MicroRNA-9a ensures the precise specification of sensory organ precursors inDrosophila (original) (raw)
Abstract
MicroRNAs (miRNAs) have been implicated in regulating various aspects of animal development, but their functions in neurogenesis are largely unknown. Here we report that loss of miR-9a function in the Drosophila peripheral nervous system leads to ectopic production of sensory organ precursors (SOPs), whereas overexpression of miR-9a results in a severe loss of SOPs. We further demonstrate a strong genetic interaction between miR-9a and senseless (sens) in controlling the formation of SOPs in the adult wing imaginal disc. Moreover, miR-9a suppresses Sens expression through its 3′ untranslated region. miR-9a is expressed in epithelial cells, including those adjacent to SOPs within proneural clusters, suggesting that miR-9a normally inhibits neuronal fate in non-SOP cells by down-regulating Sens expression. These results indicate that miR-9a ensures the generation of the precise number of neuronal precursor cells during development.
Keywords: MicroRNA, SOP, Senseless, Drosophila, PNS
The peripheral nervous system (PNS) is essential for animals to detect and relay environmental stimuli to central neurons for information processing. It is largely unknown what ensures the development of precise numbers of sensory organs for a particular external sensory cue. The Drosophila PNS has been used as an excellent model system for dissecting the genetic programs that control sensory organ formation (Ghysen and Dambly Chaudiere 1993; Jan and Jan 1993; Modolell 1997). In embryonic abdominal segments, external sensory (ES) organs and chordotonal (CH) organs contain single-dendrite neurons associated with support cells and function in receiving mechanical stimuli (Campos-Ortega and Hartenstein 1985; Ghysen et al. 1986). In contrast, multidendritic (MD) neurons elaborate highly branched dendrites underneath the epidermis (Bodmer and Jan 1987; Gao et al. 1999) that probably function as stretch, touch, or other sensory receptors (Ainsley et al. 2003; Liu et al. 2003; Tracey et al. 2003). In adult flies, most external sensory organs have one neuron, one hair cell, and a few support cells (Campos-Ortega and Hartenstein 1985).
Precise numbers of sensory organs are generated through similar developmental processes in embryos and adults (Campos-Ortega and Hartenstein 1985). For example, in embryonic/larval abdominal segments, each dorsal cluster contains four ES organs and eight MD neurons. Some ES neurons are generated from sensory organ precursors (SOPs) that also produce MD neurons (ES– MD lineage), while other lineages produce ES but not MD neurons (Brewster and Bodmer 1995; Vervoort et al. 1997). Adult flies have four macrochaetes on the notum and a well-defined number of sensory bristles on the wing margin. All cells in each adult sensillium are generated from a single SOP in two or three rounds of asymmetric cell division (Hartenstein and Posakony 1989; Bardin et al. 2004).
The selection of SOPs from early ectoderm begins with the proneural cluster, which consists of a small number of cells that express proneural genes encoding the basic helix–loop–helix (bHLH) proteins (Achaete, Scute, Asense, Atonal, and Amos), rendering those cells competent to develop into SOPs (Romani et al. 1989; Cubas et al. 1991; Skeath and Carroll 1991; Jarman et al. 1993; Ruiz-Gomez and Ghysen 1993; Goulding et al. 2000; Huang et al. 2000). It is thought that the cells with the highest level of proneural proteins are selected as SOPs, which express a higher level of Delta and activate Notch in neighboring cells (Goriely et al. 1991; Artavanis-Tsakonas et al. 1999). Activation of Notch initiates a signaling cascade in which Suppressor of hairless [_Su(H)_] and Enhancer of split [_E(spl)_] complexes are involved to suppress neuronal fate in non-SOP cells (Knust et al. 1992; Schweisguth and Posakony 1992).
One of the genes activated by bHLH proteins, senseless (sens), encodes a transcription factor with four zinc fingers whose expression is dynamically regulated within the proneural clusters (Nolo et al. 2000). Within the proneural cluster, a high level of Sens is required to up-regulate and maintain the proneural gene expression in SOPs (Nolo et al. 2000), while a low level of Sens represses the transcription of proneural genes in adjacent cells (Jafar-Nejad et al. 2003). This dual function of Sens as an activator or suppressor suggests that the proper level of Sens is required to control the formation of the precise number of SOPs. However, how differential expression levels of Sens in SOPs and adjacent cells are ensured remains unknown.
In recent years, post-transcriptional regulation by microRNAs (miRNAs) has emerged as an important mechanism for controlling gene expression in animal development (Carrington and Ambros 2003; Bartel 2004; He and Hannon 2004; Alvarez-Garcia and Miska 2005; Carthew 2006). miRNAs are endogenous noncoding small RNAs 21–23 nucleotides (nt) in length. Several miRNAs have been implicated in nervous system development, with roles ranging from regulating left/right neuronal asymmetry in Caenorhabditis elegans, photo-receptor formation in Drosophila, and brain morphogenesis in zebrafish, to neuronal differentiation in mammals (Johnston and Hobert 2003; Chang et al. 2004; Giraldez et al. 2005; Li and Carthew 2005; Vo et al. 2005; Krichevsky et al. 2006; Schratt et al. 2006). However, the precise roles of miRNAs in early neurogenesis have not been studied yet. Moreover, only a few miRNAs have been studied using loss-of-function approaches in Drosophila (e.g., Brennecke et al. 2003; Xu et al. 2003; Kwon et al. 2005; Li and Carthew 2005; Sokol and Ambros 2005; Teleman et al. 2006).
Here, through both loss-of-function and gain-of-function in vivo analyses, we found that miR-9a is required to ensure the generation of precise numbers of sensory organs in Drosophila embryos and adults. To accomplish this regulatory function, miR-9a down-regulates the expression of Sens through its 3′ untranslated region (UTR) to ensure the differential expression of Sens in SOPs and adjacent epithelial cells. These findings provide new evidence that miRNAs function at the translational level to ensure the appropriate level of gene expression during different developmental processes.
Results
Generation of miR-9a mutant flies
To understand the roles of miRNAs in neural development, we chose Drosophila miR-9a for further analysis for the following reasons. First, the Drosophila homolog of the human fragile X mental retardation protein affects the formation of terminal dendritic branches (Lee et al. 2003) and is associated with miRNAs and the RNA-induced silencing complex, implicating miRNAs in human mental disorders (Caudy et al. 2002; Ishizuka et al. 2002). Second, miR-9a is one of the miRNAs that are specifically and highly expressed in the mammalian brain, suggesting a unique role in neural development and/or function (Lagos-Quintana et al. 2002). Third, mature miR-9a is 100% conserved at the nucleotide level from flies to humans (Fig. 1A; Aravin et al. 2003), indicating an evolutionarily conserved function. Moreover, in contrast to the mouse genome, which has three miR-9a precursors located on different chromosomes (Lagos-Quintana et al. 2002), the Drosophila genome has only one (Aravin et al. 2003; Sempere et al. 2004), providing a good opportunity to study the function of this miRNA.
