Skeletrophin, a Novel Ubiquitin Ligase to the Intracellular Region of Jagged-2, Is Aberrantly Expressed in Multiple Myeloma (original) (raw)
Abstract
Recent research has indicated that ligand-dependent activation of the Notch receptor in stromal cells plays a crucial role in the tumorigenesis of multiple myeloma. Ubiquitination of intracellular regions of Notch receptor and its ligands is important for Notch signal transduction. In vitro autoubiquitination analysis using recombinant proteins identified skeletrophin as a novel RING finger-dependent ubiquitin ligase. Skeletrophin bound the intracellular regions of the Notch ligand Jagged-2, but not to those of Delta-1, -3, -4, or Jagged-1. Skeletrophin, but not its RING-mutated form, ubiquitinized the intracellular region of Jagged-2. In malignant plasma cells from 23 of 40 multiple myeloma specimens, strong skeletrophin expression was observed, especially from patients with osteolytic bone lesions. Cytoplasmic localization, which may indicate Jagged-2 internalization, was found in many skeletrophin-positive myeloma cells. In contrast, skeletrophin was only weakly expressed in a few nonneoplasmic plasma cells in chronically inflamed tissues. Interestingly, exogenous expression of skeletrophin, but not the RING-mutated form, in Jagged-2-positive P3U1 myeloma cells induced Hes-1 (Hairy and Enhancer of Split homolog-1) gene expression in Notch receptor-positive bone marrow stromal cells through direct cell-cell contact. Thus, skeletrophin is a novel ubiquitin ligase that targets the intracellular region of Jagged-2 and is aberrantly overexpressed in multiple myeloma cells, possibly activating Hes-1 on stromal cells through ligand-dependent Notch signaling.
Skeletrophin was first identified as a novel cytoplasmic protein, the expression of which was induced by a truncated SWI1 molecule, which is a component of the chromatin-remodeling complex.1 Notably, an analysis of the molecular structure revealed that the skeletrophin protein has two RING finger domains. An increasing number of proteins containing RING finger domains have been characterized as ubiquitin ligase (E3).2,3 The ubiquitination pathway, by the action of the cascade consisting of the E1 ubiquitin-activating enzyme (E1), the E2 ubiquitin-conjugating enzymes, and the E3 ubiquitin ligases, activates and transfers ubiquitin to the substrates. In addition, the ubiquitination pathway plays various biological roles including pathogenesis of various tumors.4 In most cases, E3 mediates specific recognition of substrates to transfer ubiquitin from E2 to substrate.
The ubiquitination of the intracellular region of receptors and their ligands commonly plays a pivotal role in ligand-dependent receptor activation. Of particular note, it is well recognized that ubiquitination of the Notch receptor is important for proteolytic cleavage, endocytosis, and signal transduction.5 Interestingly, endocytosis of the Notch ligand is also essential for ligand-dependent Notch activation.6,7
Recently, a skeletrophin-related molecule, DIP-1 and the (identical) mib molecule, which together with skeletrophin may constitute a new subfamily of ubiquitin ligase, was found to be involved in the endocytosis of Delta, a Notch ligand.6 Jin and colleagues8 identified a novel murine E3 ubiquitin ligase, designated DIP-1 (DAPK-interacting protein), which targets a death-associated protein kinase (DAPK). DIP-1 promotes tumor necrosis factor-induced apoptosis. Itoh and colleagues6 isolated a novel ubiquitin ligase, designated mib, which was thought to be responsible for the phenotype of the zebrafish mind bomb mutant. Interestingly, mib ubiquitin ligase mediated endocytosis of Delta by ubiquitination, thereby facilitating transendocytosis of the Notch extracellular domain, resulting in the transportation of the Notch intracellular domain to the nucleus, where it activates various genes. Further studies eventually showed that DIP-1 and mib were in fact the same molecule. Because DIP has been used for the nomenclature for the distinct molecule, mDia-interacting protein,9 before Jin and colleagues8 reporting their identified molecule, we provisionally use the name DIP-1/mib for this RING ubiquitin ligase.
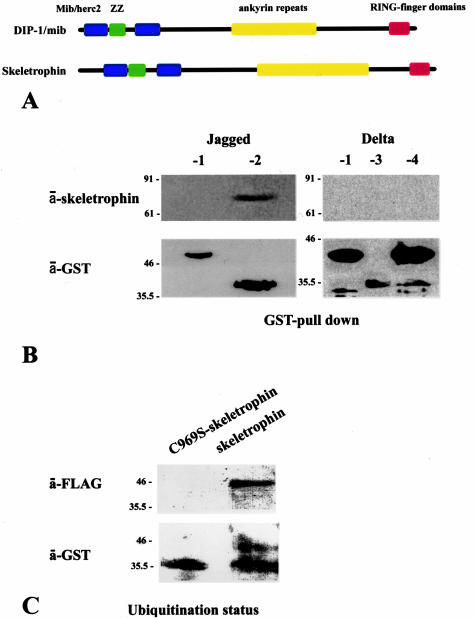
The molecular structure of skeletrophin is closely related to that of human DIP-1/mib protein. Both skeletrophin and DIP-1/mib have, from the N- to C-terminus, a Mib/Herc2 region, a ZZ zinc finger domain, ankyrin repeat domains, and RING finger domains (see schematic representation of molecular structures of DIP-1/mib and skeletrophin in Figure 2A). The significant identity between human skeletrophin and human DIP-1/mib, 36.0% by deduced amino acid sequence, clearly indicates that skeletrophin and DIP-1/mib are in a new E3 ubiquitin ligase subfamily. However, further experimental evidence is necessary to conclude whether skeletrophin is also a ubiquitin ligase, because several molecules containing the RING motif are known to lack ubiquitin ligase activity. To understand the biological and potent pathogenetic properties of skeletrophin, it is essential to identify its substrates and/or target molecules.
Figure 2.
Skeletrophin binds and enhances ubiquitination toward the intracellular region of Jagged-2. A: Schematic representation of molecular structures of skeletrophin and DIP-1/mib ubiquitin ligases, the latter zebrafish homologue appeared to target intracellular region of Delta.6 The homology of deduced amino acids of human skeletrophin and human DIP-1/mib is 36.0%. B: The GST pull-down assay demonstrated the co-precipitation of skeletrophin with the GST-fused intracellular region of Jagged-2 by GT-Sepharose beads. Note the band detected by Western immunoblotting using anti-skeletrophin antibody. This figure represents the three independent experiments, all of which demonstrated the binding of skeletrophin to Jagged-2, but not to Delta-1, -3, -4, or Jagged-1. C: Skeletrophin mediates the ubiquitination of the intracellular region of Jagged-2 in the presence of E1 and E2. Note the slowly migrating band observed by Western immunoblotting using anti-GST antibody. An identical band was also detected by anti-FLAG antibody, which highlighted the presence of the intracellular region of Jagged-2 labeled by FLAG ubiquitin (see Materials and Methods). In contrast, the only significant band that represented the nonubiquitinized intracellular region of Jagged-2 was observed with the RING-mutated skeletrophin (C969S-skeletrophin).
