Widespread Adaptive Evolution of Drosophila Genes With Sex-Biased Expression (original) (raw)
Abstract
Many genes in higher eukaryotes show sexually dimorphic expression, and these genes tend to be among the most divergent between species. In most cases, however, it is not known whether this rapid divergence is caused by positive selection or if it is due to a relaxation of selective constraint. To distinguish between these two possibilities, we surveyed DNA sequence polymorphism in 91 Drosophila melanogaster genes with male-, female-, or nonsex-biased expression and determined their divergence from the sister species D. simulans. Using several single- and multilocus statistical tests, we estimated the type and strength of selection influencing the evolution of the proteins encoded by genes of each expression class. Adaptive evolution, as indicated by a relative excess of nonsynonymous divergence between species, was common among the sex-biased genes (both male and female). Male-biased genes, in particular, showed a strong and consistent signal of positive selection, while female-biased genes showed more variation in the type of selection they experience. Genes expressed equally in the two sexes, in contrast, showed no evidence for adaptive evolution between D. melanogaster and D. simulans. This suggests that sexual selection and intersexual coevolution are the major forces driving genetic differentiation between species.
MALES and females of animal species often differ in many morphological and behavioral traits. This sexual dimorphism has long fascinated biologists and served as the inspiration for Darwin's theory of sexual selection (Darwin 1871). Recent microarray studies have revealed that sexual dimorphism is also common at the level of gene expression (Parisi et al. 2003; Ranz et al. 2003; Gibson et al. 2004). For example, ∼30% of all genes in Drosophila melanogaster show a twofold or greater difference in expression between the sexes (Parisi et al. 2004). Comparative genomic studies have shown that such sex-biased genes, particularly those with male-biased expression, are among the most rapidly evolving genes between species (Swanson et al. 2001; Zhang et al. 2004; Khaitovich et al. 2005; Richards et al. 2005). This raises the possibility that adaptive processes, such as sexual selection, may drive the evolution of a large number of genes with sexually dimorphic expression (Civetta and Singh 1999; Singh and Kulathinal 2000). An alternate possibility, however, is that sex-biased genes evolve under relaxed selective constraint, which allows them to accumulate more neutral (or nearly neutral) changes between species. For instance, the product of an autosomal gene with sex-specific expression will be visible to selection only over half of its evolutionary history when it is in the appropriate sex. The rest of the time, it will be in the sex where it is not expressed and will be invisible to selection. Thus, it may experience only half as much purifying selection as a gene expressed equally in the two sexes (Barker et al. 2005).
In some well-studied cases, the rapid evolution of male-biased genes has been attributed to positive selection (Swanson and Vacquier 2002). In particular, the male reproductive genes of Drosophila, including those encoding accessory gland proteins (Acp's), appear to be a rich source of adaptively evolving genes (Tsaur and Wu 1997; Tsaur et al. 1998; Aguadé 1998, 1999; Nurminsky et al. 1998; Ting et al. 1998; Begun et al. 2000; Betrán and Long 2003). However, the evolutionary forces affecting the vast majority of male-biased genes are unknown. Although they have been less studied than male-biased genes, there is also evidence for positive selection driving the rapid evolution of particular female-biased genes (Swanson and Vacquier 2002). In these cases, either cooperative or antagonistic coevolution between male and female reproductive proteins is thought to play an important role (Civetta and Singh 2005). For example, a survey of expressed sequence tags (ESTs) from the female reproductive tract of D. simulans uncovered a number of adaptively evolving genes that may be the female counterparts of rapidly evolving male reproductive genes (Swanson et al. 2004).
A powerful method to distinguish the selective forces influencing a gene's evolution is to use combined polymorphism and divergence. Genes that have evolved adaptively are expected to show relatively little polymorphism within species, but high divergence between species. Genes under relaxed selective constraint, in contrast, should show higher levels of polymorphism within species that are proportional to their divergence between species. We have used this approach to determine the selective forces influencing the evolution of sex-biased genes. We surveyed DNA sequence polymorphism in 91 D. melanogaster genes with male-, female-, or non-sex-biased expression and also determined their divergence from the sister species D. simulans. Using statistical tests that compare ratios of polymorphism and divergence at synonymous and nonsynonymous sites, we inferred the type and strength of selection affecting the proteins encoded by genes of the three expression classes. We find that adaptive evolution is common among sex-biased genes (both male and female), but rare among non-sex-biased genes. This suggests that sexual selection and intersexual coevolution play major roles in the genetic differentiation of species.
