encoded by the PDR5, SNQ2, and YOR1 genes. The promoters of these genes harbor one or several PDREs. A perfect PDRE regulatory element contains the consensus sequence TCCGCGGA, which displays CGG triplets in an everted repeat orientation. Importantly, the PDR3 promoter also contains two PDREs, and these elements not only make up a critical component of a positive autoregulatory loop but are also controlled by Pdr1p (51).
FIG. 4.
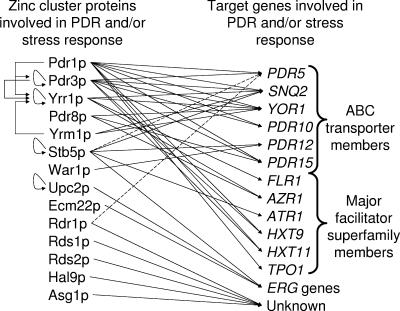
Zinc cluster proteins involved in the PDR network in S. cerevisiae. Zinc cluster proteins (left column) and their target genes involved in PDR and the stress response (right column) are represented. Dashed lines point to genes that are not known to be direct or indirect targets of zinc cluster proteins. Only the most important ABC transporters and major facilitator superfamily members are shown. ERG genes are included because they are also involved in drug resistance.
As stated above, gain-of-function mutations in Pdr1p and Pdr3p can result in drug resistance due to an increased production of the multidrug efflux pumps. More specifically, at least seven mutations acquired in the PDR1 gene are considered multidrug resistance mutations. Three of these point mutations (_pdr1_-2, _pdr1_-6, and _pdr1_-7) are within 10 amino acids of each other, located within the structural motifs I and II found in the regulatory domain of Pdr1p, supporting its role as an inhibitory domain. Two other mutations, _pdr1_-3 and _pdr1_-8, are found in or just outside the C-terminal activation domain (29). PDR5 and SNQ2 mRNA levels are highest in a _pdr1_-3 mutant, but they are also elevated in the _pdr1_-8 mutant as well. The recently identified _pdr1_-12 and _pdr1_-33 Pdr1p mutants mediate resistance to the antimicrobial compound diazaborine by overexpressing the ABC transporters Pdr5p, Snq2p, and Ycf1p, as well as the major facilitator superfamily member Flr1p (282). The same study showed that increased mRNA levels of PDR3 are also caused by the _pdr1_-12 allele. Many hyperactive Pdr3p mutants also induce increased expression of PDR5 and SNQ2, as well as PDR3 (196). Five mutants characterized by Nourani et al. (196) are also located in a short protein segment within structural motifs I and II of the regulatory domain.
The identification of another zinc cluster protein, Yrr1p (_y_east _r_everomycin A _r_esistance), as an additional regulator of PDR genes provided some of the first evidence of cross talk between regulators of PDR in S. cerevisiae (296). It also demonstrated that many Pdr1p, Pdr3p, and Yrr1p targets overlap (45, 46). Yrr1p (initially referred to as Pdr2p) was originally implicated in PDR because it bestowed resistance to sulfometuron methyl (an acetolactate synthase inhibitor) (64). It is now known that Yrr1p also confers resistance to the cell cycle inhibitor reveromycin A, to oligomycin, and to 4-nitroquinoline-1-oxide by binding to the YOR1 promoter and positively regulating the expression of the ABC transporter (45, 64). Efflux of these compounds via Yor1p results in resistance to these toxic compounds. Interestingly, Yrr1p also appears to be self-regulated; it contains a putative Yrr1p response element (YRRE) and a PDRE in its promoter (296). As stated above, expression of YRR1 is even further regulated by yet another zinc cluster protein, Yrm1p (158). Recent microarray experiments performed by Le Crom et al. provide evidence that Yrr1p positively regulates genes through YRREs containing the consensus sequence T/ACCGC/TG/TG/TA/TA/T (144). It is postulated that Yrr1p targets most likely overlap with Pdr1p/Pdr3p targets because YRRE and PDRE sequences are closely related. A gain-of-function mutant, the _yrr1_-1 mutant, provides more insight into how this transcriptional regulator functions. The mutation is a duplication of 12 amino acids located near the C terminus, and it results in a marked resistance to 4-nitroquinoline-1-oxide compared with that of the wild-type strain (46). Northern blot analyses show that SNQ2 mRNA levels are constitutively elevated in a _yrr1_-1 mutant (46).
Other regulators of drug resistance include the zinc cluster proteins Pdr8p, Stb5p, Rds1p, and Rds2p. Pdr8p binds to the promoters of certain genes implicated in PDR, such as YOR1, PDR15, and AZR1. However, this binding was demonstrated using a chimeric Pdr8p; therefore, the exact role of the wild-type protein in PDR is not clear (100). Stb5p was originally picked out of a yeast two-hybrid screen because it interacted with the Sin3p corepressor (118), while Δ_rds1_ and Δ_rds2_ deletion strains exhibit interesting drug phenotypes that may also implicate them in PDR. Deletion strains Δ_rds1_ and Δ_stb5_ are hypersensitive to cycloheximide, and a Δ_rds2_ deletion strain has severely impaired growth in the presence of the antifungal azole ketoconazole (4). The same study shows that cells lacking STB5 have reduced mRNA levels of SNQ2, PDR16, and PDR5. Further evidence that Stb5p is a direct positive regulator of SNQ2 transcription is demonstrated by binding of Stb5p to the SNQ2 promoter in vivo (138). Moreover, in the presence of diamide, Stb5p is a direct activator of two other genes encoding the drug pumps Atr1p and Pdr12p (138).
Lastly, the Rdr1p (_r_epressor of d_rug r_esistance) zinc cluster protein is characterized as a negative regulator of PDR genes (97). A Δ_rdr1 strain is resistant to cycloheximide (4, 97). Rdr1p was confirmed as a transcriptional repressor in microarray experiments that showed that mRNA levels were increased significantly for five genes in the deletion strain compared with the wild-type strain (97). Curiously, all five of these genes (PDR5, PDR15, PDR16, RSB1, and PHO84) encode membrane or membrane-associated proteins, and four of these genes (with the exception of PHO84) actually contain PDREs in their promoters. Furthermore, cycloheximide resistance exhibited in a Δ_rdr1 strain is mediated by the ABC transporter Pdr5p, and Rdr1p appears to act negatively on PDR5 through the same PDREs used by Pdr1p/Pdr3p to activate transcription (97). Whether or not Rdr1p represses its target genes by binding directly to PDREs has not yet been determined.
The studies mentioned above state that at least three different zinc cluster proteins (Pdr1p, Pdr3p, and Rdr1p) modulate transcription of the PDR5 gene by acting on the same PDREs (4, 97, 120). Therefore, these three regulators must somehow cooperate together in order to regulate this drug transporter gene. Interestingly, Pdr1p and Pdr3p are capable of heterodimerizing in vivo (167). Stb5p is found predominantly as a Pdr1p/Stb5p heterodimer, while the zinc cluster protein Yrr1p, which regulates SNQ2, prefers to form homodimers (3). These interactions describe a complex interplay among regulators of PDR genes (Fig. 5). Moreover, it is hypothesized that Pdr1p acts as the master regulator of drug resistance, because it is the only zinc cluster protein in this network that is able to heterodimerize with more than one partner. It most likely does so in order to respond to different conditions or changes in the cell's extracellular or intercellular environment, thereby coordinating an effective regulatory pathway (3).
FIG. 5.
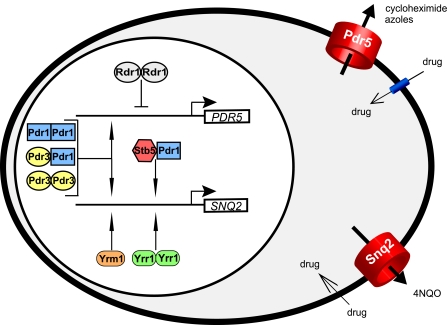
Interplay among zinc cluster proteins implicated in PDR in budding yeast. A network of characterized zinc cluster proteins cooperatively coordinate the transcriptional regulation of PDR genes in S. cerevisiae. Pdr1p (in blue) can form homodimers as well as heterodimers with Pdr3p (in yellow) and Stb5p (in red) (3, 167). Pdr3p, Rdr1p (in gray), and Yrr1p (in green) are able to form homodimers (3, 167; S. MacPherson et al., unpublished data). These different combinations may differentially regulate the expression of PDR5 and SNQ2 ABC transporters. A direct binding of Rdr1p to PDR5 has not been demonstrated, and the mode of binding (e.g., monomer or heterodimer) of Yrm1p (in orange) is not known. 4NQO, 4-nitroquinoline-1-oxide.
Regulation of Ergosterol Biosynthesis
Yeast can also become resistant to certain drugs by using another strategy, which involves effecting changes in the ergosterol biosynthetic pathway. Several zinc cluster proteins are involved in the regulation of genes in this process. Ergosterol is considered the consensus sterol in fungi because it is the major component of the fungal cell membrane. It performs many crucial roles within the cell, including maintaining membrane fluidity and integrity by generally allowing lipids, membrane-spanning proteins, or membrane-associated proteins to function properly (159). Ergosterol also contributes specifically to the regulation of cell growth and proliferation (206, 211, 217, 218). Many drugs developed to specifically inhibit fungal growth target its biosynthesis (see below).
The ergosterol biosynthetic pathway can be divided into two parts: the biochemical conversion of acetyl coenzyme A into squalene and the transformation of squalene into ergosterol (203). Its synthesis, however, is energetically expensive and oxygen dependent (203). In addition, yeast can accumulate exogenous sterols from the environment only under anaerobic conditions (aerobic sterol exclusion). Therefore, ergosterol biosynthesis, its intermediates, and/or its by-products must perform other critical functions within the yeast cell besides modulating membrane structure (203). The model organism S. cerevisiae provides an excellent basis for studying acquired resistance to antifungal drugs, as well as to other toxic compounds. For example, overexpression of the sole azole drug target, a lanosterol 14-α-demethylase encoded by the ERG11 gene, confers resistance to fluconazole (130).