In the present study, we first explored whether skeletrophin exhibits ubiquitin ligase activity, and then examined which molecule skeletrophin targets. As a result, we found that skeletrophin was a novel ubiquitin ligase, the substrate of which was an intracellular region of Jagged-2, which is one of the Notch ligands. Because Jagged-2 appeared to be overexpressed in myeloma and to play a crucial role in the tumorigenesis of multiple myeloma through stromal-myeloma cell interactions,10 we examined the skeletrophin expression in myeloma cells and nonneoplasmic plasma cells, and subsequently explored the pathogenic role of skeletrophin in multiple myeloma.
Materials and Methods
Generation of Recombinant Proteins
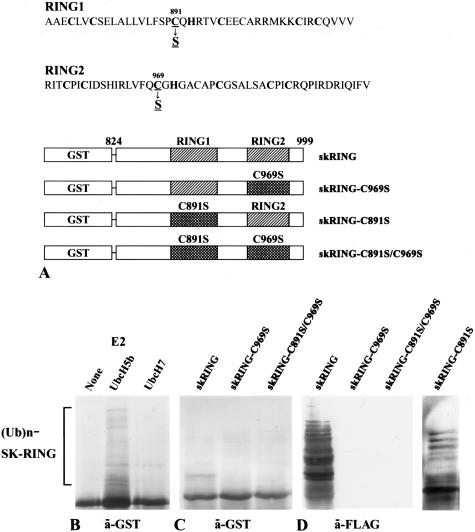
Expression of the glutathione _S_-transferase (GST) fusion RING domain of skeletrophin, the intracellular region of Jagged-1, and -2, and Delta-1, -3, and -4, was driven by isopropyl-1-thio-β-d-galactopyranoside-inducible promoter elements as previously described.11 Briefly, human skeletrophin (amino acids 824 to 998) cDNA encoding two RING-HC domains was amplified from human brain cDNAs (BD Biosciences Clontech, Palo Alto, CA) by polymerase chain reaction (PCR) with a sense (5′-CGA_CTCGAG_GGTCGCGTGCTCAAGGCCC-3′) primer (in which the _Xho_I site is underlined), and an anti-sense (5′-TCACACGAAGATCTGGATGCGG-3′) primer. The amplified RING finger domain was first cloned into pTarget-T vector (Promega, Madison, WI), where the _Not_I cut site was present at the 3′-region, followed by digestion with _Xho_I and _Not_I, and subcloned in-frame into the pGEX-5X-1 vector (Amersham Biosciences Co., Piscataway, NJ). The construct was verified by sequencing and designated skRING. The amino acid substitution, C969S, was introduced into skRING by PCR site-directed mutagenesis by using a 5′-GGGCGCAATGCGCCGTGGCACTCTGGAAC-3′ primer, verified by sequencing, and designated skRING-C969S. Subsequently, the amino acid substitution, C891S, was introduced into skRING or skRING-C969S by PCR site-directed mutagenesis with 5′-CACACACGGTGCGGTGCTGGCTAGGCGAG-3′, verified by sequencing, and designated skRING-C891S and skRING-C891S/C969S, respectively. Notably, recombinant proteins generated by using skRING-C891S or skRING-C969S contained one intact RING domain and one mutated domain, C891S and C969S, respectively, whereas both of the two RING domains of the skRING-C891S/C969S recombinant protein were mutated (Figure 1).
Figure 1.
Skeletrophin possesses autoubiquitination activity. A: Schematic illustration of GST-fused RING finger motifs and its RING motif mutants of skeletrophin (824 to 999). B: Western immunoblotting using an antibody to GST demonstrated an E2-dependent autoubiquitination of recombinant wild-type GST-fused RING fingers of skeletrophin (824 to 999). Ub and Ubc mean ubiquitin and ubiquitin-conjugating enzyme, respectively. C and D: The autoubiquitination activity in the presence of UbcH5b was also observed by Western immunoblotting with an antibody to GST (C) or to the FLAG peptide (D). The latter detected FLAG ubiquitin in polyubiquitinated recombinant protein. Notably, substitution of the critical amino acid, C969S, but not C891S, significantly decreased ubiquitin ligase activity.
cDNAs that encoded the intracellular regions of Jagged-1, -2, and Delta-1, -3, and -4 were also amplified by PCR using the appropriate primer set (summarized in Table 1), subcloned into pGEX-5X-1, and verified by sequencing. All GST-fused recombinant proteins were expressed in log phase Escherichia coli BL21-CodonPlus-RP (Stratagene, La Jolla, CA). Sufficient expression and solubilization of GST-fused recombinant proteins were confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by staining with Coomassie Brilliant Blue.
Table 1.