MATERIALS AND METHODS
Gene selection:
Genes with sex-biased expression were selected on the basis of their male/female (or testes/ovaries) expression ratios, as determined by microarray experiments that used D. melanogaster (Parisi et al. 2003; Ranz et al. 2003; Gibson et al. 2004). For male-biased genes, we required that the ratio be >2.0 (mean = 15.2), while for the female-biased genes we required a ratio <0.5 (mean = 0.23). In other words, we required at least a twofold expression difference between the sexes for a gene to be classified as sex biased. Non-sex-biased genes were required to have a male/female expression ratio between 0.75 and 1.25 (mean = 1.01). In general, the male-biased genes showed more extreme expression differences between the sexes than the female-biased genes, reflecting the pattern that is seen genomewide (Gibson et al. 2004; Parisi et al. 2004). Because the above three experiments used different microarray platforms, not all genes were represented in each experiment. However, for 44 (48%) of the genes, the sex-bias classification could be confirmed by all three experiments. An additional 43 (47%) genes were confirmed by two of the three experiments. The remaining genes (4 male-biased genes) were confirmed by additional microarray experiments (Andrews et al. 2000; Stolc et al. 2004). Because only one of the above experiments also compared male and female expression in D. simulans (Ranz et al. 2003), we could not confirm the bias of all genes in this species. However, of the 61 genes with data from both species, 60 (98%) showed the same sex-bias classification. This included 22 male-biased genes, 25 female-biased genes, and 13 non-sex-biased genes. The one conflicting gene (CG4570) was female biased in D. melanogaster, but non-sex biased in D. simulans. This gene showed no evidence for selection (supplemental Table S1 at http://www.genetics.org/supplemental/) and removing it from our analysis does not affect our results or conclusions.
In addition to the expression criteria, genes were also selected to fall within a relatively narrow size distribution and to have similar intron/exon structures. This was done to remove the influence of coding sequence or intron length on the ratio of nonsynonymous/synonymous polymorphism or divergence (Comeron and Kreitman 2002; Comeron and Guthrie 2005). The mean lengths (standard deviations) for male-, female-, and non-sex-biased genes were 1006 (325), 1098 (372), and 821 (167) bp, respectively. Because male-biased genes are known to be underrepresented on the X chromosome (Parisi et al. 2003; Ranz et al. 2003), we limited our analysis to autosomal genes. It is important to note that functional information or measures of interspecific divergence were not considered in gene selection. Thus, aside from the selection criteria outlined above, our sample represents a random collection of sex-biased (and non-sex-biased) genes that is expected to be representative of the genome as a whole.
PCR and DNA sequencing:
Oligonucleotide primers flanking the coding sequence of each gene were designed on the basis of the complete D. melanogaster genome sequence (release 4.0; http://www.flybase.org) and used for PCR with genomic DNA from 12 highly inbred D. melanogaster lines derived from Lake Kariba, Zimbabwe (Glinka et al. 2003), and one highly inbred D. simulans line derived from Chapel Hill, North Carolina (Meiklejohn et al. 2004). A complete list of the PCR primers, as well as the cycling conditions used for each gene, is provided in supplemental Table S2 at http://www.genetics.org/supplemental/. PCR products were purified with ExoSAP-IT (United States Biochemical, Cleveland, OH). Sequencing of PCR products (both strands) was carried out using BigDye chemistry and a 3730 automated sequencer (Applied Biosystems, Foster City, CA). The PCR primers were also used as sequencing primers. When necessary to get complete sequence coverage of the entire coding region, additional internal sequencing primers were used (supplemental Table S2). For some genes, we were unable to get successful PCR or DNA sequence from all 12 D. melanogaster strains (see supplemental Table S1 at http://www.genetics.org/supplemental/). The average number of strains sequenced per gene was 11. For 25 genes, we were unable to obtain a PCR product from D. simulans using our primers designed to D. melanogaster. In these cases, we used the sequence from the D. simulans genome project (Washington University School of Medicine Genome Sequencing Center) downloaded from the UCSC Genome Browser (http://genome-test.cse.ucsc.edu/).
Analysis:
Sequences were edited using either Sequencher (Gene Codes, Ann Arbor, MI) or DNAstar (Madison, WI) software with manual adjustments to the alignments. Polymorphism and divergence statistics were calculated using DnaSP 4 (Rozas et al. 2003). For McDonald–Kreitman (MK) table data, we used the number of segregating mutations (instead of the number of segregating sites), because some genes had sites with three segregating variants. In these cases, the frequency of each mutation was considered separately for calculation of Tajima's D and the identification of singleton polymorphisms. For divergence, we considered only sites with fixed differences between all D. melanogaster lines and D. simulans. The fraction of positively selected amino acid substitutions, α, its 95% confidence intervals, and a likelihood-ratio test for positive selection were calculated using the program DoFE (kindly provided by A. Eyre-Walker). The selection parameter, γ, its 95% confidence intervals, and the proportion of the distribution falling below zero were calculated using the MKPRF web server (http://cbsuapps.tc.cornell.edu/mkprf.aspx). Multilocus HKA and Tajima's D tests were performed using the program HKA, which was kindly provided by J. Hey (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm).