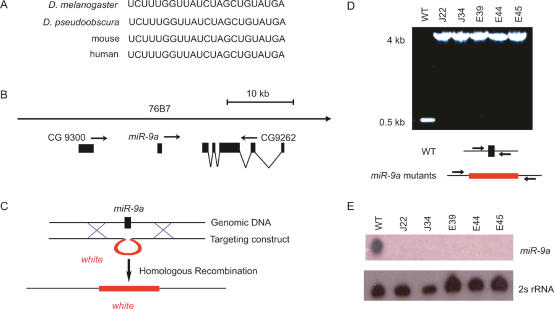
Figure 1.
Generation of _miR_-9a loss-of-function mutant flies. (A) Nucleotide sequence alignment of miR-9a among different species. (B) The cytological location of miR-9a in the Drosophila genome. (C) The ends-out recombination approach was used to replace the 78-nt long miR-9a precursor with the white gene. (D) The miR-9a mutant lines were identified by PCR analysis using primers flanking the miR-9a locus. (E) Northern blotting analysis confirmed that miR-9a was absent in the mutant lines. The same Northern membrane was probed with a 2s rRNA probe as the loading control.
The Drosophila miR-9a precursor is located on the left arm of the third chromosome 76B7, 10 kb to the right of gene CG9300 and 6.4 kb to the left of gene CG9262 (Fig. 1B). To generate loss-of-function mutants of miR-9a, we used ends-out homologous recombination (Gong and Golic 2003) to replace the 78-nt long miR-9a precursor DNA sequence with the white gene (Fig. 1C). To screen for potential miR-9a mutants, we used PCR analysis with two primers flanking miR-9a. These primers produced a 0.5-kb fragment when wild-type genomic DNA was used as the template and a 4-kb fragment if miR-9a was replaced by white (Fig. 1D). From two independent gene-targeting experiments, we generated five mutant lines in which homologous recombination occurred at the miR-9a locus (Fig. 1D). To confirm that the expression of miR-9a was indeed abolished in mutants, we performed Northern blot analysis that showed the absence of miR-9a in all five mutant lines (Fig. 1E). Lines J22 and E39, generated from two independent targeting events, were selected for further analysis.
Loss of miR-9a function results in ectopic sensory neurons in Drosophila embryos and larvae
Both miR-9aJ22 and miR-9aE39 mutant flies developed to adulthood at the expected Mendelian ratio and were fertile, suggesting that miR-9a activity is not essential for survival or fertility. However, when we examined the morphology of PNS neurons, we found unexpectedly that some of the MD neurons were duplicated. Normally, Drosophila embryos and larvae have 12 sensory neurons in each dorsal cluster in the abdominal segments (Fig. 2A), including eight MD neurons. At the larval stage, each MD neuron has a unique dendritic branching pattern that is fairly consistent from segment to segment and among different animals (Bodmer and Jan 1987; Grueber et al. 2002; Sweeney et al. 2002). Using a specific driver, Gal4221, we examined two MD neurons, ddaE and ddaF (Sweeney et al. 2002), which extend smooth dendrites toward the posterior and anterior, respectively (Fig. 2B). Twenty-nine percent of miR-9aJ22 (n = 62) and 37% of miR-9aE39 mutant third instar larvae (n = 27) contained an extra ddaE or ddaF neuron in one or two but not other hemisegments (Fig. 2C,D), while none of the wild-type larvae had this phenotype (n = 24). A similar phenotype was also seen in vpda, a ventral MD neuron that also extends simple and smooth dendritic branches (Supplementary Fig. S1).
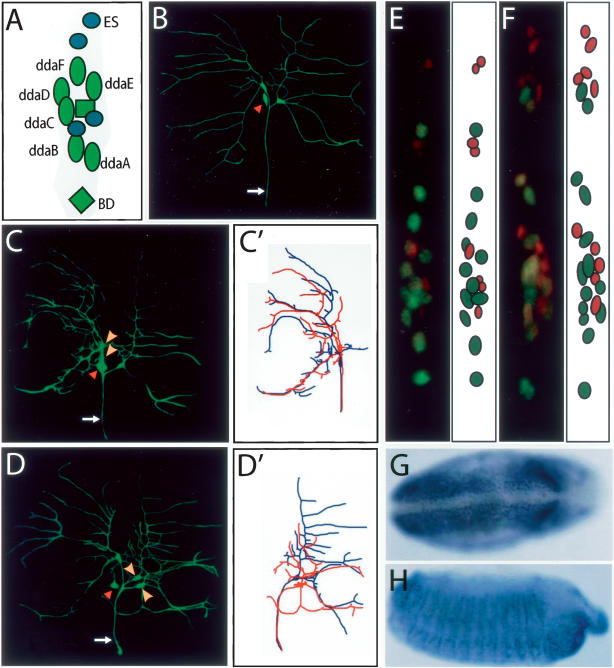
Figure 2.
Loss of miR-9a activity resulted in the formation of ectopic sensory neurons in Drosophila embryos and larvae. (A) Schematic representation of the dorsal cluster sensory neurons in each abdominal hemisegment. (B) Wild-type ddaE and ddaF neurons in third instar larvae are labeled by mCD8-GFP with Gal4221. The white arrows in B, C, and D indicate axons of these sensory neurons. The red arrowheads indicate ddaC neurons that are faintly labeled by GFP with Gal4221. In miR-9a mutant larvae, duplicated ddaF (C) and ddaE (D) neurons (yellow arrowheads) were observed with near normal dendritic branches (traced in _C_′,_D_′). (E,F) All the neurons of one dorsal cluster of stage 15 embryos were double-labeled with anti-ELAV (green) and anti-Cut (red) antibodies. (E) In wild-type embryos, there are 12 ELAV+ cells, including MD neurons and ES neurons, and eight Cut+/ELAV− support cells. (F)A miR-9aJ22 embryo with 14 ELAV+ cells and 10 Cut+/ELAV− support cells. (G,H) miR-9a expression is detected in epithelial cells in wild-type embryos with in situ hybridization. (G) After gastrulation at stage 7, miR-9a expression persists in most epithelial cells except the ventral ectoderm. (H) At stage 12–13, miR-9a is expressed in the ectoderm, with higher levels at the segmental boundaries and stomodeum.
