NSD1 is essential for early post-implantation development and has a catalytically active SET domain (original) (raw)
Abstract
The nuclear receptor-binding SET domain-containing protein (NSD1) belongs to an emerging family of proteins, which have all been implicated in human malignancy. To gain insight into the biological functions of NSD1, we have generated _NSD1_-deficient mice by gene disruption. Homozygous mutant NSD1 embryos, which initiate mesoderm formation, display a high incidence of apoptosis and fail to complete gastrulation, indicating that NSD1 is a developmental regulatory protein that exerts function(s) essential for early post-implantation development. We have also examined the enzymatic potential of NSD1 and found that its SET domain possesses intrinsic histone methyltransferase activity with specificity for Lys36 of histone H3 (H3-K36) and Lys20 of histone H4 (H4-K20).
Keywords: gastrulation/gene disruption/HMTase/nuclear receptor cofactor/SET domain
Introduction
Nuclear receptors are members of a superfamily of sequence-specific transcription factors that play diverses roles in the control of cell growth and differentiation, development and homeostasis by stimulating or repressing target gene expression (reviewed in Ciana et al., 2002). Upon binding to their cognate DNA response elements, nuclear receptors modulate transcription through the recruitment of various co-regulatory proteins, called co-repressors and co-activators (reviewed by Robyr et al., 2000; Rosenfeld and Glass, 2001). Co-repressors that are recruited in the absence of ligand, such as N-CoR and SMRT, are part of multiple histone deacetylase complexes, which stabilize chromatin structure and repress transcription. Co-activators that are recruited in a ligand-dependent fashion, such as CBP, p/CAF and members of the p160 family, possess or can recruit histone acetyltransferase and histone methyltransferase (HMTase) activities that are capable of chromatin remodeling/modification, whereas other ligand-recruited complexes such as the TRAP–DRIP–ARC–SMCC complex appear to act more directly on the basal transcriptional machinery (Rosenfeld and Glass, 2001; and references therein).
In a screen to isolate co-regulators for retinoic acid receptor, a novel nuclear receptor-binding SET domain-containing protein (NSD1) was identified (Huang et al., 1998) and subsequently shown to belong to an emerging family of proteins that includes NSD2 [also known as MMSET (Chesi et al., 1998) and WHSC1 (Stec et al., 1998)] and NSD3 (Angrand et al., 2001). The domain structure that characterizes these proteins contains a SET domain (Tschiersch et al., 1994), a PWWP domain (Stec et al., 2000) and multiple PHD fingers (Aasland et al., 1995). In addition to these conserved domains, which are present in members of the Trithorax gene family and other chromatin modulators acting positively and/or negatively on transcription, NSD1 contains two distinct nuclear receptor-interacting domains (NIDs) that bind the apo- and holo- forms of the ligand-binding domain of different subsets of nuclear receptors, with characteristics of both co-activators and co-repressors (Huang et al., 1998). Supporting the notion that NSD1 may act as a bifunctional transcriptional cofactor playing a dual role in transcription, NSD1 was also reported to possess distinct activation and repression domains (Huang et al., 1998).