Our polymorphism survey revealed a few potential annotation errors in genome release 4.0. One female-biased gene (CG17361) had a frameshift-causing insertion (relative to the annotated ORF) in some D. melanogaster lines (2 bp) and in D. simulans (1 bp). This occurred 42 bp downstream of the start codon. The ORF was otherwise intact with both _d_N/_d_S and πN/πS < 1, suggesting that it is maintained by purifying selection. Another in-frame ATG codon is present 90 bp downstream of the annotated start codon and we used this as the starting point of our alignment. Two unbiased genes (CG17404 and CG18553) had frameshift-causing deletions (1 and 2 bp, respectively) segregating in D. melanogaster. Both genes had otherwise intact ORFs with _d_N/_d_S and πN/πS < 1, suggesting functional constraint on the coding sequence. It is possible that these deletions fall within unannotated introns. For our analyses, we ignored these sites with deletions. Elimination of the three above genes from our analyses has negligible effect on our results and does not alter the conclusions of this article.
RESULTS
To investigate the type and strength of selection influencing the evolution of sex-biased genes, we surveyed DNA sequence polymorphism in 91 protein-encoding genes in a sample of 12 highly inbred D. melanogaster isofemale lines from Zimbabwe, Africa (Table 1 and supplemental Table S1 at http://www.genetics.org/supplemental/). The genes were selected on the basis of previously published microarray results (Parisi et al. 2003; Ranz et al. 2003; Gibson et al. 2004), which allowed them to be separated into three expression classes: male biased, female biased, and non-sex biased. For the sex-biased genes, we required at least a twofold difference in expression between the sexes, while for the non-sex-biased genes we required the difference to be <1.25-fold. In all cases, the expression difference was confirmed by at least two independent microarray experiments. The Zimbabwe population of D. melanogaster was chosen because it is an ancestral, near-equilibrium population that is expected to be largely free from confounding demographic factors, such as population expansion or subdivision (Glinka et al. 2003; Ometto et al. 2005). For each gene, we also determined interspecific divergence using a single sequence from D. simulans.
TABLE 1.
Summary of polymorphism and divergence statistics
| Bias | No. of genes | Significant MK testsa | Positive selectionb | _D_sc | _P_sd | _D_ne | _P_nf | _P_-valueg |
|---|---|---|---|---|---|---|---|---|
| Male | 33 | 7 | 7 | 744 | 447 | 370 | 112 | <0.0001 |
| Female | 28 | 6 | 3 | 631 | 233 | 299 | 90 | 0.15 |
| Non-sex | 30 | 1 | 0 | 436 | 267 | 121 | 83 | 0.51 |
The combination of within-species polymorphism and between-species divergence data allows the application of powerful statistical methods to detect departures from neutral evolution. For example, The HKA test (Hudson et al. 1987) compares the ratio of polymorphism to divergence at two (or more) loci. Under neutrality, these ratios are expected to be equal. A departure from the neutral expectation could be caused by selective or demographic factors. For the 91 genes in our survey, a multilocus HKA test was highly significant (χ2 = 181.1, P < 0.001). In contrast, Ometto et al. (2005) detected no significant departure from neutrality for 232 noncoding loci (introns and intergenic regions) sequenced in the same Zimbabwe population sample. This suggests that the departure observed for our genes is caused by selection and not the demographic history of the population.
The combination of polymorphism and divergence data also allows the application of powerful statistical methods to infer the type and strength of selection affecting groups of protein-encoding genes. In general, these methods are based on the MK test (McDonald and Kreitman 1991), which compares the ratio of polymorphism and divergence at synonymous sites to that at nonsynonymous sites. Under a neutral model of molecular evolution, the two ratios are expected to be equal. A relative excess of nonsynonymous divergence is indicative of positive selection favoring amino acid replacements between species. A relative excess of nonsynonymous polymorphism could be caused either by balancing selection, which maintains amino acid polymorphism within a species, or by weak purifying selection, which allows slightly deleterious nonsynonymous mutations to segregate as low-frequency polymorphisms, but not become fixed between species.