The frequent observation of two-cell clones generated by mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo 1999) that contained an ES neuron and either a ddaE neuron or a ddaF neuron (Sweeney et al. 2002) suggests that ddaE and ddaF neurons are generated from the ES–MD lineages. To examine whether the duplicated neurons in miR-9a mutants resulted from cell fate transformation within the ES–MD lineage or from an ectopic SOP, we counted neurons and support cells of A5–A6 dorsal clusters in embryos. As shown by immunostaining, each wild-type dorsal cluster contained 12 Elav+ neurons (eight MD and four ES neurons) and eight Cut+/ELAV− cells (external cells, Fig. 2E). In the miR-9a mutant embryos, more cells were observed in some dorsal clusters. Among the four extra cells, there were two Elav+ neurons accompanied by two Cut+/ ELAV− external cells (Fig. 2F), suggesting that an extra SOP was generated. These findings suggest that miR-9a affects the precise production of SOPs during Drosophila embryonic development.
To understand how miR-9a is involved in SOP specification, we performed in situ hybridization with a probe for the miR-9a precursor. The earliest expression of miR-9a was at stage 5 and was observed in all cells except most ventral cellular blastula (data not shown). After gastrulation at stage 7, miR-9a expression persisted in all epithelial cells except the ventral ectoderm (Fig. 2G). Ectodermal epithelial cells continued to express miR-9a, and at stage 10–13, strong expression of miR-9a was seen at the ectoderm of the segmental boundaries and stomodeum (Fig. 2H). During this stage, embryonic PNS SOPs are derived from the epidermal cells. Consistently, we observed a similar embryonic expression of miR-9a with a probe made out of the mature miR-9a (data not shown), which was also reported independently during the course of our work by Stark et al. (2005).
miR-9a controls the generation of sensory organs in adult flies
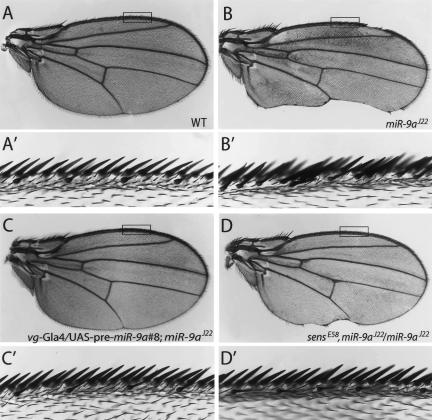
The increased production of sensory neurons in miR-9a mutant embryos/larvae prompted us to further examine the sensory organ formation in adult flies. Wild-type anterior wing margins have a fixed number of regularly spaced sensory bristles (Fig. 3A′). About 40% of miR-9a mutants flies showed ectopic sensory bristles on anterior wing margins (Fig. 3B′), while flies overexpressing miR-9a had a marked decrease in the number of bristles (data not shown). miR-9a mutant flies also exhibited a posterior wing margin defect, which was 100% penetrant (Fig. 3B). To confirm that the wing phenotype was caused by the loss of miR-9a activity, we performed a genetic rescue experiment. Expressing the miR-9a precursor by vg-Gal4, which drives gene expression in the thin strip of dorsal–ventral boundary of wing imaginal discs, in the miR-9a mutant background completely rescued the defects on posterior wing margins (n = 152, Fig. 3C). The bristle defect on the anterior margin of miR-9a mutants was also largely rescued (Fig. 3C′).
Figure 3.
Wing margin defects in miR-9a mutant flies. (A) A wild-type wing. The rectangle indicates the areas enlarged into _A_′ (same for those in B–D). (_A_′) Sensory hairs on anterior wing margin are assembled in a regular pattern with four macrochaetes flanking one microchaete. (B) All miR-9a mutant flies lose the posterior wing margin, while the veins are normal in appearance. (_B_′) Ectopic sensory hairs on the anterior wing margin in miR-9a mutants. (C,_C_′) The wing margin defects in miR-9a were rescued by expression of the wild-type miR-9a precursor (line #8) with _vg-_Gal4 (n = 152). (D,_D_′) The wing margin defects of miR-9a mutants were suppressed by removing one copy of sens. sensE58 was recombined with miR-9aJ22 onto the same chromosome.
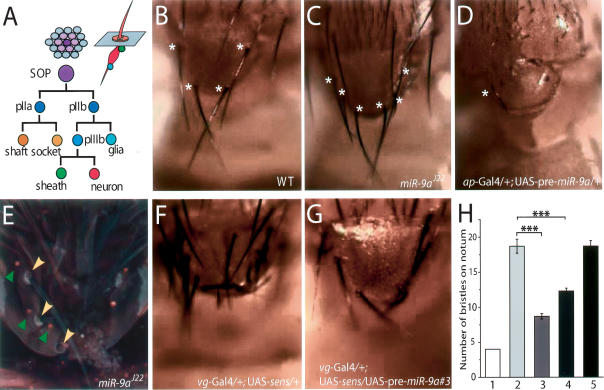
Besides the wing phenotype, miR-9a mutant flies also displayed an ectopic production of sensory organs on the notum. Each SOP generates five different cells in two to three rounds of division to form an entire sensory organ on the notum (Fig. 4A). Wild-type flies have two scutellar bristles (Fig. 4B) on each heminotum, but 14.3% of miR-9a mutant flies (14 out of 98) had three or four bristles (Fig. 4C) with accompanying shaft and socket cells (Fig. 4E). Conversely, flies overexpressing miR-9a in the notum region by ap-Gal4 had a severe loss of scutellar sensory organs (Fig. 4D). Like the wing margin phenotype, defects on the notum of miR-9a mutant could be rescued by expression of miR-9a precursors using vg-Gal4 (n = 152). To further demonstrate the bristle phenotype was due to increased number of scutellar SOPs in miR-9a mutants, we stained the notum with Elav antibody. We found that each ectopic bristle was also associated with an extra neuron (Fig. 4E), supporting our hypothesis that miR-9a regulates the specification of SOPs during sensory organ development.