Primer Sets Used for the Generation of cDNAs that Encoded the Intracytoplasmic Regions of Delta-1, -3, -4, and Jagged-1, -2
| Delta-1 |
|---|
| Sense: 5′-cgggatccaccggcccccagccgacccctg-3′ |
| _Bam_HI site |
| Anti-sense: 5′-ggaattcctcaagagaaacgggagtcttgc-3′ |
| _Eco_RI site |
| Delta-3 |
| Sense: 5′-cgaattcctcttgctggtccacgtgcgcc-3′ |
| _Eco_RI site |
| Anti-sense: 5′-ggctcgagtttcacggacagaatcgagg-3′ |
| _Xho_I site |
| Delta-4 |
| Sense: 5′-cgaattcctgcggcttcagcggccggac-3′ |
| _Eco_RI site |
| Anti-sense: 5′-ggctcgagtacctccgtggggcaatgacac-3′ |
| _Xho_I site |
| Jagged-1 |
| Sense: 5′-cgggatccaagcggcgcaagccgggcagc-3′ |
| _Bam_HI site |
| Anti-sense: 5′-gaattctaggatgtactccattgggt-3′ |
| _Eco_RI site |
| Jagged-2 |
| Sense: 5′-gaagatctcgcaggaaagagcgggagagg-3′ |
| _Bgl_II site |
| Anti-sense: 5′-ggaattcctattccttgccgacgtagcgg-3′ |
| _Eco_RI site |
cDNA, which encoded full-length skeletrophin containing the intact RING domains described previously,1 was subcloned in-frame into the pET-24d vector (Novagen, Darmstadt, Germany). A vector containing the C969S point mutation at the C-terminal RING domain was obtained by substitution of the _Eco_47III-_Not_I region from the skRING-C969S vector, subcloned in-frame into pET-24d, verified by sequencing, and designated as C969S-skeletrophin. Subsequently, recombinant full-length skeletrophin with or without the RING mutation, which was tagged with 6xHis at the C-terminus, was expressed in Rosetta2 (DE3) (Novagen) according to the manufacturer’s protocol. We noted that full-length skeletrophin was not efficiently generated, due to a mechanism not known to us, and the presence of these proteins was confirmed in cell lysates by Western immunoblotting using a specific antibody to skeletrophin, and obtained from large-scale cell lysates by a Ni-NTA column (Novagen) according to the manufacturer’s protocol. The preparation of the specific antibody to skeletrophin, which was also used in further experiments, including immunohistochemical staining, has been described previously.1
In Vitro Ubiquitination Assays
In vitro ubiquitination assays were performed according to the methods of Lorick and colleagues.12 Recombinant FLAG-tagged ubiquitin and recombinant ubiquitin carrier proteins UbcH5b and UbcH7 were purchased from Sigma-Aldrich Inc. (Saint Louis, MO). Recombinant murine ubiquitin-activating enzyme E1 was purchased from Calbiochem (La Jolla, CA). Using the UbcH5b and UbcH7 as E2s, the autoubiquitination assay was performed as follows: GST-fused RING proteins were bound to glutathione Sepharose 4B (GT) beads (Amersham Biosciences Co.), washed with wash buffer (50 mmol/L Tris-HCl, pH 7.4, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml aprotinin), and mixed with a reaction mixture (50 μl) containing 50 mmol/L Tris-HCl, pH 7.4, 5 mmol/L MgCl2, 4 mmol/L ATP, 0.5 mmol/L dithiothreitol, 15 μg of FLAG-ubiquitin, and 200 ng E1 in the presence or absence of 200 ng of E2 at 30°C for 60 minutes. The beads were washed with wash buffer. Proteins bound to beads were subjected to SDS-PAGE under reducing conditions and detected by Western immunoblotting using an antibody to the FLAG tag (Sigma-Aldrich Inc.) or GST (Amersham Biosciences).
GST Pull-Down Assays
Cell extracts including the recombinant full-length skeletrophin protein and the GST-fused intracellular region of Delta-1, -3, -4, and Jagged-1 and -2 were mixed in a binding buffer (20 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.5% Nonidet P-40, 0.5% bovine serum albumin, 0.5 mmol/L phenylmethyl sulfonyl fluoride, and 2 μg/ml aprotinin) and incubated for 6 hours at 4°C. Then, the GST-fused molecules were pulled-down by GT beads, washed four times with binding buffer containing 300 mmol/L KCl, suspended in SDS sample buffer, and analyzed by SDS-PAGE followed by Western immunoblotting with anti-skeletrophin antibody or anti-GST antibody.
Ubiquitination of the Intracellular Region of Jagged-2 by Skeletrophin
In vitro ubiquitination assays were performed by incubating FLAG-tagged ubiquitin, E1, UbcH5b, recombinant full-length skeletrophin, or C969S-skeletrophin, with the GST-fused intracellular region of Jagged-2. Briefly, GST-fused Jagged-2 proteins were bound to GT-Sepharose beads, washed with wash buffer (50 mmol/L Tris-HCl, pH 7.4, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml aprotinin), and mixed with a reaction mixture (50 μl) containing 50 mmol/L Tris-HCl, pH 7.4, 5 mmol/L MgCl2, 4 mmol/L ATP, 0.5 mmol/L dithiothreitol, 15 μg of FLAG-ubiquitin, 200 ng E1, 200 ng UbcH5b in the presence of recombinant full-length skeletrophin or C969S-skeletrophin at 30°C for 60 minutes. Proteins bound to GT beads were analyzed by Western immunoblotting with anti-GST and anti-FLAG antibodies.
Immunohistochemical Staining
Details of the immunohistochemical staining procedures including the preparation of affinity-purified rabbit antibody to skeletrophin have been described previously.1 In the present study, we newly generated a monoclonal antibody to intracellular region of Jagged-2. Briefly, the recombinant GST-fused intracellular region of Jagged-2, described above, was digested with bovine factor Xa (Seikagaku Co. Ltd., Osaka, Japan) to separate Jagged-2 from GST tag. Factor Xa was subsequently adsorbed with Xa Removal Resin (Qiagen GmbH, Hilden, Germany). BALB/c mice were immunized intraperitoneally with the recombinant intracellular region of Jagged-2. Monoclonal antibody was generated according to the modified method of Koehler and Milstein13 and Takeuchi and colleagues.14 One of the established hybridoma clones secreted an antibody, which could immunostain the intracellular domain of Jagged-2 in the routinely processed paraffin-embedded tissue sections. Western immunoblotting was also performed to evaluate the specificity of the antibody. To identify the Jagged-2 band in the Western immunoblotting, we also used commercially available rabbit antibody to Jagged-2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Extracted protein mixtures from human thymus were purchased from BD Biosciences Clontech.
In the present study, we examined archival tissue specimens comprising a spectrum of various intramedullar multiple myeloma and nonneoplasmic plasma cells (summarized in Table 2). When we examined multiple myeloma specimens, we focused on exploring the initial bone marrow biopsy specimens of patients with untreated intramedullary multiple myeloma. To demonstrate skeletrophin expression in the brain, which exhibited comparatively high levels of mRNA expression in our previous study,1 we stained a small part of the forebrain surgically resected from patients with hemorrhage due to amyloid angiopathy. Normocellular nonneoplasmic bone marrow tissues were obtained from patients with various cancers to evaluate the metastasis to bone marrow, and were used in the present study. All specimens were fixed in 10% buffered formalin, and paraffin-embedded. Deparaffinized sections were placed in plastic Coplin jars (Asone, Tokyo, Japan) filled with preheated citrate buffer (pH 6.0) and microwave-irradiated for 5 minutes at 650 W. Next, the slides were incubated for 30 minutes in fetal bovine serum. The tissues were then immunostained with affinity-purified rabbit anti-human skeletrophin peptide, monoclonal antibody to intracellular region of Jagged-2, or control antibodies using a streptavidin-biotin complex peroxidase kit (DAKO LSAB kit; Dakopatts, Kyoto, Japan). The procedures were performed according to the manufacturer’s protocol. In several experiments, anti-skeletrophin antibody or anti-Jagged-2 antibody was preadsorbed with the immunized skeletrophin peptide or recombinant intracellular domain of Jagged-2.
Table 2.