Application of individual MK tests to the genes in our survey revealed interesting selective differences among genes of the three expression classes. Strikingly, ∼20% of the genes in both the male- and the female-biased classes gave a significant MK test result (Table 1). All of the significant male-biased genes departed from neutrality in the direction of positive selection, while only half of the significant female-biased genes were indicative of positive selection (Table 2). The other half departed from neutrality in a pattern consistent with either balancing or weak purifying selection. The former should increase the frequency of polymorphic amino acids within a population and, thus, increase Tajima's D statistic (Tajima 1989) at nonsynonymous sites. However, there was no evidence for this within the female-biased genes in general (Table 3) or within the individual genes showing significant MK tests in this direction (Table 2). For the female-biased genes with a significant excess of nonsynonymous polymorphism, the average Tajima's D at nonsynonymous sites was −1.38, which is far lower than the average for all other female-biased genes of −0.39. This suggests that the observed departures from the neutral expectation are due to weak purifying selection against nonsynonymous mutations. Only one of the non-sex-biased genes showed a significant departure from neutrality by the MK test (Tables 1 and 2), and this gene was also consistent with weak purifying selection. Thus, both groups of sex-biased genes showed evidence for increased positive selection relative to non-sex-biased genes. For the genes showing significant evidence for positive selection, the average Tajima's D at nonsynonymous sites was −0.30, which is well above the average for male- and female-biased genes (see Table 3), but still lower than the average D at synonymous sites in these same genes (−0.09). Thus, it may be that some amino acid positions in these genes have been subject to weak purifying selection, while others have been subject to positive selection.
TABLE 2.
Genes with significant McDonald–Kreitman tests
| Gene | Bias | _D_s | _P_s | _D_n | _P_n | _P_-valuea | Positive selection? | TDsynb | TDnonc |
|---|---|---|---|---|---|---|---|---|---|
| CG3085 | Male | 25 | 41 | 5 | 1 | 0.028 | Yes | −0.45 | −0.07 |
| CG5565 | Male | 11 | 15 | 13 | 5 | 0.047 | Yes | −0.07 | −1.30 |
| CG6255 | Male | 28 | 25 | 10 | 1 | 0.011 | Yes | 0.57 | 1.83 |
| CG8564 | Male | 30 | 23 | 27 | 7 | 0.026 | Yes | −0.45 | −0.69 |
| CG10750 | Male | 21 | 20 | 10 | 0 | 0.004 | Yes | −0.33 | — |
| CG11475 | Male | 31 | 39 | 22 | 4 | 0.000 | Yes | 0.06 | −1.04 |
| CG18418 | Male | 27 | 33 | 13 | 5 | 0.040 | Yes | −0.01 | 0.25 |
| CG3509 | Female | 26 | 13 | 35 | 6 | 0.048 | Yes | 0.36 | −0.24 |
| CG3975 | Female | 22 | 30 | 31 | 17 | 0.029 | Yes | −0.68 | −1.11 |
| CG4973 | Female | 41 | 6 | 9 | 12 | 0.000 | No | −0.66 | −1.30 |
| CG6874 | Female | 17 | 4 | 22 | 0 | 0.048 | Yes | 0.11 | — |
| CG9273 | Female | 18 | 9 | 2 | 6 | 0.035 | No | −0.19 | −1.92 |
| CG12276 | Female | 41 | 3 | 8 | 5 | 0.008 | No | 0.02 | −0.91 |
| CG3476 | Nonsex | 17 | 16 | 0 | 6 | 0.027 | No | −0.99 | −1.56 |
TABLE 3.
Average values of Tajima's D
| Bias | Synonymous | Nonsynonymous |
|---|---|---|
| Male | −0.27 (0.10) | −0.76 (<0.001) |
| Female | −0.03 (0.59) | −0.51 (<0.001) |
| Non-sex | −0.11 (0.40) | −0.62 (<0.001) |
An MK test using the summed polymorphism and divergence values within each class of genes indicated a significant departure from neutrality in the direction of positive selection for the male-biased genes (Table 1). Female-biased genes also showed an excess of nonsynonymous divergence consistent with positive selection, although this was not significant. Non-sex-biased genes did not differ from the neutral expectation and showed a slight, though insignificant, excess of within-species nonsynonymous polymorphism.