Figure 4.
miR-9a mutant flies show a sensory bristle defect on the notum, and miR-9a genetically interacts with sens. (A) Schematic representation of the cell lineage of an adult sensory organ from a single SOP. (B) Wild-type flies have four scutellar bristles (asterisks) on the notum. (C) miR-9a mutant flies have four to seven scutellar bristles on the notum (asterisks). (D) Overexpression of miR-9a resulted in a loss of scutellar bristles (asterisk indicates the only remaining bristle). (E) In miR-9a mutant flies, all bristles (yellow arrowhead) have normal shaft and socket cells and are accompanied by ELAV+ sensory neurons (green arrowheads). Some bristles were lost during the staining process. (F) Overexpression of Sens dramatically increased the number of sensory bristles on the notum. (G) Coexpression of miR-9a suppressed the generation of ectopic bristles induced by Sens overexpression. (H) Statistical analysis of genetic interactions between sens and miR-9a. Genotypes are as follows: wild-type (n = 50) (bar 1); vg_-Gal4/+;UAS–_sens/+ (n = 20) (bar 2); vg_-Gal4/+;UAS–_sens/UAS–pre-miR-9a (line #3) (n = 20) (bar 3); vg_-Gal4/UAS–pre-miR-9a; (line #8) UAS–_sens/+ (n = 20) (bar 4); and vg_-Gal4/+;UAS–_sens/UAS–mutant–pre-miR-9a (n = 20) (bar 5). Values are means ± SEM. (***) p < 0.001.
miR-9a genetically interacts with sens
The wing margin phenotypes of miR-9a mutant flies are remarkably similar to that seen in Lyra1 mutant flies, a gain-of-function mutant of sens, and in flies with forced expression of Sens in the wing (Nolo et al. 2001). Moreover, the sens 3′UTR contains _miR-9a_-binding sites (Stark et al. 2003), raising the possibility that Sens might be misregulated in miR-9a mutant flies. To test this hypothesis, we performed genetic interaction studies. In a miR-9aJ22 homozygous mutant background, removing one copy of sens (sensE58/+) led to a substantial rescue of the wing margin defect. About 30% of these flies had wings indistinguishable from the wild type, while others lost only a smaller part of the posterior margin (Fig. 3D); the bristles on the anterior margin also resembled wild type (Fig. 3D′). In contrast, 100% of miR-9a mutant flies had posterior wing margin defects (Fig. 3). These findings suggest that up-regulation of Sens is a key downstream event caused by loss of miR-9a activity.
To provide further evidence that miR-9a regulates Sens during the sensory organ formation, we performed additional genetic interaction experiments on the notum. Overexpressing Sens with the vg_-Gal4 induced ectopic formation of numerous bristles on the notum (Fig. 4F). However, coexpressing of the miR-9a precursor and Sens largely repressed ectopic bristle formation (Fig. 4G), indicating that miR-9a can suppress Sens expression in vivo. In this experiment, the UAS–_sens construct contains the sens 3′UTR. The bristle defect was rescued to different degrees with two different UAS–pre_miR-9a_ transgenic lines (lines #3 and #8) (Fig. 4H). As a control, overexpression of the mutant miR-9a precursor with a 7-nt deletion failed to rescue the sens overexpression phenotype (Fig. 4H). These data provide further evidence that miR-9a and sens function in the same genetic pathway in controlling SOP formation.
miR-9a regulates Sens expression in vivo
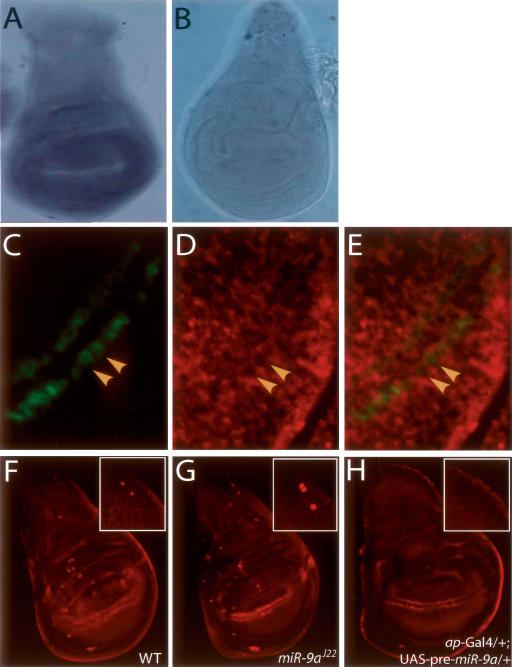
To provide mechanistic insights into the actions of miR-9a in controlling sensory organ formation, we set out to examine whether the Sens protein level could be affected by miR-9a in vivo. In the Drosophila imaginal wing disc, sens is expressed in proneural clusters and accumulates in presumptive SOPs (Nolo et al. 2000). We performed in situ hybridization on the wing discs of third instar larvae and found that miR-9a is widely expressed except for some cells in the wing margins in wild-type flies (Fig. 5A), but not miR-9a mutants (Fig. 5B). To examine miR-9a expression at the cellular level, we performed in situ hybridization of miR-9a on a _A101-_Gal4/UAS–mCD8-GFP disc in which a subset of SOPs is labeled with GFP (Fig. 5C). Interestingly, miR-9a is not expressed in SOPs, but is expressed in most, if not all, epithelial cells, including the ones adjacent to SOPs (Fig. 5D,E). Sens is dynamically regulated within proneural clusters during SOPs specification: It is up-regulated in SOPs and down-regulated in other cells (Nolo et al. 2000). The expression pattern of miR-9a is consistent with the “avoidance hypothesis” that miR-9a may control SOP formation through regulation of gene expression in non-SOP cells (Stark et al. 2005).
Figure 5.
miR-9a is expressed in epithelial cells, including cells adjacent to SOPs in wing imaginal discs. (A) miR-9a is ubiquitously expressed in wild-type wing imaginal discs except the wing margins, as shown with a Dig-labeled probe detected with an AP-conjugated anti-Dig antibody. (B)A miR-9aJ22 mutant wing imaginal disc has no miR-9a expression. (C–E) High-resolution fluorescent in situ hybridization shows that miR-9a is expressed in the epithelial cells, including cells adjacent to the SOPs in the pouch region of a wild-type wing imaginal disc. (C) At the wing margins, SOPs were visualized by mCD8-GFP driven by _A101_-Gal4 (yellow arrowheads indicate two of them). (D) A Dig-labeled miR-9a probe detected by Cy3-conjugated anti-Dig antibody (red) shows miR-9a is expressed in the epithelial cells, including ones adjacent to the SOPs. (E) Merged image of C and D showing miR-9a is excluded from the SOPs in the wing margins. (F–H) Immunostaining of Sens (red) in wing imaginal discs of early third instar larvae. (F) In the wild-type wing disc, Sens is highly expressed in SOPs and wing margins. Inset shows a magnified view of the notum region from which scutellar bristles are derived. (G) miR-9aJ22 mutant imaginal wing disc has ectopic SOPs. (Inset) A high frequency of extra SOPs was observed in the notum region. (H) When miR-9a is overexpressed with _ap-_Gal4 in the dorsal part of the wing disc, Sens expression was suppressed in most of the dorsal compartment. Inset shows the absence of Sens+ cells (SOPs) in the region that develops into the notum.