Primer Sets Used for RT-PCR
| Murine Notch-1 |
|---|
| Sense: 5′-gccacagattgaggaggcct-3′ |
| Anti-sense: 5′-accattggtgccaggaagca-3′ |
| Notch-2 |
| Sense: 5′-accagcacccctcctgctac-3′ |
| Anti-sense: 5′-tcctgttcctgctcatcagg-3′ |
| Notch-3 |
| Sense: 5′-acactgggagttctctgt-3′ |
| Antisense: 5′-acactgggagttctctgt-3′ |
| Jagged-1 |
| Sense: 5′-cttgagccttctgctcgcc-3′ |
| Anti-sense: 5′-tgcagaagccatgcttgg-3′ |
| Jagged-2 |
| Sense*: 5′-tggaagactgcaacagctgccgctgc-3′ |
| Anti-sense: 5′-gtttccaccttgacctcggt-3′ |
| Hes-1 |
| Sense: 5′-tcctccccggtggctgctaccc-3′ |
| Anti-sense: 5′-ttcatgcactcgctgaagcc-3′ |
| Skeletrophin |
| Sense: 5′-gaccgtacagttgtcgttcagtgggac-3′ |
| Anti-sense: 5′-catctgagatccaccttgcccttgtgg-3′ |
| Human Jagged-2 |
| Sense*: 5′-tggaagactgcaacagctgccgctgc-3′ |
| Anti-sense: 5′-ccttgccgacgtagcgg-3′ |
| Skeletrophin |
| Sense: 5′-cgggaacctgcgtgtagcagtcgctggtc-3′ |
| Anti-sense: 5′-cagagcggtcctgccttggttcttggtgtc-3′ |
The tissue sections were counterstained with hematoxylin and semiquantitatively scored for staining intensity of skeletrophin in myeloma cells or plasma cells as weakly positive (+, 5> to 20%), moderately positive (++, >20 to 80%), or markedly positive (+++, >80%). Several morphological and clinicopathological classifications including morphology of infiltrating myeloma cell type (the presence of cleaved and/or large nuclei, nucleated or not), growth pattern (interstitial or nodular), percentage of plasma cells in the bone marrow, the osteolytic bone regions of the patients, and the concentration of lactate dehydrogenase in the patients’ serum at the biopsy, were examined for their relationship to skeletrophin expression in multiple myeloma specimens. The relationship of skeletrophin expression and subcellular distribution of Jagged-2 was also examined.
Cell Culture, Reverse Transcriptase (RT)-PCR, Transfection, and Co-Culture Assay
A murine bone marrow-derived stromal cell line, ST-2,15 was obtained from the Riken BioSource Center (Tsukuba, Japan). A murine myeloma cell line P3U1 was maintained in our laboratory. A human myeloma cell line, RPMI8226, was obtained from the Japan Heath Science Research Resources Bank (Osaka, Japan). Another human myeloma cell line, U266, was a kind gift from Dr. Ikezoe Takayuki (KMS, Department of Hematology and Respiratory Medicine, Kochi University, Kochi, Japan). Cells were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies, Inc., Grand Island, NY) containing 10% fetal bovine serum and 50 μg/ml of gentamicin (Life Technologies, Inc.).
The construction and the method for obtaining the stable transfection of a pCI-neo expression vector (Promega) containing a human wild-type skeletrophin has been previously reported.1 A pCI-neo vector containing a RING finger mutant, C969S, was constructed by substitution of an _Eco_47III-_Not_1 part from skRING-C969S. P3U1 clones, which expressed human full-length or C969S-mutated skeletrophin, were established after the limiting dilution. Culture wells of six-well plates were seeded with 1 × 104 ST-2 cells 16 hours before co-culture assay. Then, 1 × 103 P3U1 cells were co-cultured with ST-2 cells with or without an insert for 14 hours. After incubation, P3U1 cells were removed by shaking the plates and aspiration when P3U1 was allowed to be in direct contact with ST-2, without an insert.
RT-PCR analysis was performed as previously described.16 Briefly, total cellular RNA was prepared from cell lysates using RNA-zol B (Biotex Laboratory, Houston, TX). In several experiments, serial fivefold dilutions were made from master lysates and used as a set in RT-PCR. The cDNA synthesis from total RNA and subsequent PCR were performed using an RNA LA (long and accurate) PCR kit (Takara Co. Ltd., Ohtsu, Japan). The procedure was performed according to the manufacturer’s instructions. The primer sets used for RT-PCR are summarized in Table 2. The PCR-amplified product was electrophoretically separated on a 2% agarose gel. To assure reproducibility of the results, the RT-PCR amplification was repeated by using different clones at least three times.
Results
Skeletrophin Catalyzes Autoubiquitination in Cooperation with UbcH5b
Ubiquitination requires sequential reactions of three enzymes. First, ubiquitin is activated by E1 through the formation of a thioester bond in the presence of ATP. Subsequently, ubiquitin is transferred to E2 via the thioester bond. Finally, E2 transfers ubiquitin, generally with the help of ubiquitin ligase E3, to an ε-amino group of an internal lysine residue of the substrate protein. Ubiquitin ligase is sometimes itself ubiquitinated and therefore in vitro demonstration of substrate-independent autoubiquitination is considered to be a good indicator of ligase activity.
As shown in Figure 1B, the recombinant GST-fused RING motif of skeletrophin protein promoted autoubiquitination in a UbcH5b-dependent manner. The high molecular mass of FLAG-tagged proteins, which indicates ubiquitination, was observed when ubiquitin, E1, UbcH5b, and the GST-fused RING domain of skeletrophin were incubated. In the absence of UbcH5b, these ladders were not observed. UbcH7 could not confer sufficient ubiquitination. These results demonstrate that skeletrophin has a ubiquitin ligase activity similar to that observed in DIP-1/mib.6,8
Alternation of RING motif, C969S, eliminated autoubiquitination activity, whereas C891S mutation did not affect the activity (Figure 1, C and D). These results indicated that skeletrophin is a ubiquitin ligase, the activity of which depends on a RING domain of C-terminus. In addition, we also examined autoubiquitination of GST-fused full-length skeletrophin or RING-mutated, C969S, skeletrophin. Autoubiquitination of the former, but not the latter, was observed (data not shown).
Skeletrophin Bound and Ubiquitinated the Intracellular Region of Jagged-2
Itoh and colleagues6 demonstrated that DIP-1/mib, the molecular structure of which is close to skeletrophin (Figure 2A), targets the intracellular domain of Delta, a ligand of Notch. Therefore, we speculated that skeletrophin ubiquitin ligase might also target the intracellular domain of Notch ligands. To determine whether skeletrophin can bind to human Notch ligands, we performed GST pull-down assays using recombinant Delta-1, -3, and -4, and Jagged-1 and -2.