Upc2p and Ecm22p are two highly homologous zinc cluster proteins in S. cerevisiae that regulate expression of ERG genes within the ergosterol biosynthetic pathway, including ERG2 and ERG3 (274). They positively regulate transcription by acting on sterol response elements in the promoters of their target genes. In fact, at least 11 ERG genes encoding enzymes that take part in ergosterol biosynthesis contain putative sterol response elements in their promoters (274). This suggests that regulation by Upc2p/Ecm22p is much more widespread. Upc2p also plays an important role in anaerobic exogenous sterol accumulation, as well as controlling DAN/TIR genes that encode mannoproteins involved in anaerobic restructuring of the cell wall (2). Upc2p (_up_take _c_ontrol) was initially characterized by a gain-of-function mutation in the UPC2 gene that allowed cells to uptake exogenous sterols even when grown in the presence of oxygen (44). An identical mutation in the ECM22 locus has also been described (241). This _upc2_-1 mutant contains a single amino acid change (Gly888Asp) within the activation domain of this protein (44). Interestingly, it was recently demonstrated that the _upc2_-1 mutant can upregulate transcription of the ABC transporter genes AUS1 and PDR11, DAN/TIR genes, and the UPC2 gene itself under aerobic conditions (284). This supports previous evidence of another autoregulatory loop within the Gal4p superfamily of zinc cluster proteins (2). It also alludes to a lesser role in the regulation of membrane transporters which may be involved in PDR.
Upc2p and Ecm22p (initially characterized as an e_xtra_c_ellular m_utant [160]) are 45% identical according to amino acid sequence and have many overlapping functions (241). More specifically, both zinc cluster proteins have highly similar DBDs and C-terminal activation domains, but their middle regions are quite different (48). It is hypothesized that they must carry out some essential function, as a double-knockout Δ_upc2_Δ_ecm22 strain is nonviable in some backgrounds (241). However, a phenotypic analysis of their deletion strains argues that they must also have distinct roles within the cell that may include PDR. A Δ_upc2 strain is sensitive to the antifungal azole ketoconazole, while a Δ_ecm22_ strain is sensitive to cycloheximide (4). Moreover, a recent study shows that Upc2p and Ecm22p respond differently upon induction of the ergosterol biosynthetic pathway by lovastatin. In untreated cells, Ecm22p levels are significantly higher than Upc2p levels (48). Davies et al. (48) showed that in lovastatin-treated cells Upc2p is overexpressed and present in copious amounts at the ERG3 promoter, while Ecm22p is downregulated and almost nonexistent at the same locus.
Hap1p is another zinc cluster protein that regulates expression of the ERG11 gene (encodes the azole drug target) in a heme- and oxygen-dependent manner (268, 273). The HAP1 gene is also upregulated in a _upc2_-1 strain, and it is postulated that a regulatory interaction between these two zinc cluster proteins might exist (284). A clear link between Hap1p and drug resistance has not yet been established. Moreover, Stb5p was recently identified as a novel regulator of ergosterol biosynthesis, since it is a direct activator of ERG5, ERG11, and ERG25 in the presence of oxidative stress (138).
ZINC CLUSTER PROTEINS IN CANDIDA ALBICANS
Candida albicans is typically a commensal organism that inhabits the mucosal linings of most warm-blooded animals, but it is also the major culprit in human fungal infections (231). This fungus was considered asexual for many years, until recent studies proved otherwise (reviewed in reference 17). C. albicans is also dimorphic (207). It can switch between a yeast or hyphal mode depending on specific alterations in environmental conditions. These may include changes in temperature and pH or exposure to different compounds such as serum, _N_-acetylglucosamine, or proline (207). The transition into hyphae is implicated in virulence and pathogenesis (142, 154).
From the complete C. albicans genome diploid sequence, 77 putative ORFs encode zinc cluster proteins, based on the highly conserved DBD elucidated in S. cerevisiae (23). Sequence comparison with known transcriptional regulators in budding yeast reveals many close orthologues. The identification and characterization of zinc cluster proteins in this fungal species has just started, and only a few them have been designated specific functions. Known zinc cluster proteins and their roles within the cell are summarized in Table 4. Functions include sugar metabolism, ergosterol biosynthesis, regulation of hyphal growth, and PDR. One can speculate that as the roles of more zinc cluster proteins in this species are elucidated, many of the proteins will be implicated in a variety of physiological roles similar to those displayed in budding yeast.
TABLE 4.
Characterized zinc cluster proteins in C. albicans
| Gene | orf19 | Function |
|---|---|---|
| CWT1 | orf19.5849 | Δ/Δ_cwt1_ strain is sensitive to calcofluor white and alters the composition of the cell wall (181) |
| CZF1 | orf19.3127 | Hyphal growth regulator (26, 283) |
| FCR1 | orf19.6817 | Negative regulator of drug resistance, complements an S. cerevisiae Δ_pdr1_ Δ_pdr3_ strain (256) |
| FGR17 | orf19.5729 | Regulator of filamentous growth (269) |
| FGR27 | orf19.6680 | Regulator of filamentous growth (269) |
| SEF2 | orf19.1926 | SEF2 expression is repressed by SFU1 under high-iron conditions (137) |
| SUC1 | orf19.7319 | Regulates sucrose metabolic genes (122) |
| TAC1 | orf19.3188 | Transcriptional activator of CDR1 and CDR2 multidrug transporter genes (41) |
| UPC2 | orf19.391 | Transcriptional activator of ergosterol biosynthetic genes (163, 243) |
| WAR1 | orf19.1035 | Confers resistance to sorbate (141) |
| ZNC1 | orf19.3187 | Encodes an essential protein of unknown function (41) |
| ZNC3 | orf19.3190 | Encodes an essential protein of unknown function (41) |
Transcriptional Regulators of PDR
A vast number of transcription factors in budding yeast coordinate the control of several genes involved in drug resistance. Since the genetic manipulation and study of C. albicans are slow in comparison, only two zinc cluster proteins so far are definitely linked to PDR in this species. FCR1 encodes a zinc cluster protein that was cloned from a library as a gene that was able to complement a Δ_pdr1_ Δ_pdr3_ phenotype. Overexpression of FCR1 (f_lu_c_onazole r_esistance 1) in budding yeast resulted in increased resistance to fluconazole and cycloheximide, as well as an increase in PDR5 expression (256). Surprisingly, that study also demonstrated that a C. albicans Δ_fcr1/Δ_fcr1 homozygous deletion strain is actually hyperresistant to fluconazole and other antimycotic drugs. The authors concluded that Fcr1p must be a negative regulator of drug resistance genes. The specific promoters targeted by Fcr1p and its mechanism of action have not yet been identified.
Tac1p (_t_ranscriptional _a_ctivator of Candida drug resistance genes) is the second C. albicans zinc cluster protein directly associated with the regulation of PDR genes (41). Initially, de Micheli et al. showed that a common drug response element (DRE) found in the CDR1 (DREI) and CDR2 (DREII) promoters was responsible for drug-induced upregulation of both of these ABC transporter genes (54). They observed that these DREs might be putative zinc cluster binding sites because they contained CGG triplets in a direct-repeat orientation. A genome-wide search for putative proteins containing the highly conserved Zn(II)2Cys6 motif mapped the TAC1 gene to a region near the mating-type locus that was previously linked to azole resistance in a few clinical isolates (225). A heterozygous deletion of the TAC1 gene caused a loss in CDR1 and CDR2 upregulation in response to fluphenazine, while a glutathione _S_-transferase-Tac1 fusion protein can bind these DREs in vitro (41). Furthermore, a _TAC1_-2 mutant recovered from an azole-resistant strain is responsible for the constitutive overexpression of both of these ABC transporters (41). This evidence provides substantial proof that Tac1p is a bona fide regulator of multidrug resistance.
Ergosterol Biosynthesis in C. albicans
Numerous and extensive studies encompass the ergosterol biosynthetic pathway in this opportunistic pathogen. Only one clear zinc cluster protein orthologue corresponds to both Upc2p and Ecm22p in Candida albicans (163, 243). Cells lacking UPC2 are susceptible to several antifungal compounds that target enzymes within the pathway and cell wall formation (including azoles, terbinafine, fenpropimorph, and lovastatin) (163, 243), while overexpression of the C. albicans UPC2 gene renders cells resistant to fluconazole, ketoconazole, and fluphenazine (163). The Upc2p orthologue is similarly important for aerobic sterol uptake in C. albicans, as demonstrated by a discernible reduction of [14C]cholesterol accumulation in a Δ_upc2_ strain (243).
ZINC CLUSTER PROTEINS IN OTHER SPECIES
Zinc cluster proteins are also found in other yeast species, as well as other fungi. These organisms are not as well studied, but many Zn(II)2Cys6 regulators have been characterized, and these are listed in Table 5. Some of these zinc cluster proteins have clear orthologues in S. cerevisiae. For example, the Gal4p regulator of galactose catabolism in S. cerevisiae is Lac9p in Kluyveromyces lactis. The LAC9 gene is able to complement a Δgal4 strain (290). Furthermore, the regulator of purine utilization in Aspergillus nidulans, UaY, is closely related to Ppr1p in S. cerevisiae and is able to recognize an identical DNA motif (251).
One of the most well-studied zinc cluster proteins in filamentous fungi is AlcR from Aspergillus nidulans and its role in ethanol catabolism (reviewed in reference 67). In brief, the AlcR transcriptional activator is essential (along with the coinducer acetaldehyde) for the utilization of ethanol as a carbon source in this species. It controls expression of the alcA and aldA genes (133, 200), which encode an alcohol dehydrogenase and an aldehyde dehydrogenase, respectively. These enzymes convert alcohol into acetaldehyde and, secondly, acetaldehyde into acetate. The AlcR activator also undergoes autoregulation (133, 177). In the presence of glucose, transcription of these genes is shut down by binding of the repressor CreA directly to the alcR and alcA promoters (176, 201). As stated above, AlcR binds as a monomer to inverted, everted, and direct repeats found into the promoters of alcR, alcA, and aldA (see Fig. 2).
