Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly (original) (raw)
Abstract
Although retroviruses from different genera form morphologically distinct capsids, we have proposed that all of these structures are composed of similar hexameric arrays of capsid (CA) protein subunits and that their distinct morphologies reflect different distributions of pentameric declinations that allow the structures to close. Consistent with this model, CA proteins from both HIV-1 and Rous sarcoma virus (RSV) form similar hexagonal lattices. However, recent structural studies have suggested that the Moloney murine leukemia virus (M-MuLV) CA protein may assemble differently. We now report an independent three-dimensional reconstruction of two-dimensional crystals of M-MuLV CA. This new reconstruction reveals a hexameric lattice that is similar to those formed by HIV-1 and RSV CA, supporting a generalized model for retroviral capsid assembly.
Keywords: CA/electron crystallography/Moloney murine leukemia virus (M-MuLV)/retroviral capsid assembly
Introduction
During the final stages of retroviral replication, the viral Gag polyprotein assembles into spherical particles that bud through the plasma membrane (Krausslich, 1996; Göttlinger, 2001). As the virions are released, Gag is cleaved into three discrete proteins: matrix (MA), capsid (CA) and the nucleocapsid (NC), which then rearrange to form the mature, infectious virion. In the mature virus, MA remains associated with the viral membrane, NC condenses to the center of the virion together with the RNA genome, and CA forms the capsid that surrounds the central ribonucleoprotein complex. Although all immature retroviral particles adopt spherical morphologies, the shapes of the mature viral capsids vary considerably, with different retroviral genera forming spherical, conical or cylindrical capsids (Coffin et al., 1997). For example, the capsids of HIV-1 (a lentivirus) are typically conical (Welker et al., 2000), whereas capsids of Rous sarcoma virus (RSV; an alpharetrovirus) and Moloney murine leukemia virus (M-MuLV; a gammaretrovirus) are approximately spherical (Yeager et al., 1998; Kingston et al., 2001).
High-resolution structures have now been determined for a number of CA proteins, which demonstrate that the retroviral CA tertiary structure is highly conserved (Gamble et al., 1996, 1997; Gitti et al., 1996; Momany et al., 1996; Berthet-Colominas et al., 1999; Jin et al., 1999; Khorasanizadeh et al., 1999; Campos-Olivas et al., 2000; Kingston et al., 2000). All CA proteins examined to date contain two distinct domains. The elongated N-terminal domain (NTD) is composed of a β-hairpin followed by seven α-helices, whereas the smaller, globular C-terminal domain (CTD) is composed of four α-helices. A flexible linker of ∼5 amino acids connects the two domains. HIV-1 CA exhibits a monomer–dimer equilibrium in solution (_K_d = ∼20 µM), with dimerization mediated through the CTD (Rosé et al., 1992; Gamble et al., 1997; Worthylake et al., 1999). However, no other CA proteins examined to date form stable dimers in solution (Khorasanizadeh et al., 1999).
Despite differences in CA dimerization and capsid morphologies, the conserved CA tertiary structure suggests that the underlying principles of capsid organization may be conserved across retroviruses. However, structural studies that might elucidate such principles have been impeded by a lack of symmetry within individual retroviral capsids and by heterogeneity between capsids from different viral particles (Yeager et al., 1998; Welker et al., 2000; Kingston et al., 2001). To overcome these limitations, symmetrical arrays of CA proteins from HIV-1, RSV and M-MuLV have been assembled and studied in vitro by electron microscopy (EM) and image analysis.
Helical tubes are one class of symmetrical assemblies formed by retroviral CA proteins, and the best characterized examples are the helical tubes formed by the HIV-1 CA and CA–NC proteins (Ehrlich et al., 1992; Gross et al., 1997; Ganser et al., 1999; Li et al., 2000). Three-dimensional (3D) helical reconstructions of frozen– hydrated tubes have revealed that the CA protein is organized on a hexagonal lattice, with the NTD forming external hexameric rings and the CTD forming internal dimeric contacts that connect adjacent hexamers (Li et al., 2000). Similar helical assemblies have also been reported for CA proteins of RSV and M-MuLV, but have not yet been analyzed in three dimensions (Campbell and Vogt, 1995; Kingston et al., 2000; Zuber et al., 2000).