The SET domain is an evolutionarily conserved sequence motif of 130–150 amino acids, which initially was identified in the Drosophila position effect variegation (PEV) suppressor Su(var)3-9, the Polycomb group protein Enhancer of zeste [E(z)] and the trithorax group protein Trithorax (TRX) (Tschiersch et al., 1994), and was later found in a variety of chromatin-associated proteins from yeast to mammals (reviewed in Jenuwein, 2001; Schneider et al., 2002). A growing number of SET domain proteins recently have been shown to harbor HMTase activity towards specific lysine residues along the N-terminal tail of histones (reviewed in Jenuwein, 2001; Kouzarides, 2002; Schneider et al., 2002). Among these enzymes, the mammalian homologs of Drosophila Su(var)3-9, SUV39H1 (Rea et al., 2000) and Suv39h2 (O’Carroll et al., 2000), its Schizosacharomyces pombe homolog Clr4 (Rea et al., 2000), the human G9a protein (Tachibana et al., 2002) and the mouse ESET/SETDB1 protein (Yang et al., 2002) specifically methylate histone H3 on Lys9 (and Lys27 in the case of G9a) and require the SET domain and two adjacent cysteine-rich regions [the pre-SET (also called SAC; Huang et al., 1998) and post-SET domains] for enzymatic activity (Rea et al., 2000). Importantly, it has been shown that H3 Lys9 (H3-K9) methylation by SUV39H1 creates a high-affinity binding site for the heterochromatin protein HP1 and thus contributes to heterochromatin-mediated silencing (Jenuwein, 2001; Kouzarides, 2002; and references therein). Recent biochemical purification of HMTase activities from HeLa cells has identified SET7 (also called Set9) as a novel, mammalian SET domain-containing protein that specifically methylates H3 on Lys4 (Wang et al., 2001; Nishioka et al., 2002a). In contrast to H3-K9 methylation, H3-K4 methylation by SET7/Set9 has been shown to activate transcription by inhibiting the association of the NuRD deacetylase complex with the H3 tail and precluding H3-K9 methylation by Suv39h1 (Wang et al., 2001; Nishioka et al., 2002a). In budding yeast, a H3-K4-specific methyltransferase has also been identified (Set1) that is able to catalyze both the di- and tri-methylated state of K4 (Santos-Rosa et al., 2002); interestingly, only the tri-methyl state of K4 was linked to activation of transcription, indicating that not only the site of methylation but also the methyl status of the site are important determinants for gene activity (Santos-Rosa et al., 2002). Recently, SET domain-containing proteins that methylate Lys20 of H4 (H4-K20) have also been described in Drosophila and mammals (Fang et al., 2002; Nishioka et al., 2002b), that are associated with silent chromatin. In an attempt to understand the structure and molecular basis of catalysis of the SET domain HMTases, the three-dimensional structures of diverse SET domain proteins with different sub strate specificities have been determined (Marmorstein, 2003; and references therein). Comparison of these three-dimensional structures revealed a common two-domain architecture, consisting of a conserved antiparallel β-barrel structure and a structurally variable insert, with the cofactor-binding site and the catalytic center constructed on an unusual but conserved knot-like substructure.
On the basis of sequence similarity within the SET domain, NSD1 has been defined as a SET family member of the Ash1 subclass (also called SET2; Huang et al., 1998; Schneider et al., 2002). This subfamily of SET domain proteins contains, in addition to NSD1 and the related proteins NSD2 and NSD3, the Drosophila trithorax group protein Ash1 (Beisel et al., 2002; and references therein) and the Saccharomyces cerevisiae protein Set2 (Strahl et al., 2002). The SET domain in this subclass is flanked by pre- and post-SET domains and is centrally located (Kouzarides, 2002). Ash1 recently has been demonstrated to be a multicatalytic HMTase that activates transcription by methylating Lys4 and Lys9 in H3, and Lys20 in H4 (Beisel et al., 2002). Set2, which is a nucleosomal H3-K36-selective methyltransferase (Strahl et al., 2002), methylates at the coding and promoter regions of target genes and functions through specific association with the elongating form of RNA polymerase II that is hyperphosphorylated (Xiao et al., 2003), indicating that methylation mediated by Set2 may be involved in regulating transcription elongation.
Members of the NSD family have all been implicated in human malignancy, which suggests a key role in controlling cell growth and differentiation for this subgroup of SET domain proteins. The human NSD1 gene, which is located at the chromosomal locus 5q35, has been isolated recently in the context of a fusion transcript with the nucleoporin gene (NUP98) in a recurrent translocation, t(5;11)(q35;p15.5), specifically associated with de novo childhood acute myeloid leukemia (AML; Jaju et al., 2001). More recently, NSD1 has also been implicated in Sotos syndrome, a rare growth disorder also known as cerebral gigantism (Kurotaki et al., 2002,. Through a search for genes located in the Wolf–Hirschhorn syndrome (WHS) critical region, the human NSD2 gene was localized on 4p16.3 (WHSC1; Stec et al., 1998) and was found to be disrupted by t(4;14) translocations causing lymphoid multiple myeloma (MMSET; Chesi et al., 1998). Using fluorescence in situ hybridization, NSD3 was mapped to 8p12 and was shown to be amplified in several tumor-derived cell lines and primary breast carcinomas (Angrand et al., 2001). Recently, NSD3 has also been identified as a translocation partner of NUP98 in AML (Rosati et al., 2002).