One important characteristic of filamentous fungi is the production of a wide range of secondary metabolites. Many of these natural compounds are important in medical and/or agricultural fields. Aflatoxins are secondary metabolites produced by several Aspergillus species. These compounds are highly toxic and carcinogenic in animals and humans (292). Indeed, epidemiological studies have established that aflatoxin exposure is a major risk factor for liver cancer (292). Interestingly, the aflatoxin production pathway is regulated by the zinc cluster protein AflR. Genes encoding enzymes of this pathway are located in a 70-kb gene cluster in both the Aspergillus flavus and Aspergillus parasiticus genomes (292). AflR positively regulates the expression of these genes by binding to the consensus motif TCGSWNNSCGR found in the promoters of target genes (58).
Colletotrichum lagenarium is a plant pathogen that causes anthracnose of cucumbers. Infection of host plants by this fungus requires the production of melanin for successful invasion by formation of an infection structure called the appressorium (132). Cmr1p, a Zn(II)2Cys6 protein, is a positive regulator of the expression of melanin biosynthetic structural genes SCD1 and THR1 in this species (266). ZFR1 is another example of a gene encoding a zinc cluster protein involved in secondary metabolite production. In the phytopathogenic fungus Fusarium verticillioides, the biosynthesis of fumonisin B1, a mycotoxin that causes leukoencephalomalacia in equids and pulmonary edema in swine (220), is severely impaired in zfr1 mutants (73). Interestingly, the fungus Penicillium citrinum produces ML-263B (compactin), which can act as a substrate for production of pravastatin sodium, a compound used in hypercholesterolemia therapy (1, 239). The zinc cluster protein MlcRp is a P. citrinum transcriptional regulator that is important in its production (1). The crucial roles of fungal secondary metabolites in various biological fields imply that many other zinc cluster proteins and their key functions are likely to be further investigated in the near future.
CONCLUSION
Since its isolation in 1982 (112, 140), the GAL4 gene has become the focus of numerous studies, leading to in-depth knowledge of its mechanism of action. Gal4p was one of the first eukaryotic transcription factors to be characterized, and it is now considered a classical model for eukaryotic transcription. Over the years, many other zinc cluster proteins have been characterized, and as a result, general rules for these transcription factors (and others) can now be derived.
The zinc finger regions of the members of the Gal4p family have a specific structure that is unique to fungi. X-ray and NMR analyses of the DNA-binding domains of some members of the family show that the cysteine-rich region has a remarkably similar structure that commonly recognizes CGG triplets. Residues flanking the cysteine-rich region are responsible for differences in DNA-binding specificity. For example, changing the relative orientation of the two zinc clusters of a homodimer allows recognition of inverted, direct, or everted repeats. In addition, alteration of the relative distance between the two zinc clusters further increases the repertoire of binding sites by allowing binding of a homodimer to CGG triplets with different spacings. Alternate modes of DNA recognition such as monomeric or heterodimeric binding have also been described.
Zinc cluster proteins function in a wide range of processes, including amino acid and vitamin synthesis, carbon and nitrogen metabolism, meiosis, and morphogenesis. Other roles include regulation of genes involved in the stress response and pleiotropic drug resistance, as demonstrated in budding yeast and in human fungal pathogens. While Gal4p appears to act solely as an activator, a growing number of zinc cluster proteins have been shown to have both activator and repressor capabilities. Genome-wide studies also show that many zinc cluster proteins have both distinct and overlapping functions. In addition, autoregulation and cross-regulation of the expression of zinc cluster proteins are becoming a theme that is more and more common. Mechanisms that regulate activity of zinc cluster proteins include phosphorylation (e.g., Cat8p and Sip4p), binding of a small inducer molecule to the factor (e.g., binding of proline to Put3p), and interaction with a metabolic intermediate (e.g., Leu3p). With the number of characterized zinc cluster proteins growing rapidly, it is becoming more and more apparent that they are crucial regulators of fungal physiology. Furthermore, their potential importance extends toward infectious diseases and the agricultural industry. From its beginnings as a pioneer model for eukaryotic transcription, the study of this family of transcriptional regulators has clearly evolved and reached a much broader significance.
Acknowledgments
Unfortunately, because of the broad scope of this review, many relevant articles could not be cited. We thank Ronen Marmorstein (Wistar Institute, Philadelphia, Pa.) for providing results before publication and Albert Berghuis (McGill University) for advice on use of programs to generate Fig. 2. We also thank Karen Hellauer for critical reading of the manuscript.
This work was supported by grants to B.T. from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Abe, Y., C. Ono, M. Hosobuchi, and H. Yoshikawa. 2002. Functional analysis of mlcR, a regulatory gene for ML-236B (compactin) biosynthesis in Penicillium citrinum. Mol. Genet. Genomics 268**:**352-361. [DOI] [PubMed] [Google Scholar]
- 2.Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. A. Davies, and C. V. Lowry. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157**:**1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akache, B., S. MacPherson, M. A. Sylvain, and B. Turcotte. 2004. Complex interplay among regulators of drug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 279**:**27855-27860. [DOI] [PubMed] [Google Scholar]
- 4.Akache, B., and B. Turcotte. 2002. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J. Biol. Chem. 277**:**21254-21260. [DOI] [PubMed] [Google Scholar]
- 5.Akache, B., K. Q. Wu, and B. Turcotte. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29**:**2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amar, N., F. Messenguy, M. El Bakkoury, and E. Dubois. 2000. ArgRII, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 20**:**2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson, S. F., C. M. Steber, R. E. Esposito, and J. E. Coleman. 1995. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 4**:**1832-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7**:**741-751. [DOI] [PubMed] [Google Scholar]
- 9.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276**:**24309-24314. [DOI] [PubMed] [Google Scholar]
- 10.Avila, J., C. Gonzalez, N. Brito, F. Machin, M. D. Perez, and J. M. Siverio. 2002. A second Zn(II)(2)Cys(6) transcriptional factor encoded by the YNA2 gene is indispensable for the transcriptional activation of the genes involved in nitrate assimilation in the yeast Hansenula polymorpha. Yeast 19**:**537-544. [DOI] [PubMed] [Google Scholar]
- 11.Axelrod, J. D., J. Majors, and M. C. Brandriss. 1991. Proline-independent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol. Cell. Biol. 11**:**564-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai, Y. L., and G. B. Kohlhaw. 1991. Manipulation of the ‘zinc cluster’ region of transcriptional activator LEU3 by site-directed mutagenesis. Nucleic Acids Res. 19**:**5991-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey, L. A., and D. J. Ebbole. 1998. The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 148**:**1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey-Shrode, L., and D. J. Ebbole. 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 166**:**1741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balzi, E., and A. Goffeau. 1994. Genetics and biochemistry of yeast multidrug resistance. Biochim. Biophys. Acta 1187**:**152-162. [DOI] [PubMed] [Google Scholar]
- 16.Becker, B., A. Feller, M. el Alami, E. Dubois, and A. Pierard. 1998. A nonameric core sequence is required upstream of the LYS genes of Saccharomyces cerevisiae for Lys14p-mediated activation and apparent repression by lysine. Mol. Microbiol. 29**:**151-163. [DOI] [PubMed] [Google Scholar]
- 17.Bennett, R., and A. Johnson. 2005. Mating in Candida albicans and the search for a sexual cycle. Annu. Rev. Microbiol. 59**:**233-255. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97**:**13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15**:**1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibbins, M., V. F. Crepin, N. J. Cummings, T. Mizote, K. Baker, K. H. Mellits, and I. F. Connerton. 2002. A regulator gene for acetate utilisation from Neurospora crassa. Mol. Genet. Genomics 267**:**498-505. [DOI] [PubMed] [Google Scholar]
- 21.Bornaes, C., M. W. Ignjatovic, P. Schjerling, M. C. Kielland-Brandt, and S. Holmberg. 1993. A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol. Cell. Biol. 13**:**7604-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowdish, K. S., H. E. Yuan, and A. P. Mitchell. 1995. Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol. 15**:**2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun, B. R., M. van het Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Köhler, J. Morschhäuser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1**:**36-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breunig, K. D. 2000. Regulation of transcription activation by Gal4p. Food Technol. Biotech. 38**:**287-293. [Google Scholar]
- 25.Bricmont, P. A., J. R. Daugherty, and T. G. Cooper. 1991. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 11**:**1161-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown, D. H., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34**:**651-662. [DOI] [PubMed] [Google Scholar]
- 27.Burger, G., J. Strauss, C. Scazzocchio, and B. F. Lang. 1991. nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol. Cell. Biol. 11**:**5746-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cahuzac, B., R. Cerdan, B. Felenbok, and E. Guittet. 2001. The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein. Structure 9**:**827-836. [DOI] [PubMed] [Google Scholar]