To rationalize how CA proteins of similar structure can form the distinct morphologies of different retroviral capsids, we have proposed that all capsids could be assembled on similar hexagonal lattices, and differ only in the distribution of the 12 pentagonal defects (‘pentons’) required to close the lattice (Ganser et al., 1999). Thus, tubular capsids could close via the introduction of six pentons at either end of the tube, conical capsids could be closed by an asymmetric distribution of pentons in the two non-equivalent caps and spherical capsids could be created by distributing the pentons more evenly throughout the hexagonal lattice.
In addition to helical tubes, N-terminally His-tagged CA proteins can also form 2D crystals on lipid monolayers that are doped with nickel-chelating lipids. CA proteins of RSV, HIV-1 and M-MuLV have all been crystallized in this fashion and analyzed by EM and image analysis (Barklis et al., 1997, 1998; Kingston et al., 2000; McDermott et al., 2000; Mayo et al., 2002b). Projection maps from 2D crystals of all three CA proteins have suggested hexameric arrangements of the subunits, in good agreement with the HIV-1 CA helical reconstructions and supporting our generalized model. Moreover, a recent 3D reconstruction revealed that 2D crystals of RSV CA are composed of CA hexamers that appear very similar to those observed for HIV-1 CA (Mayo et al., 2002b). However, Barklis and co-workers have produced two separate 3D reconstructions (presented in three separate publications) of the 2D crystals of M-MuLV CA that differ significantly from one another and from the RSV and HIV-1 CA reconstructions (McDermott et al., 2000; Mayo et al., 2002a, 2003). In the first reconstruction (McDermott et al., 2000), the reported M-MuLV CA lattice lacked symmetry (plane group _p_1), and the subunits were modeled as strips of asymmetric CA dimers. In the second and third reconstructions (Mayo et al., 2002a, 2003), the M-MuLV CA lattice was reportedly hexagonal (_p_6). However, as compared with the RSV and HIV CA reconstructions, the maximal density in the M-MuLV CA reconstruction was displaced further away from the six-fold axes, leading the authors to model the connections between hexamers as NTD dimer interactions (rather than CTD dimer contacts, as proposed for the other CA reconstructions). Both M-MuLV CA reconstructions were interpreted to suggest that the M-MuLV capsid assembles differently from capsids of other retroviral genera. Because this issue has important implications for our understanding of retrovirus structure, we have reinvestigated the structure of M-MuLV CA crystals on lipid monolayers to determine whether M-MuLV CA represents a true outlier that requires a new paradigm for understanding retroviral capsid assembly.
Results and discussion
Structure determination
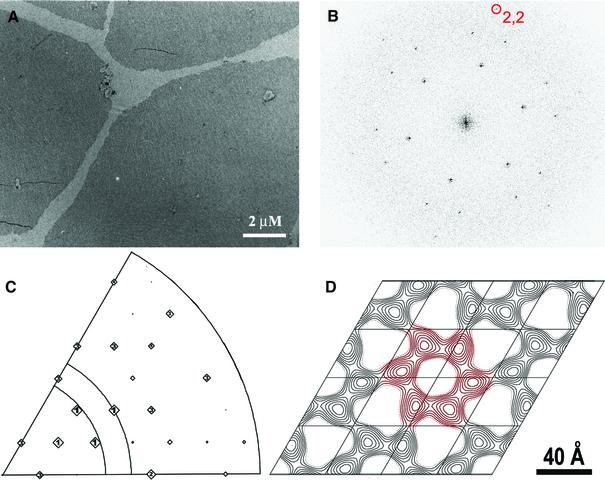
Recombinant, N-terminally His-tagged M-MuLV CA was expressed in Escherichia coli, purified using nickel chelate and cation-exchange chromatography, and crystallized on monolayers doped with nickel-chelating lipids. As reported previously (Barklis et al., 1997), large well-ordered crystals were observed under a variety of crystallization conditions. However, our initial analyses were hampered by the formation of multi-layered crystals. This problem was resolved by reducing both the protein and lipid concentrations, which produced very large single-layered crystals (see Materials and methods; Figure 1A).