As demonstrated in Figure 2B, full-length skeletrophin molecules bound to the recombinant GST-fused intracellular domain of Jagged-2, but not to the other GST-fused intracellular domains of Notch ligands tested. Subsequently, we explored whether the intracellular domain of Jagged-2 was ubiquitinized by skeletrophin. The result is shown in Figure 2C. The band corresponding to the ubiquitinated intracellular domain of Jagged-2, which migrated slower than that of the intact one, was observed by Western immunoblotting using a specific antibody to GST and the FLAG tag. These observations indicate that skeletrophin is a ubiquitin ligase that acts toward the intracellular domain of Jagged-2.
Aberrant Expression of Skeletrophin in Multiple Myeloma Cells
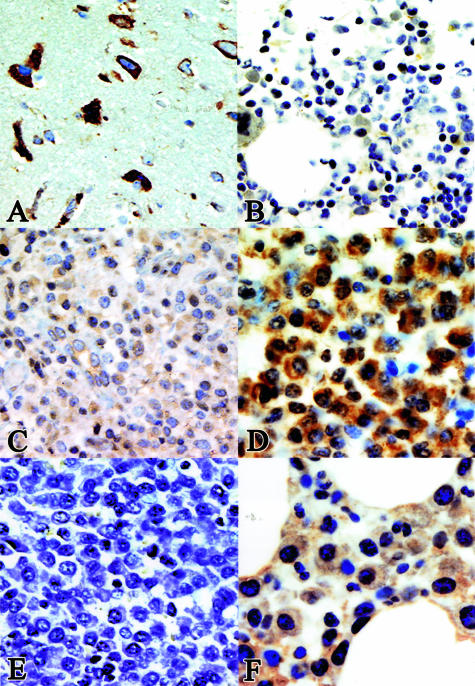
An increasing number of articles state that Jagged-1 and -2 are overexpressed in multiple myeloma and that ligand-dependent Notch activation in bone marrow stromal cells has a pivotal role in the pathogenesis of multiple myeloma, possibly by accelerating paracrine interleukin (IL)-6 secretion.10,17 Because recombinant skeletrophin was able to bind and mediate the ubiquitination of the intracellular domain of Jagged-2, we were interested in the expression of skeletrophin in multiple myeloma cells. The results of immunohistochemical staining are summarized in Tables 3 and 4 and representative results are shown in Figure 3. First, we demonstrate the staining pattern in neural cells to show the protein expression in skeletrophin mRNA-rich tissues (Figure 3A).1 As demonstrated in Figure 3B, no significant staining was observed in normocellular nonneoplastic bone marrow. This result is compatible with our preliminary findings by dot-blot analysis, in which we did not detect skeletrophin mRNA in nonneoplastic bone marrow tissues (data not shown). Compared to neural cells, skeletrophin was expressed weakly or not at all in most nonneoplasmic plasma cells in various chronically inflamed tissues. A few dispersed plasma cells were weakly stained in the cytoplasm (Figure 3C). In contrast, strong staining was observed in malignant plasma cells in 23 of 40 specimens. In skeletrophin-positive cases, skeletrophin was localized in the entire cytoplasm (Figure 3D). We could not find any significant relationship between skeletrophin expression and the morphology of the infiltrating myeloma cell type (the presence of cleaved and/or large nuclei, nucleated or not), growth pattern (interstitial or nodular), the percentage of plasma cells in the bone marrow, or the serum level of lactate dehydrogenase. However, skeletrophin expression was more commonly observed in multiple myeloma specimens from patients with osteolytic bone lesions (Table 4, Figure 3D). Preadsorption of antibody with immunized peptide diminished the staining (Figure 3E). We also demonstrated the staining of the specimens from patients without osteolytic bone lesions (Figure 3F). These data obtained by immunohistochemical staining revealed that skeletrophin was significantly overexpressed in a considerable number of malignant myeloma specimens, especially skeletrophin from patients with an osteolytic region.
Table 3.
Summary of Immunohistochemical Staining: Skeletrophin Expression in Nonneoplasmic and Malignant Plasma Cells
| Skeletrophin expression (% of positive cells in plasma cells) | ||||
|---|---|---|---|---|
| Status | − (<5%) | + (5 to 20%) | ++ (20 to 80%) | +++ (>80%) |
| Chronically inflamed tissues | ||||
| Gastritis (biopsy diagnosis) | 10 | 0 | 0 | 0 |
| Cholecystitis | 10 | 0 | 0 | 0 |
| Tonsillitis | 8 | 0 | 0 | 0 |
| Multiple myeloma* | ||||
| % of atypical plasma cells in bone marrow | ||||
| [10 to 30%] | 6 | 2 | 2 | 4 |
| [30 to 80%] | 6 | 2 | 1 | 1 |
| 80% | 5 | 1 | 0 | 10 |
Table 4.
Summary of Immunohistochemical Staining: Correlation between Osteolytic Bone Regions and Skeletrophin Expression
| Skeletrophin expression* | ||
|---|---|---|
| Negative | Positive | |
| Number of patients with an osteolytic bone lesion | 2 | 12 |
| Without an osteolytic bone lesion | 15 | 11 |
Figure 3.
Immunohistochemical staining of various tissues. A: Skeletrophin expression in the brain. The cytoplasmic region of neural cells was stained with anti-skeletrophin antibody. B: No significant staining was observed in nonneoplasmic normocellular bone marrow tissues. B represents the staining of five specimens. C: Considerable weak staining was observed throughout the cytoplasm in nonneoplasmic plasma cells in various types of chronic inflammation specimens. C represents the staining of 28 specimens of chronically inflamed tissues as summarized in Table 3. D and F represent the markedly positive and negative cases of multiple myeloma specimens, respectively. Strong staining with anti-skeletrophin antibody is demonstrated in D in a multiple myeloma specimen from a patient with an osteolytic lesion, whereas preadsorption of antibody with immunized peptide diminished the staining (E). Original magnifications: ×400 (A–E); ×640 (F).