- 29.Carvajal, E., H. B. Van denHazel, A. Cybularz Kolaczkowska, E. Balzi, and A. Goffeau. 1997. Molecular and phenotypic characterization of yeast Pdr1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256**:**406-415. [DOI] [PubMed] [Google Scholar]
- 30.Cazelle, B., A. Pokorska, E. Hull, P. M. Green, G. Stanway, and C. Scazzocchio. 1998. Sequence, exon-intron organization, transcription and mutational analysis of prnA, the gene encoding the transcriptional activator of the prn gene cluster in Aspergillus nidulans. Mol. Microbiol. 28**:**355-370. [DOI] [PubMed] [Google Scholar]
- 31.Cecchetto, G., S. Amillis, G. Diallinas, C. Scazzocchio, and C. Drevet. 2004. The AzgA purine transporter of _Aspergillus nidulans_—characterization of a protein belonging to a new phylogenetic cluster. J. Biol. Chem. 279**:**3132-3141. [DOI] [PubMed] [Google Scholar]
- 32.Chan, C. K., S. Hubner, W. Hu, and D. A. Jans. 1998. Mutual exclusivity of DNA binding and nuclear localization signal recognition by the yeast transcription factor GAL4: implications for nonviral DNA delivery. Gene Ther. 5**:**1204-1212. [DOI] [PubMed] [Google Scholar]
- 33.Chan, C. K., and D. A. Jans. 1999. Synergy of importin alpha recognition and DNA binding by the yeast transcriptional activator GAL4. FEBS Lett. 462**:**221-224. [DOI] [PubMed] [Google Scholar]
- 34.Charbon, G., K. D. Breunig, R. Wattiez, J. Vandenhaute, and I. Noel-Georis. 2004. Key role of Ser562/661 in Snf1-dependent regulation, of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell. Biol. 24**:**4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charron, M. J., E. Read, S. R. Haut, and C. A. Michels. 1989. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 122**:**307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, H., and G. R. Fink. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20**:**1150-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung, K. R., M. E. Daub, K. Kuchler, and C. Schuller. 2003. The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 302**:**302-310. [DOI] [PubMed] [Google Scholar]
- 38.Cohen, B. D., O. Sertil, N. E. Abramova, K. J. Davies, and C. V. Lowry. 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29**:**799-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collingwood, T. N., F. D. Urnov, and A. P. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23**:**255-275. [DOI] [PubMed] [Google Scholar]
- 40.Coornaert, D., S. Vissers, and B. André. 1991. The pleiotropic UGA35 (DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene 97**:**163-171. [DOI] [PubMed] [Google Scholar]
- 41.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3**:**1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Côté, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of Gal4 derivative binding to nucleosomal DNA by the yeast Swi/Snf complex. Science 265**:**53-60. [DOI] [PubMed] [Google Scholar]
- 43.Creusot, F., J. Verdiere, M. Gaisne, and P. P. Slonimski. 1988. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol. 204**:**263-276. [DOI] [PubMed] [Google Scholar]
- 44.Crowley, J. H., F. W. Leak, Jr., K. V. Shianna, S. Tove, and L. W. Parks. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180**:**4177-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui, Z., D. Hirata, and T. Miyakawa. 1999. Functional analysis of the promoter of the yeast SNQ2 gene encoding a multidrug resistance transporter that confers the resistance to 4-nitroquinoline _N_-oxide. Biosci. Biotechnol. Biochem. 63**:**162-167. [DOI] [PubMed] [Google Scholar]
- 46.Cui, Z., T. Shiraki, D. Hirata, and T. Miyakawa. 1998. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline _N_-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol. 29**:**1307-1315. [DOI] [PubMed] [Google Scholar]
- 47.Davie, J. K., D. G. Edmondson, C. B. Coco, and S. Y. R. Dent. 2003. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278**:**50158-50162. [DOI] [PubMed] [Google Scholar]
- 48.Davies, B. S. J., H. S. Wang, and J. Rine. 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25**:**7375-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis, M. A., A. J. Small, S. Kourambas, and M. J. Hynes. 1996. The tamA gene of Aspergillus nidulans contains a putative zinc cluster motif which is not required for gene function. J. Bacteriol. 178**:**3406-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Defranoux, N., M. Gaisne, and J. Verdiere. 1994. Functional analysis of the zinc cluster domain of the CYP1 (HAP1) complex regulator in heme-sufficient and heme-deficient yeast cells. Mol. Gen. Genet. 242**:**699-707. [DOI] [PubMed] [Google Scholar]
- 51.Delahodde, A., T. Delaveau, and C. Jacq. 1995. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 15**:**4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delahodde, A., R. Pandjaitan, M. Corral-Debrinski, and C. Jacq. 2001. Pse1/Kap121-dependent nuclear localization of the major yeast multidrug resistance (MDR) transcription factor Pdr1. Mol. Microbiol. 39**:**304-312. [DOI] [PubMed] [Google Scholar]
- 53.Delaveau, T., A. Delahodde, E. Carvajal, J. Subik, and C. Jacq. 1994. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 244**:**501-511. [DOI] [PubMed] [Google Scholar]
- 54.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43**:**1197-1214. [DOI] [PubMed] [Google Scholar]
- 55.Deng, Y., T. He, Y. Wu, P. Vanka, G. Yang, Y. Huang, H. Yao, and S. J. Brown. 2005. Computationally analyzing the possible biological function of _YJL103C_—an ORF potentially involved in the regulation of energy process in yeast. Int. J. Mol. Med. 15**:**123-127. [PubMed] [Google Scholar]
- 56.Dufresne, M., S. Perfect, A. L. Pellier, J. A. Bailey, and I. Langin. 2000. A Gal4-like protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common bean. Plant Cell 12**:**1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehrlich, K. C., B. G. Montalbano, D. Bhatnagar, and T. E. Cleveland. 1998. Alteration of different domains in AFLR affects aflatoxin pathway metabolism in Aspergillus parasiticus transformants. Fungal Genet. Biol. 23**:**279-287. [DOI] [PubMed] [Google Scholar]
- 58.Ehrlich, K. C., B. G. Montalbano, and J. W. Cary. 1999. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230**:**249-257. [DOI] [PubMed] [Google Scholar]
- 59.El Alami, M., A. Feller, A. Pierard, and E. Dubois. 2000. Characterisation of a tripartite nuclear localisation sequence in the regulatory protein Lys14 of Saccharomyces cerevisiae. Curr. Genet. 38**:**78-86. [DOI] [PubMed] [Google Scholar]
- 60.Empel, J., I. Sitkiewicz, A. Andrukiewicz, K. Lasocki, P. Borsuk, and P. Weglenski. 2001. arcA, the regulatory gene for the arginine catabolic pathway in Aspergillus nidulans. Mol. Genet. Genomics 266**:**591-597. [DOI] [PubMed] [Google Scholar]
- 61.Endo, H., S. Kajiwara, O. Tsunoka, and K. Shishido. 1994. A novel cDNA, pribC, encoding a protein with a Zn(II)2cys6 zinc cluster DNA-binding motif, derived from the basidiomycete Lentinus edodes. Gene 139**:**117-121. [DOI] [PubMed] [Google Scholar]
- 62.Entian, K. D., T. Schuster, J. H. Hegemann, D. Becher, H. Feldmann, U. Guldener, R. Gotz, M. Hansen, C. P. Hollenberg, G. Jansen, W. Kramer, S. Klein, P. Kotter, J. Kricke, H. Launhardt, G. Mannhaupt, A. Maierl, P. Meyer, W. Mewes, T. Munder, R. K. Niedenthal, M. R. Rad, A. Rohmer, A. Romer, M. Rose, et al. 1999. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262**:**683-702. [DOI] [PubMed] [Google Scholar]
- 63.Espelin, C. W., K. T. Simons, S. C. Harrison, and P. K. Sorger. 2003. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell 14**:**4557-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falco, S. C., and K. S. Dumas. 1985. Genetic analysis of mutants of Saccharomyces cerevisiae resistant to the herbicide sulfometuron methyl. Genetics 109**:**21-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fankhauser, H., and M. E. Schweingruber. 1994. Thiamine-repressible genes in Schizosaccharomyces pombe are regulated by a Cys(6) zinc-finger motif-containing protein. Gene 147**:**141-144. [DOI] [PubMed] [Google Scholar]
- 66.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21**:**6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Felenbok, B., M. Flipphi, and I. Nikolaev. 2001. Ethanol catabolism in Aspergillus nidulans: a model for studying gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 69**:**149-204. [DOI] [PubMed] [Google Scholar]
- 68.Feller, A., E. Dubois, F. Ramos, and A. Pierard. 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14**:**6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feller, A., F. Ramos, A. Pierard, and E. Dubois. 1999. In Saccharomyces cerevisae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem. 261**:**163-170. [DOI] [PubMed] [Google Scholar]
- 70.Feng, B., and G. A. Marzluf. 1996. The regulatory protein NIT4 that mediates nitrate induction in Neurospora crassa contains a complex tripartite activation domain with a novel leucine-rich, acidic motif. Curr. Genet. 29**:**537-548. [DOI] [PubMed] [Google Scholar]
- 71.Fernandes, M., N. P. Keller, and T. H. Adams. 1998. Sequence-specific binding by Aspergillus nidulans Aflr, a C-6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28**:**1355-1365. [DOI] [PubMed] [Google Scholar]
- 72.Fitzgerald, M. X., J. R. Rojas, J. M. Kim, G. B. Kohlhaw, and R. Marmorstein. 2006. Structure of a Leu3/DNA complex: recognition of everted CGG half sites by a Zn2Cys6 binuclear cluster protein. Structure 14**:**725-735. [DOI] [PubMed] [Google Scholar]
- 73.Flaherty, J. E., and C. P. Woloshuk. 2004. Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster-type gene, ZFR1. Appl. Environ. Microbiol. 70**:**2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flick, K. M., N. Spielewoy, T. I. Kalashnikova, M. Guaderrama, Q. Z. Zhu, H. C. Chang, and C. Wittenberg. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14**:**3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flynn, P., and R. J. Reece. 1999. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol. Cell. Biol. 19**:**882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friden, P., C. Reynolds, and P. Schimmel. 1989. A large internal deletion converts yeast Leu3 to a constitutive transcriptional activator. Mol. Cell. Biol. 9**:**4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao, C., L. M. Wang, E. Milgrom, and W. C. W. Shen. 2004. On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in _Saccharomyces cerevisiae_—evidence for enhanced recruitment of coactivators and altered nucleosome structures. J. Biol. Chem. 279**:**42677-42686. [DOI] [PubMed] [Google Scholar]