Fig. 1. Two-dimensional crystals of M-MuLV CA. (A) Single-layer crystals of M-MuLV (His6)-CA on a Ni2+-doped lipid monolayer. (B) Computed Fourier transform of a stained, nominally untilted 2D crystal of M-MuLV CA. (C) Plot of significant reflections identified in (B) following unbending and boxing. The size of the box is proportional to the ratio of the background-subtracted amplitude to the averaged background of the corresponding reflection. Reflections with the highest signal-to-noise ratio are labeled with ‘1–4’. (D) Two-dimensional Fourier-filtered projection image of M-MuLV CA crystals (_p_1 plane group). Nominally untilted images from the 11 best tilt series were used to calculate the merged projection map.
Untilted diffraction patterns appeared hexagonal and exhibited high signal-to-noise reflections that extended to ∼10 Å resolution after lattice corrections (Figure 1B), although our reconstructions appear to be dominated by reflections between 20 and 30 Å resolution (see below). Indexing in plane group _p_1 gave a unit cell that was nearly hexagonal (a = 80.2 ± 0.4 Å, b = 80.3 ± 0.5 Å, γ = 120.4 ± 0.3°). This unit cell is ∼10% larger than that originally reported for negatively stained M-MuLV CA crystals (a = 72.6 Å, b = 72.5 Å, γ = 119.5°) (McDermott et al., 2000), but matches the cell dimensions subsequently reported for negatively stained crystals and unstained crystals of M-MuLV CA in vitreous ice (Barklis et al., 1997; Mayo et al., 2002a, 2003).
A projection density map was initially calculated from merged and corrected diffraction patterns of 11 separate crystals (Figure 1C). To avoid bias, the diffraction patterns were merged without imposed symmetry. As a ∼ b, all six indexing options were calculated for every diffraction pattern (McDermott et al., 2000). Origin refinement against a reference image (6443) was performed for each indexing option, and the option that gave the lowest phase residuals was used for the merging. The resulting projection map is clearly hexameric (Figure 1D) and ALLSPACE (Crowther et al., 1996) analyses of each untilted image also suggested the presence of _p_6 symmetry (Table I). Indeed, the M-MuLV CA structure appears very similar to the projection maps calculated from 2D crystals of RSV CA and flattened helical crystals of HIV-1 CA (Li et al., 2000; Mayo et al., 2002a, 2003).
Table I. Averaged internal phase residuals for 11 untilted 2D M-MuLV CA crystals.
| Space group | Residuals (90° is random) |
|---|---|
| _P_1 | 19.2 ± 1.9 |
| _P_2 | 17.1 ± 4.1 |
| _p_12_b | 62.9 ± 10.9 |
| _p_12_a | 52.3 ± 24.3 |
| _p_121_b | 21.2 ± 13.3 |
| _p_121_a | 64.7 ± 11.0 |
| _c_12_b | 62.9 ± 10.9 |
| _c_12_a | 52.3 ± 24.3 |
| _p_222 | 51.1 ± 8.0 |
| _p_2221b | 56.1 ± 4.8 |
| _p_2221a | 24.2 ± 5.7 |
| _p_22121 | 55.2 ± 4.3 |
| _c_222 | 51.1 ± 8.0 |
| _P_3 | 10.0 ± 3.4 |
| _p_312 | 42.1 ± 2.5 |
| _p_321 | 40.2 ± 3.3 |
| _P_6 | 11.8 ± 2.9 |
| _p_622 | 39.7 ± 2.3 |
To create an unbiased 3D structure of the M-MuLV CA crystals, diffraction patterns from 40 tilted and untilted crystals were initially merged in the plane group _p_1 (i.e. without imposed symmetry) using the _p_6 phase origin. Although the _p_1 data set was incomplete, both the lattice lines (Figure 2A) and the structure itself exhibited good _p_6 symmetry, with six CA subunits clearly forming closed hexameric rings when viewed in cross-section normal to the membrane (Figure 2B). This structure is very different from the asymmetric, open lattice structure originally reported for 2D crystals of M-MuLV CA (McDermott et al., 2000).