To provide direct evidence that miR-9a plays a role in SOP formation by regulating Sens expression, we examined Sens expression in the wing imaginal disc. As shown by immunostaining of wing imaginal discs of wild-type third instar larvae, Sens is first expressed in proneural cluster cells at an early stage and is concentrated in SOPs during SOP specification. Two rows of Sens+ cells at the pouch region develop into SOPs in the late third instar stage and into sensory bristles on adult wing margins (Fig. 5F). In the region that develops into the notum, there are two Sens+ cells, which are the precursors for the two scutellar bristles on each heminotum (Fig. 5F, inset). In the miR-9a mutant, there were more SOPs in early third instar wing discs (Fig. 5G), and >60% of the discs contained three or four Sens+ SOPs in the notum region (Fig. 5G, inset). These findings are consistent with the ectopic bristles in adult miR-9a mutant flies (Fig. 4C).
Conversely, overexpression of miR-9a in the dorsal compartment of the wing disc by _ap-_Gal4 (Calleja et al. 1996) significantly reduced the number of Sens+ cells, and the two SOPs in the notum region were absent (Fig. 5H), consistent with the balding phenotype in adult mutant flies (Fig. 4D). These data provide strong evidence that the ectopic sensory organs in miR-9a mutant flies result from ectopic SOPs during neurogenesis, and that miR-9a normally suppresses Sens expression, thereby inhibiting the SOP fate in surrounding epithelial cells. miR-9a directly suppresses sens expression through its 3_′_UTR
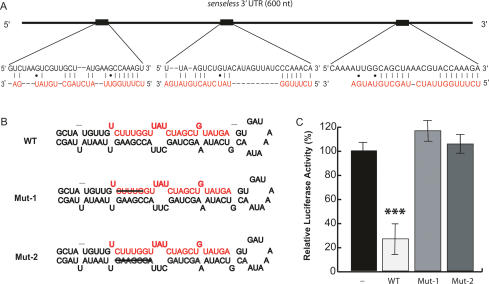
It was predicted that the sens 3′UTR contains three _miR-9a_-binding sites (Fig. 6A; Stark et al. 2003). To test whether miR-9a suppresses Sens expression directly through its 3′UTR, we performed luciferase assays. HEK293 cells were cotransfected with the wild-type miR-9a precursor and sens 3′UTR downstream from the luciferase coding region. The relative luciferase activity was 70% lower than in cells expressing the luciferase-sens 3′UTR alone (Fig. 6C). Two mutant constructs of miR-9a precursors were generated as controls, one with a 5-nt deletion in the mature miR-9a region (Mut-1) and one with a 7-nt deletion in the stem of the precursor (Mut-2) (Fig. 6B). Both of these mutant constructs failed to suppress the expression of luciferase with the sens 3′UTR (Fig. 6C). As an additional control, none of the miR-9a constructs had any effect on the expression of luciferase without the sens 3′UTR (data not shown). These findings provide strong evidence that miR-9a directly suppresses sens expression through its 3′UTR.
Figure 6.
Luciferase assay showing the suppression by miR-9a through the sens 3′UTR. (A) The sequences and locations of three predicted _miR-9a_-binding sites in the sens 3′UTR and their alignment with miR-9a (red). (B) The predicted stem-loop-stem structure of the miR-9a precursor. (WT) Wild-type precursor; (Mut-1) mutant precursor 1 with a 5-nt deletion in mature miR-9a; (Mut-2) the mutant precursor with a 7-nt deletion in the stem region complementary to mature miR-9a. Mature miR-9a is highlighted in red, and strikethroughs indicate the deleted nucleotides. (C) Wild-type (WT) but not mutant miR-9a precursors (Mut-1 and Mut-2) could suppress the expression of luciferase with the sens 3′UTR. Bars indicate the average of luciferase activity in one triplicated experiment. (***) p < 0.001.
Discussion
Drosophila miR-9a suppresses neuronal precursor specification
miR-9a is one of the miRNAs that are highly expressed in the mammalian brain and 100% conserved at the nucleotide sequence from flies to humans, suggesting an important role in brain development and/or function (Lagos-Quintana et al. 2002; Aravin et al. 2003; Sempere et al. 2004). We generated miR-9a loss-of-function alleles and found that homozygous mutant flies developed into adulthood at the expected Mendelian ratio. Adult mutant flies are grossly normal and fertile, indicating that miR-9a is not required for viability or fertility. This finding is different from the reported severe dorsal closure defects and embryonic lethal phenotype generated by antisense 2′O-methyl oligoribonucleotide-mediated depletion of miR-9a (Leaman et al. 2005).
Interestingly, Drosophila miR-9a is not expressed in mature neurons, but is expressed in epithelial cells, including the proneural clusters that give rise to SOPs (Figs. 2, 5; Stark et al. 2005). Detailed analysis of embryonic PNS development revealed an unexpected finding that miR-9a mutants have an increased number of sensory neurons (Fig. 2) that elaborate extensive dendritic arbors underneath the epithelial cell layer, such as ddaE and ddaF neurons (Sweeney et al. 2002). The duplicated neurons occupy the same dendritic field and appear to have similar dendritic branching patterns (Fig. 2C,D). Indeed, the average numbers of dendritic ends of ddaE and ddaF neurons in abdominal segments 3–5 were similar in wild-type and miR-9a mutant larvae and MARCM clones (data not shown), indicating that loss of miR-9a activity affected the number of these sensory neurons only but had no cellautonomous effect on their dendritic branching patterns.
The effect of miR-9a on the number of embryonic sensory neurons has two major features. First, the ectopic ddaE or ddaF neurons were generated as a result of ectopic SOPs and not cell fate transformation within a cell lineage (Fig. 2), suggesting miR-9a affects an early step in neurogenesis, consistent with its embryonic expression pattern (Fig. 2; Stark et al. 2005). Second, both the expressivity and penetrance of this defect were relatively low. This finding supports the idea that miRNAs, at least in this particular case, are not developmental switches, but instead function as a fine-tuning mechanism to ensure the accuracy of a particular developmental process (e.g., Farh et al. 2005; Stark et al. 2005).
In this study, we focused on the formation of SOPs in adult flies. Like embryos, only ∼14% of miR-9a mutant flies exhibited ectopic SOPs on the notum, again indicating a fine-tuning role for miR-9a in controlling SOP formation. However, our analysis of the miR-9a mutant phenotype in adult flies also indicates that miRNAs can have dramatic effects on some other developmental processes. For instance, miR-9a is widely expressed in the wing disc (Fig. 5), and 100% of miR-9a mutant flies exhibited a severe posterior wing margin defect (Fig. 3), suggesting that cell proliferation and/or survival are much more sensitive to changes in the expression levels of the proteins regulated by miR-9a.