- 78.Gardner, K. H., T. Pan, S. Narula, E. Rivera, and J. E. Coleman. 1991. Structure of the binuclear metal-binding site in the GAL4 transcription factor. Biochemistry 30**:**11292-11302. [DOI] [PubMed] [Google Scholar]
- 79.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11**:**4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goldar, M. M., H. T. Jeong, K. Tanaka, H. Matsuda, and M. Kawamukai. 2005. Moc3, a novel Zn finger type protein involved in sexual development, ascus formation, and stress response of Schizosaccharomyces pombe. Curr. Genet. 48**:**345-355. [DOI] [PubMed] [Google Scholar]
- 81.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103**:**423-433. [DOI] [PubMed] [Google Scholar]
- 82.Gomez, D., B. Cubero, G. Cecchetto, and C. Scazzocchio. 2002. PrnA, a Zn(2)Cys(6) activator with a unique DNA recognition mode, requires inducer for in vivo binding. Mol. Microbiol. 44**:**585-597. [DOI] [PubMed] [Google Scholar]
- 83.Gomi, K., T. Akeno, T. Minetoki, K. Ozeki, C. Kumagai, N. Okazaki, and Y. Iimura. 2000. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 64**:**816-827. [DOI] [PubMed] [Google Scholar]
- 84.Gorlich, D., S. Kostka, R. Kraft, C. Dingwall, R. A. Laskey, E. Hartmann, and S. Prehn. 1995. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5**:**383-392. [DOI] [PubMed] [Google Scholar]
- 85.Gorlich, D., U. K. Laemmli, Y. Adachi, and M. Kohler. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. EMBO J. 18**:**4348-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gray, W. M., and J. S. Fassler. 1996. Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol. Cell. Biol. 16**:**347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Groom, K. R., H. C. Heyman, M. C. Steffen, L. Hawkins, and N. C. Martin. 1998. Kluyveromyces lactis SEF1 and its Saccharomyces cerevisiae homologue bypass the unknown essential function, but not the mitochondrial RNase P function, of the S. cerevisiae RPM2 gene. Yeast 14**:**77-87. [DOI] [PubMed] [Google Scholar]
- 88.Ha, N., K. Hellauer, and B. Turcotte. 2000. Fusions with histone H3 result in highly specific alteration of gene expression. Nucleic Acids Res. 28**:**1026-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ha, N., K. Hellauer, and B. Turcotte. 1996. Mutations in target DNA elements of yeast HAP1 modulate its transcriptional activity without affecting DNA binding. Nucleic Acids Res. 24**:**1453-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hallstrom, T. C., D. J. Katzmann, R. J. Torres, W. J. Sharp, and W. S. Moye-Rowley. 1998. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18**:**1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hallstrom, T. C., L. Lambert, S. Schorling, E. Balzi, A. Goffeau, and W. S. Moye-Rowley. 2001. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276**:**23674-23680. [DOI] [PubMed] [Google Scholar]
- 92.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431**:**99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hasper, A. A., L. M. Trindade, D. van der Veen, A. J. J. van Ooyen, and L. H. de Graaff. 2004. Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology 150**:**1367-1375. [DOI] [PubMed] [Google Scholar]
- 94.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36**:**193-200. [DOI] [PubMed] [Google Scholar]
- 95.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. D. Muller, S. Fields, D. Baker, J. R. Yates, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12**:**1353-1365. [DOI] [PubMed] [Google Scholar]
- 96.Hedges, D., M. Proft, and K. D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15**:**1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hellauer, K., B. Akache, S. MacPherson, E. Sirard, and B. Turcotte. 2002. Zinc cluster protein Rdr1p is a transcriptional repressor of the PDR5 gene encoding a multidrug transporter. J. Biol. Chem. 277**:**17671-17676. [DOI] [PubMed] [Google Scholar]
- 98.Hellauer, K., M. H. Rochon, and B. Turcotte. 1996. A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol. Cell. Biol. 16**:**6096-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hiesinger, M., S. Roth, E. Meissner, and H. J. Schuller. 2001. Contribution of Cat8 and Sip4 to the transcriptional activation of yeast gluconeogenic genes by carbon source-responsive elements. Curr. Genet. 39**:**68-76. [DOI] [PubMed] [Google Scholar]
- 100.Hikkel, I., A. Lucau-Danila, T. Delaveau, P. Marc, F. Devaux, and C. Jacq. 2003. A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J. Biol. Chem. 278**:**11427-11432. [DOI] [PubMed] [Google Scholar]
- 101.Hiltunen, J. K., A. M. Mursula, H. Rottensteiner, R. K. Wierenga, A. J. Kastaniotis, and A. Gurvitz. 2003. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEBS Microbiol. Rev. 27**:**35-64. [DOI] [PubMed] [Google Scholar]
- 102.Holmberg, S., and P. Schjerling. 1996. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144**:**467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95**:**717-728. [DOI] [PubMed] [Google Scholar]
- 104.Hon, T., H. C. Lee, Z. Z. Hu, V. R. Iyer, and L. Zhang. 2005. The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae. Genetics 169**:**1343-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang, H. L., and M. C. Brandriss. 2000. The regulator of the yeast proline utilization pathway is differentially phosphorylated in response to the quality of the nitrogen source. Mol. Cell. Biol. 20**:**892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425**:**686-691. [DOI] [PubMed] [Google Scholar]
- 107.Idicula, A. M., G. L. Blatch, T. G. Cooper, and R. A. Dorrington. 2002. Binding and activation by the zinc cluster transcription factors of Saccharomyces cerevisiae—redefining the UAS(GABA) and its interaction with Uga3p. J. Biol. Chem. 277**:**45977-45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iraqui, I., S. Vissers, B. André, and A. Urrestarazu. 1999. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 19**:**3360-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ito, T., T. Chiba, and M. Yoshida. 2001. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol. 19**:**S23-S27. [DOI] [PubMed] [Google Scholar]
- 110.Jackson, J. C., and J. M. Lopes. 1996. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 24**:**1322-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnston, M., and J. Dover. 1987. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc. Natl. Acad. Sci. USA 84**:**2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnston, S. A., and J. E. Hopper. 1982. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc. Natl. Acad. Sci. USA 79**:**6971-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jungwirth, H., and K. Kuchler. 2006. Yeast ABC transporters—a tale of sex, stress, drugs and aging. FEBS Lett. 580**:**1131-1138. [DOI] [PubMed] [Google Scholar]
- 114.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89**:**365-371. [DOI] [PubMed] [Google Scholar]
- 115.Karpichev, I. V., Y. Luo, R. C. Marians, and G. M. Small. 1997. A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of beta-oxidation enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol. 17**:**69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karpichev, I. V., and G. M. Small. 2000. Evidence for a novel pathway for the targeting of a Saccharomyces cerevisiae peroxisomal protein belonging to the isomerase/hydratase family. J. Cell Sci. 113**:**533-544. [DOI] [PubMed] [Google Scholar]
- 117.Karpichev, I. V., and G. M. Small. 1998. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 18**:**6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kasten, M. M., and D. J. Stillman. 1997. Identification of the Saccharomyces cerevisiae genes _STB1_-STB5 encoding Sin3p binding proteins. Mol. Gen. Genet. 256**:**376-386. [DOI] [PubMed] [Google Scholar]
- 119.Katzmann, D. J., P. E. Burnett, J. Golin, Y. Mahe, and W. S. Moye-Rowley. 1994. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14**:**4653-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Katzmann, D. J., T. C. Hallstrom, Y. Mahe, and W. S. Moye-Rowley. 1996. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 271**:**23049-23054. [DOI] [PubMed] [Google Scholar]
- 121.Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson, and A. D. Johnson. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68**:**709-719. [DOI] [PubMed] [Google Scholar]
- 122.Kelly, R., and K. J. Kwon-Chung. 1992. A zinc finger protein from Candida albicans is involved in sucrose utilization. J. Bacteriol. 174**:**222-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim, J. H., J. Polish, and M. Johnston. 2003. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol. Cell. Biol. 23**:**5208-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.King, D. A., L. Zhang, L. Guarente, and R. Marmorstein. 1999. Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein. Nat. Struct. Biol. 6**:**64-71. [DOI] [PubMed] [Google Scholar]
- 125.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13**:**2339-2352. [DOI] [PubMed] [Google Scholar]
- 126.Kirkpatrick, C. R., and P. Schimmel. 1995. Detection of leucine-independent DNA site occupancy of the yeast Leu3p transcriptional activator in vivo. Mol. Cell. Biol. 15**:**4021-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kohlhaw, G. B. 2003. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67**:**1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kolaczkowska, A., and A. Goffeau. 1999. Regulation of pleiotropic drug resistance in yeast. Drug Res. Updates 2**:**403-414. [DOI] [PubMed] [Google Scholar]
- 129.Kolaczkowski, M., A. Kolaczkowska, B. Gaigg, R. Schneiter, and W. S. Moye-Rowley. 2004. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot. Cell 3**:**880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kontoyiannis, D. P., N. Sagar, and K. D. Hirschi. 1999. Overexpression of Erg11p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 43**:**2798-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kren, A., Y. M. Mamnun, B. E. Bauer, C. Schuller, H. Wolfger, K. Hatzixanthis, M. Mollapour, C. Gregori, P. Piper, and K. Kuchler. 2003. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 23**:**1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kubo, Y., and I. Furusawa. 1991. Melanin biosynthesis: prerequisite for successful invasion of the plant host by appressoria of Colletotrichum and Pyricularia, p. 205-217. In G. T. Cole and H. C. Hoch (ed.), The fungal spore and disease initiation in plants and animals. Plenum Publishing, New York, N.Y.