Fig. 2. 3D reconstruction of 2D crystals of M-MuLV CA. (A) Comparison of the phases (upper panels) and amplitudes (lower panels) of the (2,0), (2,–2) and (0,2) reflections. These reflections are all at 34.5 Å resolution and should be equivalent for a _p_6 crystal. (B) Cross-section of the _p_1 3D map of M-MuLV CA crystals (1 Å thick slab parallel to the membrane). (C) Phases (upper panel) and amplitudes (lower panel) of the (2,0) reflection assuming _p_6 symmetry. (D) Cross-section of the _p_6 3D map (1 Å thick slab parallel to the membrane).
As our analyses demonstrated that the M-MuLV CA crystals actually had _p_6 symmetry, final maps were calculated in this plane group using a total of 47 different corrected images (Table II). The final resolution of the structure is 15 Å in the xy plane, and the point spread function indicates a resolution of ∼25 Å in the z direction (Unger and Schertler, 1995). A cross-section of the resulting 3D map viewed normal to the membrane is given in Figure 2D, and lattice line fittings for reflections at 15.1 Å resolution are shown in Figure 3.
Table II. Statistics of electron crystallographic image analysis.
| 2D parameters | |
|---|---|
| Two-sided plane group | _p_6 |
| Unit cell dimensions (Å) | _p_1 (a = 80.2 ± 0.4 Å, b = 80.3 ± 0.5 Å, γ = 120.4 ± 0.3°, c ∼ 50 Å) |
| _p_6 (a = b = 80.3 ± 0.5 Å, c ∼ 50 Å) | |
| 3D reconstruction parameters (_p_6 plane group) | |
| No. of images | 47 |
| Range of defocus (µm) | 0.4–1.5 |
| IQ cut-off of data | 5 |
| Maximum tilt (‘nominal’) (°) | 50 |
| No. of observed amplitudes and phases | 1267 |
| No. of unique reflections | 121 |
| Percent completeness at 15 Å to 50° tilt angle | 81 |
| In-plane resolution cut-off (Å) | 15 |
| Estimated resolution normal to the bilayer (Å) | 25a |
| Overall phase residual (°) (_R_-factor) | 20.6 (0.243) |
| Overall weighted phase residual (°) (_R_-factor) | 12.2 (0.191) |
Fig. 3. Phase (upper) and amplitude (lower) variation along the (4,1) and (1,4) lattice lines at 15.1 Å resolution, after merging data from tilted crystals in the two-sided plane group _p_6.
Overview of the structure
The M-MuLV CA lattice is an array of linked hexameric rings, with inter-ring spacings of 80 Å (corresponding to the unit cell dimension) and interior holes ∼40 Å in diameter (Figures 2D and 4). As compared with the previous reconstructions of M-MuLV CA (Mayo et al., 2002a, 2003), the regions of strongest density occur closer to the six-fold axes (within the hexamers), consistent with the independent RSV and HIV CA reconstructions. In our reconstruction, the six CA subunits that comprise each ring adopt elongated, bi-lobed structures, and the larger, membrane-proximal lobes self-associate to form the rings (red in Figure 4A and C). Further from the lipid monolayer, the smaller lobes (colored yellow) project away from the six-fold axes with a left-handed twist as viewed from membrane, and their densities connect each hexameric ring to its six adjacent neighbors. Interestingly, the hand of the twist of the smaller lobes is opposite from that seen for HIV-1 and RSV CA.