MiR-9a inhibits Sens expression in non-SOP cells
How does miR-9a exert its effect on SOP formation? Sens is a zinc finger transcription factor required to maintain high-level expression of proneural gene in SOPs and to suppress their expression in non-SOP cells (Nolo et al. 2000; Jafar-Nejad et al. 2003). Several findings in this study demonstrate that Sens is a key target of miR-9a regulation and is essential for mediating miR-9a function in SOP formation. (1) The wing margin defects in miR-9a mutant flies were remarkably similar to that caused by overexpression of Sens by the UAS-Gal4 system or in Lyra1 mutants (Nolo et al. 2000, 2001). (2) miR-9a was expressed at a much lower level in SOPs than in adjacent epithelial cells, correlating with the high level of Sens expression in SOPs and the low level of Sens in non-SOP cells in proneural clusters. The inability to use immunostaining to detect subtle changes of Sens expression level in non-SOP cells due to miR-9a loss of function could be attributed to the following reasons: Sens expression is primarily down-regulated at the transcriptional level in the non-SOP proneural cells, and _miR-9a_’s function is limited to preventing translation of the leaky/residual sens mRNA, consistent with the model proposed by Stark et al. (2005). The alteration in Sens level, due to loss of miR-9a function in the non-SOP cells, is sufficient to initiate the production of ectopic SOPs, but it may not be dramatic enough to be detected by immunostaining. (3) The sens 3′UTR contains three _miR-9a_-binding sites and is the best predicted target of miR-9a (Stark et al. 2003). (4) Wild-type but not mutant miR-9a precursors down-regulated reporter gene expression through the sens 3′UTR in transfected cells. (5) Overexpression of miR-9a in vivo inhibited Sens expression. We observed that Sens expression along the wing margin in the dorsal compartment was lower than in the ventral compartment in some wing discs when miR-9a was expressed by _ap_-Gal4 (data not shown). The failure to completely suppress Sens expression in the dorsal compartment (Fig. 5H) is probably due to the fact that, at this developmental stage, Sens expression in the wing margins is controlled by proneural genes, unlike SOPs in the notum region where Sens expression is maintained by itself (Jafar-Nejad et al. 2003). (6) miR-9a and sens showed strong genetic interactions in controlling SOP formation.
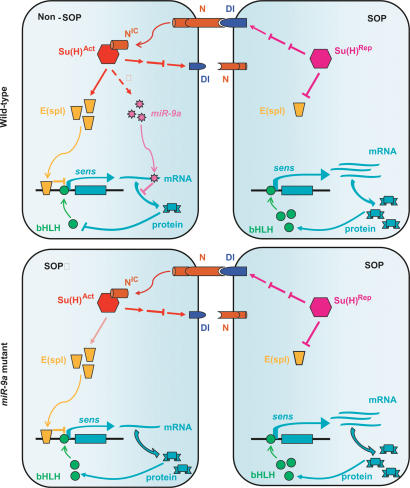
These findings provide an experimental example to support the notion that miRNAs and their mRNA targets are often expressed in cells adjacent to each other (Stark et al. 2005). The differential expression of Sens in SOPs and adjacent neuroepithelial cells is essential for the production of a precise number of SOPs during development (Nolo et al. 2000; Jafar-Nejad et al. 2003). We propose a model in which miR-9a functions in non-SOPs cells to further suppress Sens expression at the translational level, as a complementary mechanism to the transcriptional inhibition of Sens expression by E(spl) (Fig. 7). Loss of miR-9a function increases Sens protein level, not so dramatically but just enough to convert Sens in some neuroepithelial cells from a transcription repressor into an activator of proneural genes, therefore resulting in the formation of a small number of ectopic SOPs. However, unlike many other genes essential for neurogenesis, such as Notch and Delta, miR-9a does not function as an absolute switch. Instead, it only ensures the accurate differential Sens expression and fine-tunes this developmental process.
Figure 7.
A model for the role of miR-9a in regulating SOP specification. Within one proneural cluster, the cell selected to become the SOP expresses a high level of Delta to activate Notch in the adjacent cells (non-SOPs). Upon activation, Notch releases the intracellular domain (NIC), which translocates to nuclei and binds to Suppressor of Hairless, switching it to the active form [Su(H)Act]. The NIC/Su(H)Act complex activates Enhancer of Split [E(spl)], and consequently represses sens transcription. In SOPs, without the activation of Notch, Su(H) functions as a repressor [Su(H)Rep] to suppress the E(spl). Therefore, sens could be continuously transcribed, allowing high-level accumulation of Sens protein, which is required for further transcription of neuron-specific genes. miR-9a functions in non-SOP cells to further suppress Sens translation through its 3′UTR to ensure the precise production of SOPs.
Overexpression of miR-9a in the wing imaginal disc could dramatically inhibit the formation of sensory organs on the notum (Fig. 4D), suggesting that misregulation of miR-9a expression itself could potentially have severe developmental consequences. Since both miR-9a and E(spl) have similar functions in non-SOP cells, it is possible that both genes may be regulated by a similar transcriptional mechanism. Indeed, we noticed that binding sites for the Achaete–Scute complex and Su(H) are present in the regulatory region of miR-9a. Taken together, our studies presented here have uncovered another layer of gene regulation during early neurogenesis in the Drosophila PNS. _miR_-9a is 100% conserved at the nucleotide level from flies to humans. Moreover, the human miR-9a is highly expressed in fetal but not in adult brains (Nelson et al. 2006). Therefore, a similar mechanism of miR-9a function may operate during mammalian neurogenesis as well.
Materials and methods
Fly strains and genetics
All the flies were maintained at 22°C–25°C on standard medium. The w1118 strain was used as a wild-type control. Gal4221 combined with UAS–mCD8::GFP was used to visualize individual ddaE and ddaF neurons in the larval PNS. It was recombined with miR-9a mutants to generate the homozygous stock Gal4221, UAS–mCD8::GFP, miR-9a. For studies of the wing and the notum, the following lines were used: _vg-_Gal4, _ap-Gal4 (Calleja et al. 1996), and UAS–_sens (Nolo et al. 2000). The presence of sens 3′UTR in the UAS–sens construct was confirmed by PCR. sensE58 was used as the sens loss-of-function mutant allele in genetic interaction experiments.