- 133.Kulmburg, P., N. Judewicz, M. Mathieu, F. Lenouvel, D. Sequeval, and B. Felenbok. 1992. Specific binding sites for the activator protein, AlcR, in the alcA promoter of the ethanol regulon of Aspergillus nidulans. J. Biol. Chem. 267**:**21146-21153. [PubMed] [Google Scholar]
- 134.Kwast, K. E., P. V. Burke, and R. O. Poyton. 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201**:**1177-1195. [DOI] [PubMed] [Google Scholar]
- 135.Laity, J. H., B. M. Lee, and P. E. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11**:**39-46. [DOI] [PubMed] [Google Scholar]
- 136.Lamb, H. K., C. F. Roberts, and A. R. Hawkins. 1992. A second gene (qutH) within the _Aspergillus nidulans_-quinic-acid utilisation gene cluster encodes a protein with a putative zinc-cluster motif. Gene 112**:**219-224. [DOI] [PubMed] [Google Scholar]
- 137.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53**:**1451-1469. [DOI] [PubMed] [Google Scholar]
- 138.Larochelle, M., S. Drouin, F. Robert, and B. Turcotte. 2006. Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol. Cell. Biol. 26**:**6690-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15**:**1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laughon, A., and R. F. Gesteland. 1982. Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc. Natl. Acad. Sci. USA 79**:**6827-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lebel, K., S. MacPherson, and B. Turcotte. 2006. New tools for phenotypic analysis in Candida albicans: the WAR1 gene confers resistance to sorbate. Yeast 23**:**249-259. [DOI] [PubMed] [Google Scholar]
- 142.Leberer, E., K. Ziegelbauer, A. Schmidt, D. Harcus, D. Dignard, J. Ash, L. Johnson, and D. Y. Thomas. 1997. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7**:**539-546. [DOI] [PubMed] [Google Scholar]
- 143.Lechner, J. 1994. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 13**:**5203-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Le Crom, S., F. Devaux, P. Marc, X. T. Zhang, W. S. Moye-Rowley, and C. Jacq. 2002. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 22**:**2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee, M. S., G. P. Gippert, K. V. Soman, D. A. Case, and P. E. Wright. 1989. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 245**:**635-637. [DOI] [PubMed] [Google Scholar]
- 146.Lemieux, K., and L. Gaudreau. 2004. Targeting of Swi/Snf to the yeast GAL1 UASG requires the mediator, TAFIIs, and RNA polymerase II. EMBO J. 23**:**4040-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leonardo, J. M., S. M. Bhairi, and R. C. Dickson. 1987. Identification of upstream activator sequences that regulate induction of the beta-galactosidase gene in Kluyveromyces lactis. Mol. Cell. Biol. 7**:**4369-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282**:**1900-1904. [DOI] [PubMed] [Google Scholar]
- 149.Levesley, I., G. H. Newton, H. K. Lamb, E. Vanschothorst, R. W. M. Dalgleish, A. C. R. Samson, C. F. Roberts, and A. R. Hawkins. 1996. Domain structure and function within the QUTA protein of _Aspergillus nidulans_—implications for the control of transcription. Microbiology 142**:**87-98. [DOI] [PubMed] [Google Scholar]
- 150.Li, D. X., and P. E. Kolattukudy. 1997. Cloning of cutinase transcription factor 1, a transactivating protein containing Cys(6)Zn(2) binuclear cluster DNA-binding motif. J. Biol. Chem. 272**:**12462-12467. [DOI] [PubMed] [Google Scholar]
- 151.Li, D. X., T. Sirakova, L. Rogers, W. F. Ettinger, and P. E. Kolattukudy. 2002. Regulation of constitutively expressed and induced cutinase genes by different zinc finger transcription factors in Fusarium solani f. sp. pisi (Nectria haematococca). J. Biol. Chem. 277**:**7905-7912. [DOI] [PubMed] [Google Scholar]
- 152.Li, L., S. He, J. M. Sun, and J. R. Davie. 2004. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 82**:**460-471. [DOI] [PubMed] [Google Scholar]
- 153.Liang, S. D., R. Marmorstein, S. C. Harrison, and M. Ptashne. 1996. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Biol. 16**:**3773-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lo, H. J., J. R. Kohler, B. Didomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90**:**939-949. [DOI] [PubMed] [Google Scholar]
- 155.Lodish, H., D. Baltimore, A. Berk, S. L. Zipursky, P. Matsudaira, and J. Darnell. 1995. Molecular cell biology, 3rd ed., vol. 1. Scientific American Books, Inc., New York, N.Y.
- 156.Lohr, D., P. Venkov, and J. Zlatanova. 1995. Transcriptional regulation in the yeast GAL gene family—a complex genetic network. FASEB J. 9**:**777-787. [DOI] [PubMed] [Google Scholar]
- 157.Losson, R., and F. Lacroute. 1981. Cloning of a eukaryotic regulatory gene. Mol. Gen. Genet. 184**:**394-399. [DOI] [PubMed] [Google Scholar]
- 158.Lucau-Danila, A., T. Delaveau, G. Lelandais, F. Devaux, and C. Jacq. 2003. Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J. Biol. Chem. 278**:**52641-52650. [DOI] [PubMed] [Google Scholar]
- 159.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8**:**76-81. [DOI] [PubMed] [Google Scholar]
- 160.Lussier, M., A. M. Sdicu, E. Winnett, D. H. Vo, J. Sheraton, A. Dusterhoft, R. K. Storms, and H. Bussey. 1997. Completion of the Saccharomyces cerevisiae genome sequence allows identification of KTR5, KTR6 and KTR7 and definition of the nine-membered KRE2/MNT1 mannosyltransferase gene family in this organism. Yeast 13**:**267-274. [DOI] [PubMed] [Google Scholar]
- 161.Ma, J., and M. Ptashne. 1987. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 51**:**113-119. [DOI] [PubMed] [Google Scholar]
- 162.MacIsaac, K. D., T. Wang, D. B. Gordon, D. K. Gifford, G. D. Stormo, and E. Fraenkel. 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7**:**113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.MacPherson, S., B. Akache, S. Weber, X. De Deken, M. Raymond, and B. Turcotte. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49**:**1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Madhani, H. D. 2000. Interplay of intrinsic and extrinsic signals in yeast differentiation. Proc. Natl. Acad. Sci. USA 97**:**13461-13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mahe, Y., A. Parle McDermott, A. Nourani, A. Delahodde, A. Lamprecht, and K. Kuchler. 1996. The ATP-binding cassette multidrug transporter Snq2 of _Saccharomyces cerevisiae_—a novel target for the transcription factors Pdr1 and Pdr3. Mol. Microbiol. 20**:**109-117. [DOI] [PubMed] [Google Scholar]
- 166.Mamane, Y., K. Hellauer, M. H. Rochon, and B. Turcotte. 1998. A linker region of the yeast zinc cluster protein Leu3p specifies binding to everted repeat DNA. J. Biol. Chem. 273**:**18556-18561. [DOI] [PubMed] [Google Scholar]
- 167.Mamnun, Y. M., R. Pandjaitan, Y. Mahe, A. Delahodde, and K. Kuchler. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 46**:**1429-1440. [DOI] [PubMed] [Google Scholar]
- 168.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356**:**408-414. [DOI] [PubMed] [Google Scholar]
- 169.Marmorstein, R., and S. C. Harrison. 1994. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 8**:**2504-2512. [DOI] [PubMed] [Google Scholar]
- 170.Martens, J. A., J. Genereaux, A. Saleh, and C. J. Brandl. 1996. Transcriptional activation by yeast Pdr1p is inhibited by its association with Ngg1p/Ada3p. J. Biol. Chem. 271**:**15884-15890. [DOI] [PubMed] [Google Scholar]
- 171.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429**:**571-574. [DOI] [PubMed] [Google Scholar]
- 172.Martens, J. A., P. Y. J. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19**:**2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marui, J., N. Kitamoto, M. Kato, T. Kobayashi, and N. Tsukagoshi. 2002. Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett. 528**:**279-282. [DOI] [PubMed] [Google Scholar]
- 174.Marui, J., A. Tanaka, S. Mimura, L. H. de Graaff, J. Visser, N. Kitamoto, M. Kato, T. Kobayashi, and N. Tsukagoshi. 2002. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet. Biol. 35**:**157-169. [DOI] [PubMed] [Google Scholar]
- 175.Masloff, S., S. Poggeler, and U. Kuck. 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152**:**191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mathieu, M., and B. Felenbok. 1994. The Aspergillus nidulans CreA protein mediates glucose repression of the ethanol regulon at various levels through competition with the AlcR-specific transactivator. EMBO J. 13**:**4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mathieu, M., S. Fillinger, and B. Felenbok. 2000. In vivo studies of upstream regulatory cis-acting elements of the alcR gene encoding the transactivator of the ethanol regulon in Aspergillus nidulans. Mol. Microbiol. 36**:**123-131. [DOI] [PubMed] [Google Scholar]
- 178.Mendizabal, I., G. Rios, J. M. Mulet, R. Serrano, and I. F. de Larrinoa. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425**:**323-328. [DOI] [PubMed] [Google Scholar]
- 179.Miller, J., A. D. McLachlan, and A. Klug. 1985. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4**:**1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mizuguchi, G., T. Tsukiyama, J. Wisniewski, and C. Wu. 1997. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell 1**:**141-150. [DOI] [PubMed] [Google Scholar]
- 181.Moreno, I., Y. Pedreno, S. Maicas, R. Sentandreu, E. Herrero, and E. Valentin. 2003. Characterization of a Candida albicans gene encoding a putative transcriptional factor required for cell wall integrity. FEMS Microbiol. Lett. 226**:**159-167. [DOI] [PubMed] [Google Scholar]
- 182.Mosley, A. L., J. Lakshmanan, B. K. Aryal, and S. Ozcan. 2003. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278**:**10322-10327. [DOI] [PubMed] [Google Scholar]
- 183.Moye-Rowley, W. S. 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acid Res. Mol. Biol. 73**:**251-279. [DOI] [PubMed] [Google Scholar]
- 184.Muratani, M., C. Kung, K. M. Shokat, and W. R. Tansey. 2005. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell 120**:**887-899. [DOI] [PubMed] [Google Scholar]
- 185.Mylin, L. M., M. Johnston, and J. E. Hopper. 1990. Phosphorylated forms of GAL4 are correlated with ability to activate transcription. Mol. Cell. Biol. 10**:**4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Naar, A. M., S. Ryu, and R. Tjian. 1998. Cofactor requirements for transcriptional activation by Sp1. Cold Spring Harbor Symp. Quant. Biol. 63**:**189-199. [DOI] [PubMed] [Google Scholar]
- 187.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99**:**677-690. [DOI] [PubMed] [Google Scholar]
- 188.Narendja, F., S. P. Goller, M. Wolschek, and J. Strauss. 2002. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol. Microbiol. 44**:**573-583. [DOI] [PubMed] [Google Scholar]
- 189.Needleman, R. 1991. Control of maltase synthesis in yeast. Mol. Microbiol. 5**:**2079-2084. [DOI] [PubMed] [Google Scholar]