Fig. 4. Hexameric assemblies of retroviral CA proteins. Structures of the M-MuLV CA (A) and HIV-1 CA (B) hexamers viewed along the six-fold axes, with the central NTDs (red) oriented toward the viewer and the CTD dimers (yellow) further away. This orientation highlights the similarities between the structures, and the maps are contoured to emphasize the domain organization within the CA molecules. In each case, six NTDs comprise the hexameric rings and six CTDs make dimeric connections to neighboring hexamers (in gray). The size difference between the two structures is probably caused by differences in contouring levels, since other HIV-1 CA helical reconstructions (not shown) have hexamer dimensions similar to the M-MuLV CA hexamer. (C) Slab of the M-MuLV CA hexamer between the white lines in (A), as viewed perpendicular to the lines and parallel to the lipid monolayer (on top in this figure). This view emphasizes the two-domain structure of the CA protein and the CTD dimer linkages that connect the hexamers. (D) Possible model to explain the unit cell polymorphisms found in retroviral CA assemblies. The relative positions of the NTD (red) and CTD (yellow) reflect changes in the flexible linker (orange), which allows the hexamer–hexamer distance to vary while maintaining the same inter- and intra-hexamer interactions.
As noted above, retroviral CA proteins are composed of two distinct domains: a larger, elongated NTD (∼50 Å in the longest dimension) and a smaller, globular CTD (20–30 Å in all dimensions). We therefore believe that the larger lobe of the reconstructed density corresponds to the NTD of M-MuLV CA. Thus, our model indicates that six NTDs associate to form the hexameric rings, and each hexamer is connected to six other hexamers via dimeric interactions of the CTD.
Comparisons with other retroviral CA protein assemblies
The structure and hexameric arrangement of the M-MuLV CA protein are strikingly similar to those seen previously for the HIV-1 CA protein (Li et al., 2000) (compare Figure 4A and B) and for the RSV CA protein (Mayo et al., 2002b) (not shown). We therefore conclude that different retroviral CA proteins, including M-MuLV CA, form similar hexameric arrays, supporting the idea that all retroviral capsids assemble on similar lattices. There are, however, significant differences in the unit cell dimensions of the RSV, M-MuLV and HIV CA assemblies, ranging between 80 and 110 Å (Li et al., 2000; Mayo et al., 2002a, 2003). Some of these differences may be a consequence of heavy metal staining and dehydration during preparation of the RSV and M-MuLV CA specimens, but significant crystal polymorphism was even observed between different helical crystal forms of the HIV-1 CA protein preserved in vitreous ice (Li et al., 2000). Although the contouring and noise levels varied, inspection of the M-MuLV and RSV reconstructions as well as several HIV-1 helical reconstructions indicates that the crystallographic polymorphisms appear to be accommodated, to a first approximation, by movement of the C-terminal domain toward the six-fold axes in the smaller unit cells, and away from the six-fold axes in larger unit cells. This motion, which can be accomplished by translation and/or rotation of the CTD with respect to the NTD, is presumably allowed by flexibility in the linker between the two domains. The flexible linker probably also allows for the different sense of CTD twist observed between the different retroviral CA structures. Hinge motions between the two domains of CA are likely to be important for retroviral assembly, and indeed similar interdomain motions are used for the assembly of many icosahedral viruses (Casjens, 1985).