Target homologous recombination constructs
miR-9a loss-of-function mutations were generated with the ends-out technology (Gong and Golic 2003). The targeting construct contains the w+ gene flanked by two arms of genomic DNA fragments at the miR-9a locus. The two arms were prepared by PCR from the genomic DNA using primers 5′-AGCGATCGTCGTCGGACTAC-3′ and 5′-ACACTGCAGATGGTTGAAAG-3′ for the left arm, and primers 5′-ACTCGAGCCAAAAACGAGGCCCACA-3′ and 5′-AGGTACCGAGACAG CAAAATCGTAGAA-3′ for the right arm. Two independent transgenic fly lines carrying the targeting construct were crossed to hs-FLP, I-SceI (Bloomington Stock Center), and the progenies at late second to third instar stages were heat-shocked for 45 min at 38°C. Virgin female flies with mosaic eyes were collected and crossed to p{70FLP}10 (Bloomington Stock Center), and at the next generation, individual males with solid-color eyes were crossed to TM3 Ser/TM6 Tb as candidate lines. For the replacement of the miR-9a by w+, a PCR screen was performed with genomic DNAs isolated from each line. Northern blot was used to confirm the absence of miR-9a expression in mutant flies.
Transgenic constructs
The 78-base-pair (bp) miR-9a precursor was cloned from wild-type genomic DNA with primers 5′-GAATTCTATACAGGGTGCTATGTTG-3′ and 5′-TCTAGACGCTGGGCAGACGCTAATATTAACTTCGG-3′. The mutant precursor with a 7-bp deletion on the stem was generated with primers 5′-GAATTCTATACAGGGTGCTATGTTG-3′ and 5′-TCTAGACGCT GGCAGACGCTAATATTAAAAGCTAGCTTTATGACGT-3′. Both fragments were subcloned into the pUAST vector and injected into embryos to generate transgenic fly stocks that express either wild-type or mutant miR-9a precursor under the control of UAS elements.
Northern blot
Total RNA was extracted from larvae with Trizol, according to the manufacturer's instructions (Invitrogen). For each sample, 20 μg of total RNA was run on a 12% polyacrylamide gel (Sequagel, National Diagnostics) and transferred to a Nytran SuperCharge Signal membrane (Schleicher & Schuell). miR-9a was detected with a purified 32P-end-labeled oligo probe, 5′-TCATACAGCTAGATAACCAAAGA-3′, which is complementary to the mature miR-9a sequence.
In situ hybridization
Embryos and wing imaginal discs were fixed and stained according to standard procedures (Tautz and Pfeifle 1989). A genomic fragment of 1 kb flanking the miR-9a precursor was obtained by PCR and cloned into the pBSK vector. A Digoxigenin (Dig)-labeled riboprobe was generated by in vitro transcription of this construct with T7 RNA polymerase. A Drosophila miR-9a locked nucleic acid (LNA) probe was obtained from Exiqon (Vedbaek), end-labeled with a Dig oligonucleotide 3′-end-labeling kit (Roche), and purified on a G-25 Microspin column (Amersham Biosciences). In situ hybridization with both probes was detected by AP-conjugated cy3-conjugated anti-Dig antibody according to standard procedures (Tautz and Pfeifle 1989) with the following modifications: After post-fixation and washes, the discs were washed with water and treated twice for 5 min each with 0.25% (v/v) acetic anhydride in 0.1 M triethanolamine (pH 8.0). The tissues were prehybridized and hybridized at 50°C in hybridization buffer (pH 6.0, adjusted with citric acid).
Antibody staining
Embryos were fixed according to the standard protocol. Third instar larval wing imaginal discs were fixed in 4% formaldehyde in PBT (PBS and 0.1% Triton X-100) for 30 min at room temperature. Pupae were dissected 48 h after pupa formation and fixed in 4% formaldehyde in PBT overnight at 4°C. The following pairs of primary and secondary antibodies were used for all experiments: anti-Sens (1:1000) (Nolo et al. 2000) and anti-guinea pig-Cy3 (1:500; Molecular Probes); anti-ELAV (1:100; Developmental Hybridoma Study Bank) and anti-mouse-Cy3 or anti-mouse-FITC (1:300; Jackson Laboratory); anti-Cut (1:1000; Developmental Hybridoma Study Bank) and anti-rabbit-Cy3 (1:300; Jackson Laboratory).
Microscopy and imaging
All samples with fluorescent signals were imaged by confocal microscopy (Nikon, D-Eclipse C1), including GFP-labeled DA neurons in third instar larvae; immunostaining on embryos, wing discs, and the pupa notum; and fluorescence-labeled in situ hybridization on wing discs. Images of adult wing and notum and Dig-labeled in situ hybridization on wing discs were obtained with an Olympus BX60 microscope and AxioCam Hrc camera.
Luciferase reporter assays
The wild-type and two mutant miR-9a precursors were obtained by PCR and cloned into the pSUPER vector under the control of the H1 promoter (OligoEngine). A 600-bp fragment of the sens 3′UTR was cloned into the pGL3-promoter vector (Promega) downstream from the firefly luciferase gene. The Renilla luciferase plasmid served as the transfection control. HEK293 cells were cotransfected in 12-well plates with three vectors (0.5 μg of each vector per well): the luciferase-sens 3′UTR reporter plasmid, the Renilla luciferase plasmid, and the pSUPER vector containing miR-9a precursor (wild type, Mut-1, or Mut-2) or the vector only. Three independent transfections with triplicates each time were performed, and cell lysates were collected 28 h after transfection. Relative luciferase activity was measured with Dual-luciferase assays (Promega) according to the manufacturer's protocol.
ACKNOWLEDGMENTS
We thank H. Bellen, the Bloomington Stock Center, and the Developmental Hybridoma Bank for reagents and fly lines. We thank S. Ordway and G. Howard for editorial assistance, K. Nyuyen for manuscript preparation, and L.-P. Chang for help with the UAS–pre-_miR_-9a construct. We also thank T. Kornberg, Y. Hong, and laboratory members for discussions and comments during the course of this work. This study was supported by an NIH training grant (T32 AG00278 to F.W.), the Korea Research Foundation (KRF: M01-2005-000-10157-0 to J.L.), and the Esther A. and Joseph Klingenstein Fund, the McKnight Endowment Fund for Neuroscience, FRAXA Foundation, Pfizer/ AFAR, and the NIH (to F.-B.G.).