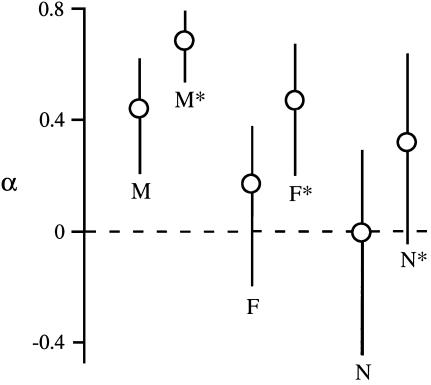
The MK test framework can be expanded to multilocus polymorphism and divergence data to estimate the average type and strength of selection affecting groups of genes. We used a maximum-likelihood method (Bierne and Eyre-Walker 2004) to estimate α, the fraction of amino acid replacements between species that can be attributed to positive selection, within each class of genes (Figure 1). For the male-biased genes, we estimate that 44% of all amino acid replacements were driven by positive selection, while for female-biased genes the estimate is 13%. This fraction is significantly greater than zero for the male-biased genes (likelihood-ratio test, P < 0.001), but not for the female-biased genes. Non-sex-biased genes, in contrast, showed no evidence for positive selection and, if anything, showed evidence for weak purifying selection (α < 0). If weak purifying selection is common in all classes of genes, then the above values of α will be underestimates. Indeed, the observation that nonsynonymous polymorphisms segregate at lower frequency than synonymous polymorphisms, as indicated by Tajima's _D_-statistic (Table 3), suggests that weak purifying selection affects all three classes of genes. To reduce the effect of weak purifying selection, we repeated the above analysis after removing all low-frequency (singleton) polymorphisms at both synonymous and nonsynonymous sites (Figure 1). This led to α-estimates of 69, 47, and 33% for male-, female-, and non-sex-biased genes, respectively. These fractions are significantly greater than zero for the male- and female-biased genes (likelihood-ratio test, P < 0.001 and P < 0.01, respectively), but not for the non-sex-biased genes. The removal of singleton polymorphisms had a particularly strong effect on the female-biased genes, where the ratio of nonsynonymous to synonymous singletons (56/94 = 0.60) was greater than that for male-biased genes (75/195 = 0.38; χ2 = 4.1, P = 0.04) and non-sex-biased genes (44/103 = 0.43; χ2 = 1.8, P = 0.18).
Figure 1.—
The fraction of positively selected amino acid replacements between species, α, for genes with male-biased (M), female-biased (F), and non-sex-biased (N) expression. The corresponding estimates for each group with all low-frequency (singleton) polymorphisms excluded are indicated by asterisks. Error bars represent 95% confidence intervals.
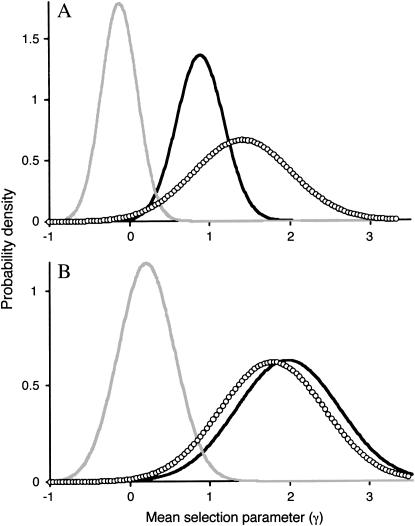
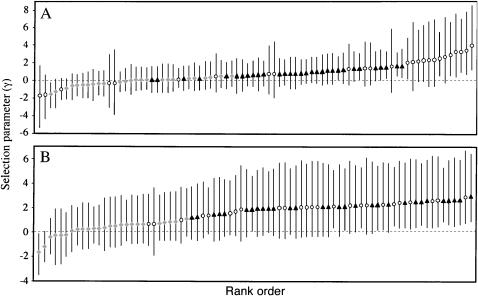
We also estimated the average strength of selection for amino acid replacements within each group of genes using a Bayesian analysis method (Bustamante et al. 2002). With this approach, the MK table data are used to estimate a selection parameter, γ = 2_N_e_s_, where _N_e is the effective population size and s is the selection coefficient. The estimated selection parameters were greater than zero for both male- and female-biased genes, with mean values of 0.9 and 1.4, respectively (Figures 2A and 3A). For both male- and female-biased genes, the proportion of the distribution of mean γ falling below zero was <1% [P(y<0) < 0.001 and P(y<0) < 0.01, respectively]. This indicates positive selection favoring amino acid replacements, with the strongest selection occurring in female-biased genes. However, the variance in the mean γ was quite large for female-biased genes and its distribution showed considerable overlap with that of the male-biased genes (Figure 2A). Non-sex-biased genes had a mean γ that was slightly (but not significantly) less than zero (γ = −0.1), again suggesting that there is weak purifying selection against nonsynonymous mutations. As above, we repeated our analysis after excluding all low-frequency polymorphisms (Figures 2B and 3B). This resulted in more similar estimates of the mean γ for male- and female-biased genes (2.0 and 1.8, respectively), and in both cases the proportion of the distribution of mean γ falling below zero was <0.01%. Non-sex-biased genes had a positive value of γ (0.2), although this was not significantly greater than zero. Removal of singleton polymorphisms had the largest effect on the mean γ of the male-biased genes, which increased by over twofold, and also had a major effect on the distribution of the sex-biased and non-sex-biased genes. After removal of the singletons, there was an almost complete separation of sex-biased and non-sex-biased genes (Figure 3B), with all of the sex-biased genes having γ-values greater than zero.