Implications for retroviral capsid structure
Many icosahedral viruses are composed of pentameric and hexameric protein rings. When the number of subunits in the viral shell exceeds 180, however, the hexamers must occupy at least two structurally non-equivalent positions (Caspar and Klug, 1962). Spherical and conical retroviral capsids represent an even more extreme example of such structural non-equivalence because every hexamer in each capsid occupies a non-equivalent site. This is most easily visualized by considering that in conical lentiviral capsids, which lack any formal symmetry, the radius of curvature (and therefore the angle between adjacent hexamer planes) gradually changes in going from the narrow end of the cone to the wide end. The hexamers in the ‘spherical’ capsids of M-MuLV and RSV are similarly non-equivalent because the capsids lack strict icosahedral symmetry (Yeager et al., 1998; Kingston et al., 2001). In principle, the different hexamer environments could be accommodated using multiple different types of intersubunit interfaces, and there is precedence for this in many icosahedral viruses (Johnson and Speir, 1997). However, as the number of non-equivalent hexamers increases, it presumably becomes increasingly difficult to evolve proteins that can adopt the required number of different protein–protein interfaces. Thus, the most parsimonious solution to the problem is instead to keep all subunit interfaces constant, and allow their relative orientations to change through semi-flexible connectors. It appears that retroviral capsids are built using just these principles, i.e. by keeping their binding interfaces constant and by using the flexible linker to adjust the relative positions of the N- and C-terminal domains to allow local environment changes. (However, subtle changes in inter- and intra-hexamer interactions cannot yet be ruled out in the current low-resolution reconstructions.) The T=16 herpes virus assembles using similar principles, with interdomain movements allowing different hexamers to occupy slightly different local environments (He et al., 2001).
In conclusion, our work demonstrates that His-tagged M-MuLV CA assembles into crystalline arrays of hexamers on a monolayer doped with nickel-chelating lipids. Our structure of M-MuLV CA does not agree with previously reported M-MuLV CA reconstructions but is remarkably similar to the hexamers observed for HIV-1 and RSV CA proteins (Li et al., 2000; Mayo et al., 2002b). The similarity of these three independent reconstructions supports the idea that all retroviral capsids are built on conserved hexameric lattices and that different distributions of pentameric declinations produce the wide variety of capsid shapes observed in nature.
Materials and methods
The complete coding sequence of M-MuLV CA was cloned into the unique _Nde_I and _Bam_HI restriction sites of _p_ET15b (Novagen), with a stop codon introduced at the end of the gene. His-tagged M-MuLV CA was expressed in E.coli BL21 (DE3) and purified by nickel chelate and cation-exchange chromatography. Purified protein fractions were pooled, dialyzed against distilled deionized water (ddH2O) with 5 mM β-mercaptoethanol, concentrated to ∼5 mg/ml and flash frozen in liquid nitrogen. For monolayer crystallization, 0.2 mg/ml pure CA in 25 mM sodium phosphate pH 7.8, 5 mM sodium acetate pH 7.6, 250 mM NaCl, 5 mM β-mercaptoethanol and 20% glycerol was overlaid with a 1:1 chloroform/hexane solution containing 50 µg/ml l-α-phosphatidyl choline, doped with 12.5 µg/ml nickel-charged 1,2-dioleoyl-_sn_-glycero-3-_N_-(5-amino-1-carboxypentyl)iminodiacetic acid succinyl (Barklis et al., 1997). Crystallization trials were kept in a humid environment for 4–6 h at room temperature, and crystals were lifted onto carbon-overlaid EM grids, which had been briefly treated with chloroform. Grids were rinsed with ddH2O and stained with 1% uranyl acetate prior to analysis in a Philips CM120 electron microscope operated at 100 kV. Images were recorded at a nominal magnification of 45 000× on Kodak SO-163 film, which was developed according to the manufacturer’s instructions.
Crystals that displayed weak first-order reflections and strong second- and third-order reflections, as observed by optical diffraction, were scanned (4000 × 4000 pixels) using a Zeiss SCAI densitometer at a step size of 7 µm (corresponding to 1.5 Å on the real image). Image files were converted to the MRC format (Crowther et al., 1996) and Fourier transforms were calculated. XIMDISP was used to manually index the crystals, and the MRC suite of programs was used to box and unbend the best area of each crystal (Crowther et al., 1996). Finally, each image was corrected for the contrast transfer function.