Footnotes
References
- Ainsley, J.A., Pettus, J.M., Bosenko, D., Gerstein, C.E., Zinkevich, N., Anderson, M.G., Adams, C.M., Welsh, M.J., Johnson, W.A. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 2003;13:1557–1563. doi: 10.1016/s0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia, I., Miska, E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., Rand, M.D., Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bardin, A.J., Le Borgne, R., Schweisguth, F. Asymmetric localization and function of cell-fate determinants: A fly's view. Curr. Opin. Neurobiol. 2004;14:6–14. doi: 10.1016/j.conb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bodmer, R., Jan, Y.N. Morphological differentiation of the embryonic peripheral neurons in. Drosophila. Rouxs Arch. Dev. Biol. 1987;196:69–77. doi: 10.1007/BF00402027. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in. Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brewster, R., Bodmer, R. Origin and specification of type II sensory neurons in. Drosophila. Development. 1995;121:2923–2936. doi: 10.1242/dev.121.9.2923. [DOI] [PubMed] [Google Scholar]
- Calleja, M., Moreno, E., Pelaz, S., Morata, G. Visualization of gene expression in living adult. Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J.A., Hartenstein, V. Springer-Verlag; Heidelberg, New York, Tokyo: 1985. The embryonic development of Drosophila melanogaster. [Google Scholar]
- Carrington, J.C., Ambros, V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Carthew, R.W. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., Hammond, S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S., Johnston, R.J., Jr., Frokjaer-Jensen, C., Lockery, S., Hobert, O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Cubas, P., de Celis, J.F., Campuzano, S., Modolell, J. Proneural clusters of achaete–scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes & Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- Farh, K.K.-W., Grimson, A., Jan, C., Benjamin, P., Lewis, B.P., Wendy, K., Johnston, W.K., Lim, L.P., Christopher, B., Burge, C.B., et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Gao, F.-B., Brenman, J.E., Jan, L.Y., Jan, Y.N. Genes regulating dendritic outgrowth, branching, and routing in Drosophila . Genes& Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen, A., Dambly-Chaudiere, C. The specification of sensory neuron identity in. Drosophila. Bioessays. 1993;15:293–298. doi: 10.1002/bies.950150502. [DOI] [PubMed] [Google Scholar]
- Ghysen, A., Dambly-Chaudiere, C., Aceves, E., Jan, L.Y., Jan, Y.N. Sensory neurons and perihperal pathways in Drosophila embryos. Rouxs Arch. Dev. Biol. 1986;195:281–289. doi: 10.1007/BF00376060. [DOI] [PubMed] [Google Scholar]
- Giraldez, A.J., Cinalli, R.M., Glasner, M.E., Enright, A.J., Thomson, J.M., Baskerville, S., Hammond, S.M., Bartel, D.P., Schier, A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gong, W.J., Golic, K.G. Ends-out, or replacement, gene targeting in. Drosophila. Proc. Natl. Acad. Sci. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely, A., Dumont, N., Dambly-Chaudiere, C., Ghysen, A. The determination of sense organs in. Effect of the neurogenic mutations in the embryo. Development . Drosophila. 1991;113:1395–1404. doi: 10.1242/dev.113.4.1395. [DOI] [PubMed] [Google Scholar]
- Goulding, S.E., zur Lage, P., Jarman, A.P. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by. lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Grueber, W.B., Jan, L.Y., Jan, Y.N. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Hartenstein, V., Posakony, J.W. Development of adult sensilla on the wing and notum of Drosophila melanogaster . Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- He, L., Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Huang, M.L., Hsu, C.H., Chien, C.T. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron. 2000;25:57–67. doi: 10.1016/s0896-6273(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., Siomi, M.C., Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafar-Nejad, H., Acar, M., Nolo, R., Lacin, H., Pan, H., Parkhurst, S.M., Bellen, H.J. Senseless acts as a binary switch during sensory organ precursor selection. Genes & Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, Y.N., Jan, L.Y. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- Jarman, A.P., Brand, M., Jan, L.Y., Jan, Y.N. The regulation and function of the helix–loop–helix gene, asense, in Drosophila neural precursors. Development. 1993;119:19–29. doi: 10.1242/dev.119.Supplement.19. [DOI] [PubMed] [Google Scholar]
- Johnston, R.J., Hobert, O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans . Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Knust, E., Schrons, H., Grawe, F., Campos-Ortega, J.A. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix–loop–helix proteins. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., Sonntag, K.-C., Isacson, O., Kosik, K.S. Specific MicroRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C., Han, Z., Olson, E.N., Srivastava, D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Leaman, D., Chen, P.Y., Fak, J., Yalcin, A., Pearce, M., Unner-stall, U., Marks, D.S., Sander, C., Tuschl, T., Gaul, U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lee, T., Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee, A., Li, W., Xu, K., Bogert, B.A., Su, K., Gao, F.-B. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Li, X., Carthew, R.W. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Liu, L., Yermolaieva, O., Johnson, W.A., Abboud, F.M., Welsh, M.J. Identification and function of thermosensory neurons in Drosophila larvae. Nat. Neurosci. 2003;6:267–273. doi: 10.1038/nn1009. [DOI] [PubMed] [Google Scholar]
- Modolell, J. Patterning of the adult peripheral nervous system of. Drosophila. Perspect. Dev. Neurobiol. 1997;4:285–296. [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Kloosterman, W.P., Kauppinen, S., Plasterk, R.H., Mourelatos, Z. RAKE and LNA ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo, R., Abbott, L.A., Bellen, H.J. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in. Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Nolo, R., Abbott, L.A., Bellen, H.J. Drosophila Lyra mutations are gain-of-function mutations of senseless. Genetics. 2001;157:307–315. doi: 10.1093/genetics/157.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani, S., Campuzano, S., Macagno, E.R., Modolell, J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes & Dev. 1989;3:997–1007. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., Ghysen, A. The expression and role of a proneural gene, achaete, in the development of the larval nervous system of. Drosophila. EMBO J. 1993;12:1121–1130. doi: 10.1002/j.1460-2075.1993.tb05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt, G.M., Tuebing, F., Nigh, E.A., Kane, C.G., Sabatini, M.E., Kiebler, M., Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F., Posakony, J.W. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensoryorgan cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-ex-pressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath, J.B., Carroll, S.B. Regulation of achaete–scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes & Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- Sokol, N.S., Ambros, V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., Cohen, S.M. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Bushati, N., Russell, R.B., Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Sweeney, N.T., Li, W., Gao, F.B. Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev. Biol. 2002;247:76–88. doi: 10.1006/dbio.2002.0702. [DOI] [PubMed] [Google Scholar]
- Tautz, D., Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Teleman, A.A., Maitra, S., Cohen, S.M. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes & Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey, W.D., Jr., Wilson, R.I., Laurent, G., Benzer, S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- Vervoort, M., Merritt, D.J., Ghysen, A., Dambly-Chaudiere, C. Genetic basis of the formation and identity of type I and type II neurons in Drosophila embryos. Development. 1997;124:2819–2828. doi: 10.1242/dev.124.14.2819. [DOI] [PubMed] [Google Scholar]
- Vo, N., Klein, M.E., Varlamova, O., Keller, D.M., Yamamoto, T., Goodman, R.H., Impey, S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., Hay, B.A. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;29:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]