Figure 2.—
Bayesian posterior distribution of the mean selection parameter (γ) for male-biased (solid line), female-biased (open circles), and non-sex-biased (shaded line) genes. (A) Distribution of mean γ using all polymorphic sites. (B) Distribution of mean γ after excluding all low-frequency (singleton) polymorphisms.
Figure 3.—
Estimated selection parameter (γ) for each gene. Male-biased genes are indicated by solid triangles, female-biased genes by open circles, and non-sex-biased genes by shaded diamonds. Error bars represent 95% confidence intervals. (A) Estimated γ for each gene using all polymorphic sites. (B) Estimated γ for each gene after excluding all low-frequency (singleton) polymorphisms.
DISCUSSION
Our analyses of polymorphism and divergence indicate that adaptive evolution occurs more frequently in sex-biased genes (both male and female) than in non-sex-biased genes. Male-biased genes, in particular, appear to be consistent targets of positive selection. Female-biased genes show more variance in the type of selection they experience, with positive selection affecting some genes and purifying selection affecting others. Non-sex-biased genes appear to evolve primarily under purifying selection and have undergone relatively little adaptive evolution since the split of D. melanogaster and D. simulans. These results argue against the hypothesis that the rapid evolution of sex-biased genes is the result of relaxed selective constraint (see Introduction). This hypothesis predicts that the ratio of nonsynonymous to synonymous polymorphism within species should equal the ratio of nonsynonymous to synonymous divergence between species. However, we find a general excess of nonsynonymous divergence in the sex-biased genes that is reflected in their positive values of the selection parameters α and γ (Figures 1 and 2) and indicates that positive selection has driven their evolution at the protein level.
The finding that male-biased genes show high rates of adaptive evolution is consistent with previous reports that looked at interspecific divergence and the relationship between protein divergence and local recombination rate (Zhang et al. 2004; Zhang and Parsch 2005). However, those studies did not find evidence for adaptive evolution in female-biased genes. A possible explanation for this is that the previous studies used a set of genes cloned from an EST survey (Domazet-Loso and Tautz 2003) that was enriched for highly expressed genes. The female-biased genes, in particular, showed exceptionally high levels of both absolute expression and synonymous codon usage bias (Hambuch and Parsch 2005). This suggests that the EST collection was composed of an unusually constrained set of female-biased genes subject to strong purifying selection. A further difference between the present and the previous studies is that the latter did not include extensive within-species polymorphism data. Thus, the previous studies had less power to detect adaptive evolution and could not account for differences in selective constraint among genes.
Although the selection parameters α and γ are defined differently (the former as the fraction of positively selected amino acid substitutions and the latter as their average scaled selection coefficient), both are calculated from the same MK table data. Thus, one would expect the two measures to be highly correlated. However, we observe a marked difference between the two with respect to the female-biased genes, where the relative level of positive selection is greater when measured by γ (compare Figures 1 and 2). The reason for this appears to be in the way the two methods are implemented. Both assume that synonymous sites evolve neutrally and use the ratio of divergence to polymorphism at these sites to determine a neutral standard. In the method of Bierne and Eyre-Walker (2004), α is calculated separately for each group of genes (male-, female-, and non-sex biased), using only the synonymous sites from that particular group, while in the method of Bustamante et al. (2002), γ is calculated for each group of genes using the combined synonymous sites of all genes as the neutral standard. In our data, the female-biased genes have a higher ratio of divergence to polymorphism at synonymous sites (631/233 = 2.71) than both the male-biased (744/447 = 1.66) and the non-sex-biased genes (436/267 = 1.63). This can explain the observed discordance in selection parameter between the two methods. If we recalculate γ using the synonymous sites of each group of genes separately, we obtain estimates of 1.23, 0.62, and −0.02 for male-, female-, and non-sex-biased genes, respectively, which agrees well with the estimates of α. Using this approach, γ for female-biased genes is no longer significantly greater than zero (P = 0.065). When all singleton polymorphisms are removed, the γ-estimates increase to 2.39, 0.89, and 0.36, for male-, female-, and non-sex-biased genes, respectively, and γ for the female-biased genes is significantly greater than zero (P = 0.002).
It is not clear why the ratio of polymorphism to divergence at synonymous sites is elevated in the female-biased genes relative to the other two groups. One possibility is that the three groups of genes experience differential selection for synonymous codon usage. On a genomewide scale, significant differences in codon bias have been observed among groups of sex-biased genes (Hambuch and Parsch 2005). However, it was male-biased genes that differed significantly from female- and non-sex-biased genes, while the latter two groups showed equal levels of codon bias. This pattern does not correspond to the pattern seen for polymorphism and divergence at synonymous sites. Furthermore, for the genes included in the present study the frequencies of optimal codon usage (_F_op) (Ikemura 1981) are 0.51, 0.53, and 0.56, for the male-, female-, and non-sex-biased genes, respectively, which also do not correspond to the pattern seen for polymorphism and divergence at synonymous sites.