Preliminary 3D maps were calculated without imposed symmetry. Initially, the best indexing option was defined for each untilted image (see text), and then used for each of the subsequent images in the tilt series. The 3D origin for each image was defined using ORIGINTILTD and the tilt parameters were calculated using EMTilt followed by necessary adjustments depending on the indexing option used for merging. Once merged, lattice lines were fit to the 3D data, and 3D maps were created using the CCP4 suite of programs (CCP4, 1994). For the six-fold averaged maps, the 3D origin and tilt parameters were defined as described above, but the symmetry-related reflections were combined by vector averaging. Finally, the correct handedness of the structure was confirmed by referencing the known tilt and scanning geometry to the correct assignment of the tilt angle of the reference crystal, connexin 43 (Unger et al., 1999). The images shown in Figure 4 were rendered in AVS (Sheehan et al., 1996).
Acknowledgments
Acknowledgements
We thank Brian Adair for helpful suggestions on improving crystallization conditions and Owen Pornillos for help with programming. This work was supported by NIH grants to W.I.S. and M.Y. M.Y. is the recipient of a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund.
References
- Barklis E. et al. (1997) Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J., 16, 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barklis E., McDermott,J., Wilkens,S., Fuller,S. and Thompson,D. (1998) Organization of HIV-1 capsid proteins on a lipid monolayer. J. Biol. Chem., 273, 7177–7180. [DOI] [PubMed] [Google Scholar]
- Berthet-Colominas C., Monaco,S., Novelli,A., Sibaï,G., Mallet,F. and Cusack,S. (1999) Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J., 18, 1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. and Vogt,V.M. (1995) Self-assembly in vitro of purified CA–NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol., 69, 6487–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Olivas R., Newman,J.L. and Summers,M.F. (2000) Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol., 296, 633–649. [DOI] [PubMed] [Google Scholar]
- Casjens S. (1985) Virus Structure and Assembly. Jones and Bartlett, Portola Valley, CA.
- Caspar D.L.D. and Klug,A. (1962) Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol., 27, 1–24. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Coffin J.M., Hughes,S.H. and Varmus,H.E. (eds) (1997) Retroviruses. Cold Spring Harbor Press, Plainview, NY.
- Crowther R.A., Henderson,R. and Smith,J.M. (1996) MRC image processing programs. J. Struct. Biol., 116, 9–16. [DOI] [PubMed] [Google Scholar]
- Ehrlich L.S., Agresta,B.E. and Carter,C.A. (1992) Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol., 66, 4874–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T.R., Vajdos,F.F., Yoo,S., Worthylake,D.K., Houseweart,M., Sundquist,W.I. and Hill,C.P. (1996) Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell, 87, 1285–1294. [DOI] [PubMed] [Google Scholar]
- Gamble T.R., Yoo,S., Vajdos,F.F., von Schwedler,U.K., Worthylake,D.K., Wang,H., McCutcheon,J.P., Sundquist,W.I. and Hill,C.P. (1997) Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science, 278, 849–853. [DOI] [PubMed] [Google Scholar]
- Ganser B.K., Li,S., Klishko,V.Y., Finch,J.T. and Sundquist,W.I. (1999) Assembly and analysis of conical models for the HIV-1 core. Science, 283, 80–83. [DOI] [PubMed] [Google Scholar]
- Gitti R.K., Lee,B.M., Walker,J., Summers,M.F., Yoo,S. and Sundquist,W.I. (1996) Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science, 273, 231–235. [DOI] [PubMed] [Google Scholar]
- Göttlinger H.G. (2001) The HIV-1 assembly machine. AIDS, 15, S13–S20. [DOI] [PubMed] [Google Scholar]
- Gross I., Hohenberg,H. and Kräusslich,H.-G. (1997) In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem., 249, 592–600. [DOI] [PubMed] [Google Scholar]
- He J., Schmid,M.F., Zhou,Z.H., Rixon,F. and Chiu,W. (2001) Finding and using local symmetry in identifying lower domain movements in hexon subunits of the herpes simplex virus type 1 B capsid. J. Mol. Biol., 309, 903–914. [DOI] [PubMed] [Google Scholar]
- Jin Z., Jin,L., Peterson,D.L. and Lawson,C.L. (1999) Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol., 286, 83–93. [DOI] [PubMed] [Google Scholar]
- Johnson J.E. and Speir,J.A. (1997) Quasi-equivalent viruses: a paradigm for protein assemblies. J. Mol. Biol., 269, 665–675. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S., Campos-Olivas,R. and Summers,M.F. (1999) Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol., 291, 491–505. [DOI] [PubMed] [Google Scholar]
- Kingston R.L., Fitzon-Ostendorp,T., Eisenmesser,E.Z., Schatz,G.W., Vogt,V.M., Post,C.B. and Rossmann,M.G. (2000) Structure and self-association of the Rous sarcoma virus capsid protein. Structure Fold Des., 8, 617–628. [DOI] [PubMed] [Google Scholar]
- Kingston R.L., Olson,N.H. and Vogt,V.M. (2001) The organization of mature Rous sarcoma virus as studied by cryoelectron microscopy.J. Struct. Biol., 136, 67–80. [DOI] [PubMed] [Google Scholar]
- Krausslich H.G., Oldstone,M.B., Vogt,P.K. and Potter,M. (eds) (1996) Morphogenesis and Maturation of Retroviruses. Springer-Verlag, Berlin, Germany.