Correlation of Skeletrophin Expression to Subcellular Localization of Jagged-2
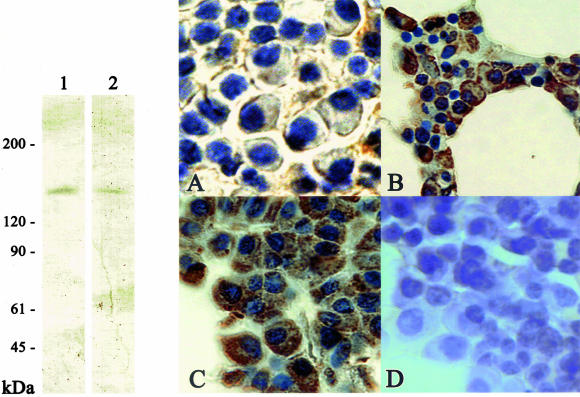
Next, we investigated the correlation of skeletrophin and Jagged-2 expression in malignant plasma cells. To perform this study, we newly generated a monoclonal antibody to explore the Jagged-2 expression in routinely processed archival tissues. Western immunoblotting verified the specificity of this monoclonal antibody (Figure 4, left). Most myeloma cells expressed Jagged-2, as recently reported by Houde and colleagues.10 Malignant plasma cells in 37 of 40 multiple myeloma specimens examined in this study appeared to express Jagged-2. Of particular interest, Jagged-2 tended to be localized in the cell surface membrane in skeletrophin-negative myeloma cases (2 of 16 specimens; Figure 4A), whereas Jagged-2 was often found in both surface, perisurface cytoplasm, and/or entire cytoplasm in skeletrophin-positive cases (14 of 21; Figure 4, B and C). In five skeletrophin-positive myeloma specimens, we observed strong Jagged-2 expression in the entire cytoplasm (Figure 4C). The result is summarized in Table 5 and representative staining is shown in Figure 4, right.
Figure 4.
Western immunoblotting and immunohistochemical staining with anti-Jagged antibody. Left: Western immunoblotting demonstrates the specificity to Jagged-2 of a monoclonal antibody generated in the present study. Note the single band of recombinant GST-fused intercellular region of Jagged-2 and a band corresponding to 150-kd Jagged-2 molecules in cell extracts of the human thymus (lane 1). For comparison, we also used commercially available rabbit antibody to the intracellular region of Jagged-2 (lane 2). Right: Jagged-2 localization in malignant plasma cells. In 14 of 16 skeletrophin-negative specimens, Jagged-2 was localized in malignant plasma cells (representative staining is shown in A). By contrast, in 9 of 21 skeletrophin-positive specimens, Jagged-2 was found in both the cell surface and the perisurface cytoplasm (B). In 5 of 21 skeletrophin-positive specimens, Jagged-2 was found in the entire cytoplasm (C), whereas preadsorption of antibody with immunized recombinant intracellular region of Jagged-2 diminished the staining (D). Original magnifications: ×640 (A), ×400 (B), ×500 (C and D).
Table 5.
Correlation of Skeletrophin Expression to Jagged-2 Localization
| Skeletrophin | Cell surface | Cell surface and cytoplasm |
|---|---|---|
| Negative (<5%) | 14 | 2 |
| Positive (>5%) | 7 | 14* |
Skeletrophin in Cultured Myeloma Cell Enhanced the Expression of Hes-1, a Notch Target Gene, in Bone Marrow-Derived Stromal Cells through Cell-Cell Contact
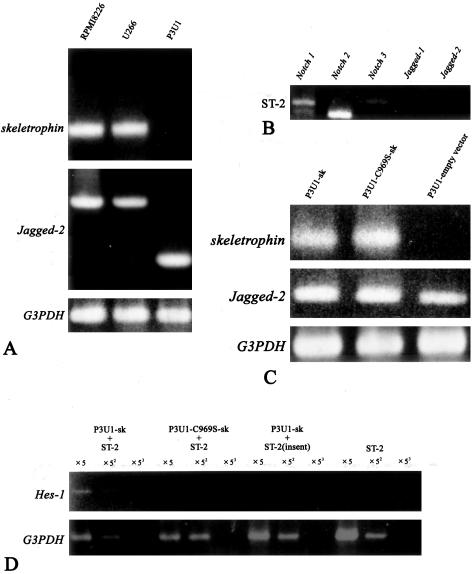
Finally, we investigated whether skeletrophin expression in myeloma cells could activate ligand-dependent Notch receptor activation in bone marrow stromal cells. To perform this experiment, we first examined the skeletrophin expression in several cultured myeloma cells. As shown in Figure 5A, both RPMI8226 and U266 appeared to express abundant skeletrophin mRNA by RT-PCR. We also examined Jagged-2 expression and found that both RPMI8226 and U266 expressed Jagged-2 mRNA (Houde and colleagues10 had already reported the Jagged-2 expression in RPMI8226 and U266). Interestingly, a murine myeloma cell line, P3U1, did express Jagged-2, but did not express the detectable skeletrophin by RT-PCR (Figure 5A). A murine bone marrow-derived stromal cell line, ST-2, expressed Notch1, Notch2, and Notch3 as previously described by Yamada and colleagues,18 but did not express Jagged-1 or -2 (Figure 5B). Lack of Jagged-2 expression in ST-2 stromal cells is also consistent with the report from Nefedova and colleagues,19 who demonstrated that Jagged-2 was not detectable in bone marrow stromal cells from healthy donors.
Figure 5.
Skeletrophin in P3U1 myeloma cells induced Hes-1 in co-culture assay with bone marrow stromal cell by direct cell-cell contact. A: RPMI8226 and U266 expressed both skeletrophin and Jagged-2 mRNAs. By contrast, P3U1 strongly expressed _Jagged-_2, but not skeletrophin mRNA. The data represents the three independent experiments. B: RT-PCR demonstrated that ST-2 bone marrow-derived stromal cells expressed Notch 1, 2, and 3 mRNA, but not Jagged-1 and -2. The data represents the three independent experiments. C: Stable P3U1 clones were prepared by transfection with expression vector containing skeletrophin, RING-mutated C891S-skeletrophin, or empty vector. Note the intense band corresponding to Jagged-2 mRNA in these transfectants. The data represents the three independent clones of each transfectant. D: Skeletrophin-expressing P3U1 induced Hes-1, a downstream gene of ligand-dependent Notch activation, in ST-2 bone marrow derived by direct P3U1 and ST-2 cell contact. By contrast, RING mutation, which diminished ubiquitin ligase activity, did not induce Hes-1 (see also Figure 1). When P3U1 transfectants and ST-2 cells were separated by insert, any significant _Hes-1_mRNA band was not observed.
Notably, skeletrophin, which was exogenously expressed in P3U1 (Figure 5C), induced Hes-1 gene expression in ST-2 stromal cells, by direct cell-cell contact (Figure 5D). By contrast, RING-mutated skeletrophin, indicated as C969S-sk, did not enhance Hes-1 expression. Notably, P3U1, in which exogenous skeletrophin or RING-mutated skeletrophin was overexpressed, expressed Jagged-2 as well as original P3U1 cells (Figure 5C). Figure 5 shows the representative results, which were obtained by using three independent clones. Hes-1 has been shown to be a target molecule of the activated Notch receptor.20 Therefore, we think that skeletrophin expression in myeloma cells could enhance ligand-dependent Notch activation in bone marrow stromal cells through direct cell-cell contact.