Why does adaptive evolution occur so frequently in sex-biased genes? We first consider the male-biased genes. In a highly polygamous species, such as D. melanogaster, in which there is no paternal investment in offspring and females are able to store the sperm from a single mating to fertilize a lifetime's worth of eggs, sexual selection among males is expected to be very strong. This is evident in the intense sperm competition that occurs among males, which is influenced by accessory gland proteins and other male-expressed genes (Clark et al. 1995, 1999). Indeed, some of these proteins are known to affect a male's reproductive output and show clear signs of adaptive evolution (Herndon and Wolfner 1995; Tsaur and Wu 1997; Aguadé 1998, 1999; Tsaur et al. 1998; Begun et al. 2000; Chapman et al. 2000). Acp's, however, represent only a small fraction (<10%) of genes with male-biased expression (Swanson et al. 2001), and none of the genes in the current study are known Acp's. This suggests that many other male-biased genes may be either directly or indirectly involved in determining reproductive success and, thus, subject to sexual selection (Zhang and Parsch 2005). Indeed, laboratory evolution experiments have shown that, when subject to strong male–male competition (or released from it), Drosophila males show heritable changes in many aspects of their reproductive biology and behavior (Rice 1996; Holland and Rice 1999), which are presumably controlled by a wide variety of genes.
What drives the adaptive evolution of female-biased genes? Because there is less variation in reproductive success among Drosophila females than males, sexual selection is expected to be much weaker in females. However, sexual selection on male traits may lead to rapid coevolution of female reproductive traits or vice versa. In some cases, the coevolution may be considered cooperative, with males and females sharing the same evolutionary interests. One possible example is the correlated evolution of male sperm length and female seminal receptacle length in Drosophila species (Pitnick et al. 1999; Miller and Pitnick 2002). However, it is also possible that in this and many other cases, conflict between male and female reproductive interests drives coevolution. For example, it may be that the strong selection pressure on males to maximize paternity leads to the fixation of traits that are harmful to females, which, in turn, leads to selection for females that can counteract their effect. Indeed, components of male seminal fluid, including Acp's, are known to have deleterious effects on mated females (Chapman et al. 1995; Wigby and Chapman 2005). Furthermore, sexually antagonistic (or “arms race”) coevolution has been demonstrated in laboratory populations of D. melanogaster, where sexually selected males are known to shorten the life span of their naive female mates (Rice 1996). Females that have coevolved with males, however, are able to avoid these damaging consequences, indicating that they adapt in response to the males in their environment. Although the genes underlying these coadapted female traits are unknown, several female-expressed genes showing the molecular hallmarks of sexually antagonistic coevolution, including a significant excess of nonsynonymous divergence between species, have been recently identified (Swanson et al. 2004).
In summary, we propose that the increased signal of positive selection seen for genes with sex-biased expression results from the combined action of sexual selection and intersexual coevolution. The former should affect primarily males, while the latter will affect both males and females. This provides a biological explanation for why the signal of selection is stronger and more consistent for male-biased genes, but weaker and more variable for the female-biased genes. An alternate explanation is that only a subset of the genes with female-biased expression may be free to evolve in response to male traits, while another subset is under strong selective constraint to perform essential functions during development. This would also lead to increased variance in the selection parameter estimate for female-biased genes. In addition, it may be that many female counterparts of rapidly evolving male reproductive genes are expressed in both sexes and/or in nonreproductive tissues and, thus, would not be identified as female biased from the microarray expression data. If this is the case, then the role of sexual antagonism in molecular evolution may be greater than suggested by our results.
Acknowledgments
We thank H. Gebhart and Y. Cämmerer for excellent technical assistance in the laboratory; A. Eyre-Walker and J. Hey for kindly providing software; the Genome Sequencing Center at Washington University School of Medicine for providing D. simulans sequence data; and J. Baines, L. Rose, W. Stephan, D. Rand, and two anonymous reviewers for comments on the manuscript. This work was supported by grant PA 903/2 from the Deutsche Forschungsgemeinschaft.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AM293861–AM294919.