- Li S., Hill,C.P., Sundquist,W.I. and Finch,J.T. (2000) Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature, 407, 409–413. [DOI] [PubMed] [Google Scholar]
- Mayo K., McDermott,J. and Barklis,E. (2002a) Hexagonal organization of Moloney murine leukemia virus capsid proteins. Virology, 298, 30–38. [DOI] [PubMed] [Google Scholar]
- Mayo K., Vana,M.L., McDermott,J., Huseby,D., Leis,J. and Barklis,E. (2002b) Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J. Mol. Biol., 316, 667–678. [DOI] [PubMed] [Google Scholar]
- Mayo K., Huseby,D., McDermott,J., Arvidson,B., Finlay,L. and Barklis,E. (2003) Retrovirus capsid protein assembly arrangements. J. Mol. Biol., 325, 225–237. [DOI] [PubMed] [Google Scholar]
- McDermott J., Mayo,K. and Barklis,E. (2000) Three-dimensional organization of retroviral capsid proteins on a lipid monolayer. J. Mol. Biol., 302, 121–133. [DOI] [PubMed] [Google Scholar]
- Momany C. et al. (1996) Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol., 3, 763–770. [DOI] [PubMed] [Google Scholar]
- Rosé S., Hensley,P., O’Shannessy,D.J., Culp,J., Debouck,C. and Chaiken,I. (1992) Characterization of HIV-1 p24 self-association using analytical affinity chromatography. Proteins, 13, 112–119. [DOI] [PubMed] [Google Scholar]
- Sheehan B., Fuller,S.D., Pique,M.E. and Yeager,M. (1996) AVS software for visualization in molecular microscopy. J. Struct. Biol., 116, 99–106. [DOI] [PubMed] [Google Scholar]
- Unger V.M. and Schertler,G.F.X. (1995) Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys. J., 68, 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger V.M., Kumar,N.M., Gilula,N.B. and Yeager,M. (1999) Three-dimensional structure of a recombinant gap junction membrane channel. Science, 283, 1176–1180. [DOI] [PubMed] [Google Scholar]
- Welker R., Hohenberg,H., Tessmer,U., Huckhagel,C. and Kräusslich, H.-G. (2000) Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol., 74, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake D.K., Wang,H., Yoo,S., Sundquist,W.I. and Hill,C.P. (1999) Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D, 55, 85–92. [DOI] [PubMed] [Google Scholar]
- Yeager M., Wilson-Kubalek,E.M., Weiner,S.G., Brown,P.O. and Rein,A. (1998) Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl Acad. Sci. USA, 95, 7299–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber G., McDermott,J., Karanjia,S., Zhao,W., Schmid,M.F. and Barklis,E. (2000) Assembly of retrovirus capsid–nucleocapsid proteins in the presence of membranes or RNA. J. Virol., 74, 7431–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]