Discussion
The pathogenesis of multiple myeloma remains obscure; however, it is known that prolonged antigenic stimulation and elevated IL-6 levels play an important role in developing multiple myeloma.21,22 A cascade of interactions between premyeloma cells and stromal cells may also be important in the development of multiple myeloma. Interestingly, researchers have recently shown that myeloma cells induce IL-6 expression in stromal cells in a primarily cell contact-dependent manner.23 More recently, Jagged-2 appeared to be overexpressed in multiple myeloma,10 thus Notch Jagged-2 ligation, after proteolysis of Notch and subsequent internalization of the Notch intracellular subunit, is thought to activate numerous genes including IL-6. Interestingly, the ubiquitination of the intracellular domain of Notch ligands, as well as that of the Notch receptors, appeared to be critical for ligand-dependent Notch activation.6,7
In the present study, we first characterized skeletrophin as a novel ubiquitin ligase, which targeted the intracellular region of Jagged-2. Subsequently, we demonstrated that skeletrophin was overexpressed in malignant plasma cells in many multiple myeloma specimens. Premyeloma or myeloma cells may adhere to stromal cells, and then skeletrophin may mediate ligand-dependent Notch activation by ubiquitination, followed by endocytosis of the intracellular domain of Jagged-2. In line with this speculation, significant cytoplasmic localization of Jagged-2 was observed in skeletrophin-positive myeloma specimens. However, we also found that Jagged-2 was localized in the cytoplasm in two skeletrophin-negative specimens. In seven skeletrophin-positive cases, Jagged-2 was only found in the cell surface membrane. These data indicate that molecular events other than skeletrophin overexpression may be involved in Jagged-2 localization in malignant plasma cells.
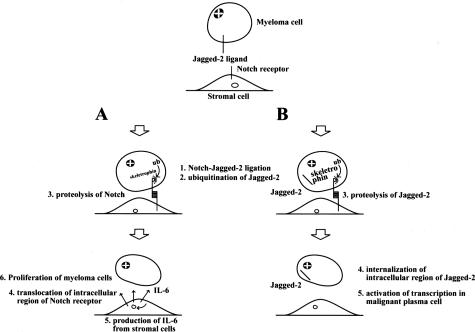
Next, exogenously expressed skeletrophin in P3U1 cells induced a Notch downstream effector gene, Hes-1, through direct cell-cell contact. Very interestingly, Houde and colleagues10 reported that Jagged-2 was overexpressed in multiple myeloma cells and induced stromal cells to secrete IL-6. Skeletrophin may therefore be involved in the endocytosis of Jagged-2 to facilitate ligand-dependent Notch signal activation to increase IL-6 production from stromal cells. The finding that skeletrophin expression was related to the presence of osteolytic lesions in patients may also be related to IL-6 secretion, because patients with multiple myeloma with high IL-6 levels have a higher incidence of osteolytic bone lesions.24 However, further study, including the expression pattern of the Notch receptor in the stromal cells of bone marrow, is necessary to verify our working hypothesis, which is summarized in Figure 6A.
Figure 6.
Hypothetical model of the pathogenic role of skeletrophin in multiple myeloma. A: Putative indirect effect of skeletrophin. Premyeloma or myeloma cells adhere to stromal cells. Subsequently overexpressed skeletrophin facilitates ligand-dependent Notch activation by ubiquitination of the intracellular domain of Jagged-2. After ligand-dependent Notch activation in stromal cells activates various genes including IL-6 to promote multiple myeloma. B: Putative direct mechanism of skeletrophin. Skeletrophin facilitates the presenilin-dependent γ-secretase cleavage of Jagged-2 to produce the soluble intracellular region of Jagged-2. As a result, the processing intracellular Jagged-2 domain is transported into nuclei to activate various gene transcriptions to achieve the development of malignant myeloma.
What is the molecular mechanism responsible for skeletrophin overexpression in malignant plasma cells? We very recently identified the promoter region of skeletrophin (GenBank accession number, AB166789) and found that the promoter region of skeletrophin is matched to the standard CpG island criteria (%GC = 68.3% > 50%, ObsCpG/ExpCpG = 0.915 > 0.6, length = 496 > 200 bp). We speculate that skeletrophin overexpression might be driven by promoter hypomethylation in malignant plasma cells in a manner similar to the molecular mechanism, which is responsible for Jagged-2 overexpression.10 Although further studies are necessary to verify this hypothesis, it seems likely that hypomethylation of the promoter region of both skeletrophin and Jagged-2 genes resulted in dual overexpression of a ubiquitin ligase, skeletrophin, and its substrates, Jagged-2, in multiple myeloma.
Recently, Ikeuchi and Sisodia25 revealed that Delta-1 and Jagged-2 are subject to presenilin-dependent, intramembranous γ-secretase processing. They also demonstrated that nuclear transport of intracellular region of Delta-1 mediated transcriptional stimulation. Therefore, Delta-1 and Jagged-2 can play dual roles as activators of Notch receptor signaling and as receptors that mediate nuclear signaling events via γ-secretase processing in the intracellular region. The intracellular region of Jagged-2 contains the putative nuclear localization signals, RKRR and KRRK. Similarly, the Delta-1 intracellular region also contains PDRKRPE and RKRP putative nuclear localization signals. From this perspective, it is particularly interesting that Jagged-2 was found in the nuclei of a few malignant plasma cells in skeletrophin-positive myeloma specimens (Figure 5C). Matsuda and colleagues26 recently performed a large-scale identification and characterization of the human genes that activate the nuclear factor (NF)-κB pathway. One of the molecules that strongly activated the NF-κB pathway is a splicing form of skeletrophin. This finding may indicate that skeletrophin is involved in the pathogenesis of multiple myeloma, not only by activating the Notch-Jagged-2 interaction in myeloma cells and stromal cells, but also directly by suppressing apoptosis by activating the NF-κB pathway in malignant plasma cells. We speculate that skeletrophin may also facilitate the production of the soluble intracellular region of Jagged-2, which might activate the transcriptional events, including the induction of a molecule, which could activate the NF-κB pathway (Figure 6B).