- 190.Ness, F., S. Bourot, M. Regnacq, R. Spagnoli, T. Berges, and F. Karst. 2001. SUT1 is a putative Zn(II)2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur. J. Biochem. 268**:**1585-1595. [PubMed] [Google Scholar]
- 191.Niedenthal, R., L. Riles, U. Guldener, S. Klein, M. Johnston, and J. H. Hegemann. 1999. Systematic analysis of S. cerevisiae chromosome VIII genes. Yeast 15**:**1775-1796. [DOI] [PubMed] [Google Scholar]
- 192.Nikolaev, I., M. F. Cochet, and B. Felenbok. 2003. Nuclear import of zinc binuclear cluster proteins proceeds through multiple, overlapping transport pathways. Eukaryot. Cell 2**:**209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nikolaev, I., F. Lenouvel, and B. Felenbok. 1999. Unique DNA binding specificity of the binuclear zinc AlcR activator of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem. 274**:**9795-9802. [DOI] [PubMed] [Google Scholar]
- 194.Nishimura, H., Y. Kawasaki, Y. Kaneko, K. Nosaka, and A. Iwashima. 1992. Cloning and characteristics of a positive regulatory gene, THI2 (PHO6), of thiamin biosynthesis in Saccharomyces cerevisiae. FEBS Lett. 297**:**155-158. [DOI] [PubMed] [Google Scholar]
- 195.Noel, J., and B. Turcotte. 1998. Zinc cluster proteins Leu3p and Uga3p recognize highly related but distinct DNA targets. J. Biol. Chem. 273**:**17463-17468. [DOI] [PubMed] [Google Scholar]
- 196.Nourani, A., D. Papajova, A. Delahodde, C. Jacq, and J. Subik. 1997. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol. Gen. Genet. 256**:**397-405. [DOI] [PubMed] [Google Scholar]
- 197.Ozcan, S., and M. Johnston. 1995. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15**:**1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ozcan, S., T. Leong, and M. Johnston. 1996. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 16**:**6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pan, T., and J. E. Coleman. 1990. The DNA binding domain of GAL4 forms a binuclear metal ion complex. Biochemistry 29**:**2023-2029. [DOI] [PubMed] [Google Scholar]
- 200.Panozzo, C., V. Capuano, S. Fillinger, and B. Felenbok. 1997. The zinc binuclear cluster activator AlcR is able to bind to single sites but requires multiple repeated sites for synergistic activation of the alcA gene in Aspergillus nidulans. J. Biol. Chem. 272**:**22859-22865. [DOI] [PubMed] [Google Scholar]
- 201.Panozzo, C., E. Cornillot, and B. Felenbok. 1998. The CreA repressor is the sole DNA-binding protein responsible for carbon catabolite repression of the alcA gene in Aspergillus nidulans via its binding to a couple of specific sites. J. Biol. Chem. 273**:**6367-6372. [DOI] [PubMed] [Google Scholar]
- 202.Park, H. D., R. M. Luche, T. G. Cooper, and H. J. Park. 1992. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 20**:**1909-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Parks, L. W., and W. M. Casey. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49**:**95-116. [DOI] [PubMed] [Google Scholar]
- 204.Parsons, L. M., M. A. Davis, and M. J. Hynes. 1992. Identification of functional regions of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol. Microbiol. 6**:**2999-3007. [DOI] [PubMed] [Google Scholar]
- 205.Pfeifer, K., K. S. Kim, S. Kogan, and L. Guarente. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56**:**291-301. [DOI] [PubMed] [Google Scholar]
- 206.Pinto, W. J., R. Lozano, B. C. Sekula, and W. R. Nes. 1983. Stereochemically distinct roles for sterol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 112**:**47-54. [DOI] [PubMed] [Google Scholar]
- 207.Pla, J., C. Gil, L. Monteoliva, F. Navarrogarcia, M. Sanchez, and C. Nombela. 1996. Understanding Candida albicans at the molecular level. Yeast 12**:**1677-1702. [DOI] [PubMed] [Google Scholar]
- 208.Polish, J. A., J. H. Kim, and M. Johnston. 2005. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 169**:**583-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Punt, P. J., J. Strauss, R. Smit, J. R. Kinghorn, C. A. van den Hondel, and C. Scazzocchio. 1995. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell. Biol. 15**:**5688-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Qui, H. F., E. Dubois, and F. Messenguy. 1991. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol. Cell. Biol. 11**:**2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ramgopal, M., and K. Bloch. 1983. Sterol synergism in yeast. Proc. Natl. Acad. Sci. USA 80**:**712-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Randez-Gil, F., N. Bojunga, M. Proft, and K. D. Entian. 1997. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol. Cell. Biol. 17**:**2502-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rank, G. H., A. J. Robertson, and K. L. Phillips. 1975. Modification and inheritance of pleiotropic cross resistance and collateral sensitivity in Saccharomyces cerevisiae. Genetics 80**:**483-493. [PMC free article] [PubMed] [Google Scholar]
- 214.Reece, R. J., and M. Ptashne. 1993. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science 261**:**909-911. [DOI] [PubMed] [Google Scholar]
- 215.Régnacq, M., P. Alimardani, B. E. Moudni, and T. Berges. 2001. Sut1p interaction with Cyc8p(Ssn6p) relieves hypoxic genes from Cyc8p-Tup1p repression in Saccharomyces cerevisiae. Mol. Microbiol. 40**:**1085-1096. [DOI] [PubMed] [Google Scholar]
- 216.Rerngsamran, P., M. B. Murphy, S. A. Doyle, and D. J. Ebbole. 2005. Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene. Mol. Microbiol. 56**:**282-297. [DOI] [PubMed] [Google Scholar]
- 217.Rodriguez, R. J., C. Low, C. D. Bottema, and L. W. Parks. 1985. Multiple functions for sterols in Saccharomyces cerevisiae. Biochim. Biophys. Acta 837**:**336-343. [DOI] [PubMed] [Google Scholar]
- 218.Rodriguez, R. J., F. R. Taylor, and L. W. Parks. 1982. A requirement for ergosterol to permit growth of yeast sterol auxotrophs on cholestanol. Biochem. Biophys. Res. Commun. 106**:**435-441. [DOI] [PubMed] [Google Scholar]
- 219.Rohde, J. R., J. Trinh, and I. Sadowski. 2000. Multiple signals regulate GAL transcription in yeast. Mol. Cell. Biol. 20**:**3880-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ross, P. F., P. E. Nelson, J. L. Richard, G. D. Osweiler, L. G. Rice, R. D. Plattner, and T. M. Wilson. 1990. Production of fumonisins by Fusarium moniliforme and Fusarium proliferatum isolates associated with equine leukoencephalomalcia and a pulmonary edema syndrome in swine. Appl. Environ. Microbiol. 56**:**3225-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Roth, S., J. Kumme, and H. J. Schuller. 2004. Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr. Genet. 45**:**121-128. [DOI] [PubMed] [Google Scholar]
- 222.Rottensteiner, H., A. J. Kal, M. Filipits, M. Binder, B. Hamilton, H. F. Tabak, and H. Ruis. 1996. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 15**:**2924-2934. [PMC free article] [PubMed] [Google Scholar]
- 223.Rottensteiner, H., A. J. Kal, B. Hamilton, H. Ruis, and H. F. Tabak. 1997. A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur. J. Biochem. 247**:**776-783. [DOI] [PubMed] [Google Scholar]
- 224.Rubin-Bejerano, I., S. Mandel, K. Robzyk, and Y. Kassir. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16**:**2518-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rustad, T. R., D. A. Stevens, M. A. Pfaller, and T. C. White. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148**:**1061-1072. [DOI] [PubMed] [Google Scholar]
- 226.Rutzler, M., A. Reissaus, M. Budzowska, and W. Bandlow. 2004. SUT2 is a novel multicopy suppressor of low activity of the cAMP/protein kinase A pathway in yeast. Eur. J. Biochem. 271**:**1284-1291. [DOI] [PubMed] [Google Scholar]
- 227.Sa-Correia, I., and S. Tenreiro. 2002. The multidrug resistance transporters of the major facilitator superfamily, 6 years after disclosure of Saccharomyces cerevisiae genome sequence. J. Biotechnol. 98**:**215-226. [DOI] [PubMed] [Google Scholar]
- 228.Sadowski, I., C. Costa, and R. Dhanawansa. 1996. Phosphorylation of Gal4p at a single C-terminal residue is necessary for galactose-inducible transcription. Mol. Cell. Biol. 16**:**4879-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sadowski, I., D. Niedbala, K. Wood, and M. Ptashne. 1991. GAL4 is phosphorylated as a consequence of transcriptional activation. Proc. Natl. Acad. Sci. USA 88**:**10510-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sanglard, D., F. Ischer, D. Calabrese, M. Demicheli, and J. Bille. 1998. Multiple resistance mechanisms to azole antifungals in yeast clinical isolates. Drug Res. Updates 1**:**255-265. [DOI] [PubMed] [Google Scholar]
- 231.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2**:**73-85. [DOI] [PubMed] [Google Scholar]
- 232.Saunders, G. W., and G. H. Rank. 1982. Allelism of pleiotropic drug resistance in Saccharomyces cerevisiae. Can. J. Genet. Cytol. 24**:**493-503. [DOI] [PubMed] [Google Scholar]
- 233.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24**:**4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43**:**139-160. [DOI] [PubMed] [Google Scholar]
- 235.Sekito, T., J. Thornton, and R. A. Butow. 2000. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 11**:**2103-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sellick, C. A., and R. J. Reece. 2005. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem. Sci. 30**:**405-412. [DOI] [PubMed] [Google Scholar]
- 237.Sellick, C. A., and R. J. Reece. 2003. Modulation of transcription factor function by an amino acid: activation of Put3p by proline. EMBO J. 22**:**5147-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sequeval, D., and B. Felenbok. 1994. Relationship between zinc content and DNA-binding activity of the DNA-binding motif of the transcription factor ALCR in Aspergillus nidulans. Mol. Gen. Genet. 242**:**33-39. [DOI] [PubMed] [Google Scholar]
- 239.Serizawa, N., S. Serizawa, K. Nakagawa, K. Furuya, T. Okazaki, and A. Terahara. 1983. Microbial hydroxylation of ML-236B (compactin). Studies on microorganisms capable of 3 beta-hydroxylation of ML-236B. J. Antibiot. 36**:**887-891. [DOI] [PubMed] [Google Scholar]
- 240.Shaikh, S., B. Burke, G. Dreyfuss, and S. Nakielny. 1999. Transport of proteins and RNAs in and out of the nucleus. EMBO J. 18**:**1982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Shianna, K. V., W. D. Dotson, S. Tove, and L. W. Parks. 2001. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J. Bacteriol. 183**:**830-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Siddiqui, A. H., and M. C. Brandriss. 1989. The Saccharomyces cerevisiae Put3 activator protein associates with proline-specific upstream activation sequences. Mol. Cell. Biol. 9**:**4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Silver, P. M., S. G. Oliver, and T. C. White. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3**:**1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sirenko, O. I., B. Ni, and R. B. Needleman. 1995. Purification and binding properties of the Mal63p activator of Saccharomyces cerevisiae. Curr. Genet. 27**:**509-516. [DOI] [PubMed] [Google Scholar]