References
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150**:** 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152**:** 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, J., G. G. Bouffard, C. Cheadle, J. Lü, K. G. Becker et al., 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10**:** 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, M. S., J. P. Demuth and M. J. Wade, 2005. Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genet. 1**:** e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156**:** 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán, E., and M. Long, 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164**:** 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2004. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 21**:** 1350–1360. [DOI] [PubMed] [Google Scholar]
- Bustamante, C. D., R. Nielsen, S. A. Sawyer, K. M. Olsen, M. D. Purugganan et al., 2002. The cost of inbreeding in Arabidopsis. Nature 416**:** 531–534. [DOI] [PubMed] [Google Scholar]
- Chapman, T., L. F. Liddle, J. M. Kalb, M. F. Wolfner and L. Partridge, 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373**:** 241–244. [DOI] [PubMed] [Google Scholar]
- Chapman, T., D. M. Neubaum, M. F. Wolfner and L. Partridge, 2000. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc. Biol. Sci. 267**:** 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1999. Broad-sense sexual selection, sex gene pool evolution, and speciation. Genome 42**:** 1033–1041. [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 2005. Rapid evolution of sex-related genes: sexual conflict or sex-specific adaptations? pp. 13–21 in Selective Sweep, edited by D. Nurminsky. Landes Bioscience, Georgetown, TX.
- Clark, A. G., M. Aguadé, T. Prout, L. G. Harshman and C. H. Langley, 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics 139**:** 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., D. J. Begun and T. Prout, 1999. Female X male interactions in Drosophila sperm competition. Science 283**:** 217–220. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., and T. B. Guthrie, 2005. Intragenic Hill-Robertson interference influences selection intensity on synonymous mutations in Drosophila. Mol. Biol. Evol. 22**:** 2519–2530. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., and M. Kreitman, 2002. Population, evolutionary and genomic consequences of interference selection. Genetics 161**:** 389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C., 1871. The Descent of Man, and Selection in Relation to Sex. John Murray, London.
- Domazet-Loso, T., and D. Tautz, 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13**:** 2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, G., R. Riley-Berger, L. Harshman, A. Kopp, S. Vacha et al., 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167**:** 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and D. De Lorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics 165**:** 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambuch, T. M., and J. Parsch, 2005. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics 170**:** 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon, L. A., and M. F. Wolfner, 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc. Natl. Acad. Sci. USA 92**:** 10114–10118.7479736 [Google Scholar]
- Holland, B., and W. R. Rice, 1999. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc. Natl. Acad. Sci. USA 96**:** 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M.A Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116**:** 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura, T., 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151**:** 389–409. [DOI] [PubMed] [Google Scholar]
- Khaitovich, P., I. Hellmann, W. Enard, K. Nowick, M. Leinweber et al., 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309**:** 1850–1854. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351**:** 652–654. [DOI] [PubMed] [Google Scholar]
- Meiklejohn, C. D., Y. Kim, D. L. Hartl and J. Parsch, 2004. Identification of a locus under complex positive selection in Drosophila simulans by haplotype mapping and composite-likelihood estimation. Genetics 168**:** 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G. T., and S. Pitnick, 2002. Sperm-female coevolution in Drosophila. Science 298**:** 1230–1233. [DOI] [PubMed] [Google Scholar]
- Nurminsky, D. I., M. V. Nurminskaya, D. De Aguiar and D. L. Hartl, 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396**:** 572–575. [DOI] [PubMed] [Google Scholar]
- Ometto, L., S. Glinka, D. De Lorenzo and W. Stephan, 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22**:** 2119–2130.15987874 [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299**:** 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, P. Edwards, J. Minor, D. Naiman et al., 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5**:** R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick, S., T. Markow and G. S. Spicer, 1999. Evolution of multiple kinds of sperm storage organs in Drosophila. Evolution 53**:** 1804–1822. [DOI] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300**:** 1742–1745. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381**:** 232–234. [DOI] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and _cis_-element evolution. Genome Res. 15**:** 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19**:** 2496–2497. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., and R. J. Kulathinal, 2000. Sex gene pool evolution and speciation: a new paradigm. Genes Genet. Syst. 75**:** 119–130. [DOI] [PubMed] [Google Scholar]
- Stolc, V., Z. Gauhar, C. Mason, G. Halasz, M. F. van Batenburg et al., 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306**:** 655–660. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3**:** 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98**:** 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., A. Wong, M. F. Wolfner and C. F. Aquadro, 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics 168**:** 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123**:** 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur, M. L. Wu and C. I. Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282**:** 1501–1504. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., and C. I. Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14**:** 544–549. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C. I. Wu, 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol. Biol. Evol. 15**:** 1040–1046. [DOI] [PubMed] [Google Scholar]
- Wigby, S., and T. Chapman, 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15**:** 316–321. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and J. Parsch, 2005. Positive correlation between evolutionary rate and recombination rate in Drosophila genes with male-biased expression. Mol. Biol. Evol. 22**:** 1945–1947. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., T. M. Hambuch and J. Parsch, 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21**:** 2130–2139. [DOI] [PubMed] [Google Scholar]