Skeletrophin and DIP-1/mib ubiquitin ligase may play a complementary role in Notch-ligand interaction by binding to each Notch-ligand target, Jagged-2 and Delta, respectively. They may also be involved in the generation of soluble cytoplasmic protein by facilitating the presenilin-dependent γ-secretase of their substrates. Further extensive studies will be required to reveal the relationship between the biological properties of skeletrophin and DIP-1/mib ubiquitin ligase, especially with respect to their ligands and the role in presenilin-dependent γ-secretase cleavage of Jagged-2 and Delta-1. However, it is likely that skeletrophin and DIP-1/mib, which have very similar molecular structures (see Figure 2A and the Introduction), may constitute a unique tribe of the RING-HC-dependent ubiquitin ligase family.
Lastly, skeletrophin did not down-regulate the expression of its substrate, Jagged-2, in myeloma cells. This indicated that the effect of skeletrophin on the Jagged-2 molecule is incessant in many malignant plasma cells. Therefore, therapeutic approaches intended to disturb skeletrophin function may have a potent therapeutic effect in patients with multiple myeloma. In the present study, we focused on specimens from untreated patients; however, we also did a preliminary exploration of skeletrophin expression in three patients, whose bone marrow tissues at initial diagnosis were not stained with anti-skeletrophin. With time, malignant plasma cells became resistant to various chemotherapy agents. Notably, skeletrophin expression was increasingly observed in these progressing malignant plasma cells. Further study to unravel the relationship between skeletrophin and resistance to chemotherapy is ongoing.
In conclusion, skeletrophin appears to be a novel ubiquitin ligase targeted toward the intracellular region of Jagged-2, which is aberrantly overexpressed in multiple myeloma cells. A precise understanding of the molecular mechanism that is involved in Notch and its ligand activation is likely to be beneficial to the development of specific therapeutic approaches for malignant myeloma.27,28
Acknowledgments
We thank Mr. Takuya Yamaguchi, Ms. Naoyo Nakamura (Department of Pathology, Kochi Medical School), and Ms. Rumi Matumura (Division of Molecular Biology, Kochi Medical School) for their skilled assistance.
Footnotes
Address reprint requests to Tamotsu Takeuchi, Department of Pathology, Kochi Medical School, Nankoku, Kochi, Japan 783-8505. E-mail: takeutit@med.kochi-ms.ac.jp.
Supported by the Ministry of Education of Japan (grants KAKEN12670165, 13670177), the Medical Research Fund of the Kochi Medical School, and the Vice-Chancellor of the Kochi Medical School (project grant for overseas study).
References
- Takeuchi T, Heng HHQ, Ye C-J, Liang SB, Sonobe H, Ohtsuki Y. Down-regulation of a novel actin-binding molecule skeletrophin, in malignant melanoma. Am J Pathol. 2003;163:1395–1404. doi: 10.1016/S0002-9440(10)63497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Freemont PS. Ubiquitination: RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- Fang S, Lorick KL, Jensen JP, Weissman AM. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol. 2003;13:5–14. doi: 10.1016/s1044-579x(02)00095-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind Bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Notch signaling: endocytosis makes delta signal better. Curr Biol. 2003;13:R273–R275. doi: 10.1016/s0960-9822(03)00199-4. [DOI] [PubMed] [Google Scholar]
- Jin Y, Blue EK, Dixon S, Shao Z, Gallagher PJ. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular levels of DAPK. J Biol Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Tominaga T. mDia-interacting protein acts downstream of Rho-mDia and modifies Src activation and stress fiber formation. J Biol Chem. 2001;276:39290–39294. doi: 10.1074/jbc.M107026200. [DOI] [PubMed] [Google Scholar]
- Houde C, Li Y, Song L, Barton K, Zhang Q, Godwin J, Nand S, Toor A, Alkan S, Smadja NV, Avet-Loiseau H, Lima CS, Miele L, Coignet LJ. Over-expression of the Notch ligand Jag2 in malignant plasma cells from multiple myeloma patients and cell line. Blood. 2004;104:3697–3704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Chen BK, Qiu Y, Sonobe H, Ohtsuki Y. Molecular cloning and expression of a novel human cDNA containing CAG repeats. Gene. 1997;204:71–77. doi: 10.1016/s0378-1119(97)00525-8. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, Ohtsuki Y. Expression of T-cadherin (CDH13, H-cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000;74:1489–1497. doi: 10.1046/j.1471-4159.2000.0741489.x. [DOI] [PubMed] [Google Scholar]
- Sudo T, Ito M, Ogawa Y, Iizuka M, Kodama H, Kunisada T, Hayashi S, Ogawa M, Sakai K, Nishikawa S. Interleukin7 production and function in stromal cell-dependent B cell development. J Exp Med. 1989;170:333–338. doi: 10.1084/jem.170.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Nicole S, Misaki A, Furihata M, Iwata J, Sonobe H, Ohtsuki Y. Expression of SMARCF1, a truncated from of SWI1, in neuroblastoma. Am J Pathol. 2001;158:663–672. doi: 10.1016/S0002-9440(10)64008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jundt F, Pröbsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, Bargou RC, Manz R, Stein H, Dörken B. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101:2227–2234. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signaling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kawano M, Hirano T, Matsuda T, Taga T, Horii Y, Iwato K, Asaoku H, Tang B, Tanabe O, Tanaka H, Kuramoto A, Kishimoto T. Autocrine generation and requirement of BSF-2/IL-6 for human multiple myelomas. Nature. 1988;332:83–85. doi: 10.1038/332083a0. [DOI] [PubMed] [Google Scholar]
- Zhang XG, Bataille R, Widjenes J, Klein B. Interleukin-6 dependence of advanced malignant plasma cell dyscrasias. Cancer. 1992;69:1373–1376. doi: 10.1002/1097-0142(19920315)69:6<1373::aid-cncr2820690612>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Vidriales MB, Anderson KC. Adhesion of multiple myeloma cells to the bone marrow microenvironment: implications for future therapeutic strategies. Mol Med Today. 1996;2:425–431. doi: 10.1016/1357-4310(96)84846-5. [DOI] [PubMed] [Google Scholar]
- Pelliniemi TT, Irjala K, Mattila K, Pulkki K, Rajamaki A, Tienhaara A, Laakso M, Lahtinen R. Immunoreactive interleukin-6 and acute phase proteins as prognostic factors in multiple myeloma. Finnish Leukemia Group. Blood. 1995;85:765–771. [PubMed] [Google Scholar]
- Ikeuchi T, Sisodia SS. The notch ligands, Delta-1 and Jagged-2, are substrates for presenilin-dependent “γ-secretase” cleavage. J Biol Chem. 2003;278:7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E, Hayashi H, Sugano S. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- Milner LA, Bigas A. Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation. Blood. 1999;93:2431–2448. [PubMed] [Google Scholar]
- Milner LA. Notch signaling: a key to the pathogenesis of multiple myeloma? Inside Blood. Blood. 2004;103:3253–3254. [Google Scholar]