- 245.Sprague, G. F., and S. C. Winans. 2006. Eukaryotes learn how to count: quorum sensing by yeast. Genes Dev. 20**:**1045-1049. [DOI] [PubMed] [Google Scholar]
- 246.St. Georgiev, V. 2000. Membrane transporters and antifungal drug resistance. Curr. Drug Targets 1**:**261-284. [DOI] [PubMed] [Google Scholar]
- 247.Strauss, J., M. I. Muropastor, and C. Scazzocchio. 1998. The regulator of nitrate assimilation in ascomycetes is a dimer which binds a nonrepeated, asymmetrical sequence. Mol. Cell. Biol. 18**:**1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Strich, R., R. T. Surosky, C. Steber, E. Dubois, F. Messenguy, and R. E. Esposito. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8**:**796-810. [DOI] [PubMed] [Google Scholar]
- 249.Struhl, K. 1995. Yeast transcriptional regulatory mechanisms. Annu. Rev. Genet. 29**:**651-674. [DOI] [PubMed] [Google Scholar]
- 250.Strunnikov, A. V., J. Kingsbury, and D. Koshland. 1995. Cep3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 128**:**749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Suarez, T., M. V. de Queiroz, N. Oestreicher, and C. Scazzocchio. 1995. The sequence and binding specificity of UaY, the specific regulator of the purine utilization pathway in Aspergillus nidulans, suggest an evolutionary relationship with the PPR1 protein of Saccharomyces cerevisiae. EMBO J. 14**:**1453-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Subik, J., S. Ulaszewski, and A. Goffeau. 1986. Genetic mapping of nuclear mucidin resistance mutations in Saccharomyces cerevisiae. A new PDR locus on chromosome II. Curr. Genet. 10**:**665-670. [DOI] [PubMed] [Google Scholar]
- 253.Swaminathan, K., P. Flynn, R. J. Reece, and R. Marmorstein. 1997. Crystal structure of a Put3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn(2)Cys(6) binuclear cluster. Nat. Struct. Biol. 4**:**751-759. [DOI] [PubMed] [Google Scholar]
- 254.Sze, J. Y., M. Woontner, J. A. Jaehning, and G. B. Kohlhaw. 1992. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258**:**1143-1145. [DOI] [PubMed] [Google Scholar]
- 255.Talibi, D., M. Grenson, and B. André. 1995. Cis- and trans-acting elements determining induction of the genes of the gamma-aminobutyrate (GABA) utilization pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 23**:**550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Talibi, D., and M. Raymond. 1999. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 181**:**231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tang, C. S., A. Bueno, and P. Russell. 1994. _ntf1_+ encodes a 6-cysteine zinc finger-containing transcription factor that regulates the nmt1 promoter in fission yeast. J. Biol. Chem. 269**:**11921-11926. [PubMed] [Google Scholar]
- 258.Tani, S., Y. Katsuyama, T. Hayashi, H. Suzuki, M. Kato, K. Gomi, T. Kobayashi, and N. Tsukagoshi. 2001. Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr. Genet. 39**:**10-15. [DOI] [PubMed] [Google Scholar]
- 259.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272**:**408-411. [DOI] [PubMed] [Google Scholar]
- 260.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21**:**388-405. [DOI] [PubMed] [Google Scholar]
- 261.Todd, R. B., A. Andrianopoulos, M. A. Davis, and M. J. Hynes. 1998. FacB, the Aspergillus nidulans activator of acetate utilization genes, binds dissimilar DNA sequences. EMBO J. 17**:**2042-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Todd, R. B., R. L. Murphy, H. M. Martin, J. A. Sharp, M. A. Davis, M. E. Katz, and M. J. Hynes. 1997. The acetate regulatory gene FACB of Aspergillus nidulans encodes a Zn(II)2Cys6 transcriptional activator. Mol. Gen. Genet. 254**:**495-504. [DOI] [PubMed] [Google Scholar]
- 263.Traven, A., B. Jelicic, and M. Sopta. 2006. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 7**:**496-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tsai, H.-F., A. A. Krol, K. E. Sarti, and J. E. Bennett. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50**:**1384-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90**:**109-119. [DOI] [PubMed] [Google Scholar]
- 266.Tsuji, G., Y. Kenmochi, Y. Takano, J. Sweigard, L. Farrall, I. Furusawa, O. Horino, and Y. Kubo. 2000. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 38**:**940-954. [DOI] [PubMed] [Google Scholar]
- 267.Turcotte, B., B. Akache, and S. MacPherson. 2004. The zinc cluster proteins: a yeast family of transcriptional regulators, p. 233-249. In S. G. Pandalai (ed.), Recent developments in nucleic acid research, vol. 1. Transworld Research Network, Trivandrum, Kerala, India. [Google Scholar]
- 268.Turi, T. G., and J. C. Loper. 1992. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J. Biol. Chem. 267**:**2046-2056. [PubMed] [Google Scholar]
- 269.Uhl, M. A., M. Biery, N. Craig, and A. D. Johnson. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 22**:**2668-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Urnov, F. D. 2002. A feel for the template: zinc finger protein transcription factors and chromatin. Biochem. Cell Biol. 80**:**321-333. [DOI] [PubMed] [Google Scholar]
- 271.van Vuuren, H. J., J. R. Daugherty, R. Rai, and T. G. Cooper. 1991. Upstream induction sequence, the _cis_-acting element required for response to the allantoin pathway inducer and enhancement of operation of the nitrogen-regulated upstream activation sequence in Saccharomyces cerevisiae. J. Bacteriol. 173**:**7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Vashee, S., H. Xu, S. A. Johnston, and T. Kodadek. 1993. How do “Zn2 Cys6” proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J. Biol. Chem. 268**:**24699-24706. [PubMed] [Google Scholar]
- 273.Verdiere, J., M. Gaisne, and R. Labbe-Bois. 1991. CYP1 (HAP1) is a determinant effector of alternative expression of heme-dependent transcribed genes in yeast. Mol. Gen. Genet. 228**:**300-306. [DOI] [PubMed] [Google Scholar]
- 274.Vik, A., and J. Rine. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21**:**6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vincent, K., Q. Wang, S. Jay, K. Hobbs, and B. C. Rymond. 2003. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics 164**:**895-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Vincent, O., and M. Carlson. 1998. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17**:**7002-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Vissers, S., B. André, F. Muyldermans, and M. Grenson. 1990. Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur. J. Biochem. 187**:**611-616. [DOI] [PubMed] [Google Scholar]
- 278.Walters, K. J., K. T. Dayie, R. J. Reece, M. Ptashne, and G. Wagner. 1997. Structure and mobility of the Put3 dimer. Nat. Struct. Biol. 4**:**744-750. [DOI] [PubMed] [Google Scholar]
- 279.Wang, H., and D. J. Stillman. 1993. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol. Cell. Biol. 13**:**1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang, Q., and B. C. Rymond. 2003. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol. Cell. Biol. 23**:**7339-7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. X. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14**:**2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wehrschutz-Sigl, E., H. Jungwirth, H. Bergler, and G. Hogenauer. 2004. The transporters Pdr5p and Snq2p mediate diazaborine resistance and are under the control of the gain-of-function allele _PDR1_-12. Eur. J. Biochem. 271**:**1145-1152. [DOI] [PubMed] [Google Scholar]
- 283.Whiteway, M., D. Dignard, and D. Y. Thomas. 1992. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc. Natl. Acad. Sci. USA 89**:**9410-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Wilcox, L. J., D. A. Balderes, B. Wharton, A. H. Tinkelenberg, G. Rao, and S. L. Sturley. 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277**:**32466-32472. [DOI] [PubMed] [Google Scholar]
- 285.Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis, C. S. de Menthiere, R. W. Davis, and R. E. Esposito. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99**:**13431-13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Witte, M. M., and R. C. Dickson. 1988. Cysteine residues in the zinc finger and amino acids adjacent to the finger are necessary for DNA binding by the LAC9 regulatory protein of Kluyveromyces lactis. Mol. Cell. Biol. 8**:**3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wolfe, S. A., L. Nekludova, and C. O. Pabo. 2000. DNA recognition by Cys(2)His(2) zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29**:**183-212. [DOI] [PubMed] [Google Scholar]
- 288.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2004. The yeast Pdr15p ATP-binding cassette (ABC) protein is a general stress response factor implicated in cellular detoxification. J. Biol. Chem. 279**:**11593-11599. [DOI] [PubMed] [Google Scholar]
- 289.Woloshuk, C. P., K. R. Foutz, J. F. Brewer, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 1994. Molecular characterization of Aflr, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60**:**2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wray, L., Jr., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7**:**1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wu, J. S., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7**:**117-126. [DOI] [PubMed] [Google Scholar]
- 292.Yabe, K., and H. Nakajima. 2004. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 64**:**745-755. [DOI] [PubMed] [Google Scholar]
- 293.Yuan, G. F., Y. H. Fu, and G. A. Marzluf. 1991. nit-4, a pathway-specific regulatory gene of Neurospora crassa, encodes a protein with a putative binuclear zinc DNA-binding domain. Mol. Cell. Biol. 11**:**5735-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zhang, L., and L. Guarente. 1994. The yeast activator Hap1—a Gal4 family member—binds DNA in a directly repeated orientation. Genes Dev. 8**:**2110-2119. [DOI] [PubMed] [Google Scholar]
- 295.Zhang, L., and A. Hach. 1999. Molecular mechanism of heme signalling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56**:**415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Zhang, X. T., Z. F. Cui, T. Miyakawa, and W. S. Moye-Rowley. 2001. Cross-talk between transcriptional regulators of multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276**:**8812-8819. [DOI] [PubMed] [Google Scholar]
- 297.Zhou, K. M., and G. B. Kohlhaw. 1990. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of the activation domain tryptophans. J. Biol. Chem. 265**:**17409-17412. [PubMed] [Google Scholar]
- 298.Zhu, Y., and W. Xiao. 2004. Pdr3 is required for DNA damage induction of MAG1 and DDI1 via a bi-directional promoter element. Nucleic Acids Res. 32**:**5066-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]