We demonstrate here that NSD1 is a developmental regulatory protein that exerts cellular function(s) essential for early post-implantation mouse development. We also provide biochemical evidence that the NSD1 SET domain functions as an HMTase to methylate H3-K36 and H4-K20 in vitro. We discuss the implications of these data for mechanistic models of NSD1 function in mammals.
Results
Targeted disruption of the mouse NSD1 gene
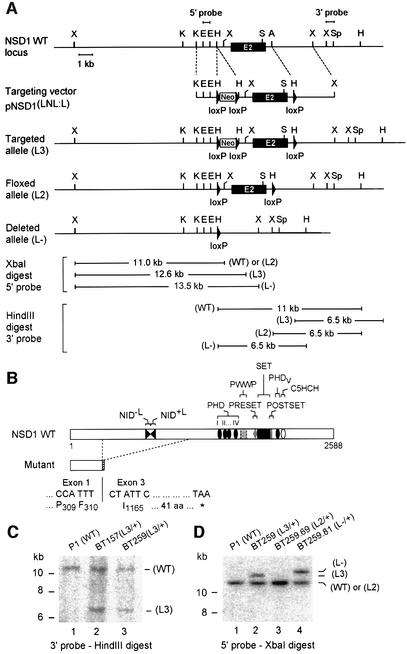
Using two genomic clones that contain a portion of the NSD1 gene (see Materials and methods), we generated a targeting vector, pNSD1(LNL:L), in which a neomycin resistance selection cassette (Neo) flanked by two loxP sites was introduced into intron 1 and a loxP site was inserted into intron 2 (Figure 1A; see Materials and methods). This targeting vector was designed with the expectation that upon homologous recombination and subsequent Cre recombinase-mediated excision, exon 2 of NSD1 together with the Neo cassette would be deleted, thereby causing a frameshift mutation with a premature termination codon in exon 3 (Figure 1B). The putative product of this deleted gene would correspond to a truncated NSD1 protein, lacking the NIDs and all conserved domains (Figure 1B).
Fig. 1. Targeted disruption of NSD1 using the Cre-loxP strategy. (A) Diagram showing a partial map of the genomic locus surrounding the NSD1 exon 2 (E2), the targeting construct, and the targeted allele before (L3) and after (L2 and _L_–) Cre-mediated excision of the neomycin resistance selection marker (Neo). The 5′ and 3′ probes used for Southern blot analyses and the fragment sizes detected with the 5′ probe upon _Xba_I digestion and with the 3′ probe upon _Hin_dIII digestion are indicated. Relevant restriction sites: X, _Xba_I; K, _Kpn_I; E, _Eco_RI; H, _Hin_dIII; S, _Sma_I; A, _Afl_II; Sp, _Spe_I. (B) Schematic representation of wild-type (WT) and mutant NSD1 proteins. The structural and functional domains are indicated. The putative product of the deleted NSD1 gene corresponds to a C-terminally truncated protein consisting of the first 310 amino acids of NSD1. (C) Southern blot analysis of DNAs derived from wild-type (P1) and targeted (BT157 and BT259) ES cells. Genomic DNA was digested with _Hin_dIII, blotted and hybridized with the 3′ probe. (D) Southern blot analysis of ES cell subclones using the 5′ probe, after Cre-mediated excision in BT259 _NSD1_L3/+ ES cells.
The targeting vector pNSD1(LNL:L) was electroporated into 129/Sv P1 embryonic stem (ES) cells, and 280 G418-resistant clones were screened for homologous recombination by Southern blot analysis using an ‘outside’ probe corresponding to a sequence 3′ of the recombination site (3′ probe; see Figure 1A). Two positive clones, BT157 and BT259, were obtained (Figure 1C), which were confirmed to carry a single-copy integration at the NSD1 locus as revealed by hybridization with a Neo probe (data not shown). One of these targeted cell lines (BT259) was transiently transfected with a Cre-encoding expression plasmid (PIC-Cre) to test whether a Cre-mediated excision of the targeted allele (L3) could be achieved. Clones harboring either a partially excised L2 allele without the _loxP_-flanked Neo cassette (BT259-69; Figure 1D) or a completely excised _L_– allele lacking the DNA sequences between the three loxP sites (BT259.81) were identified by Southern blotting (Figure 1D).
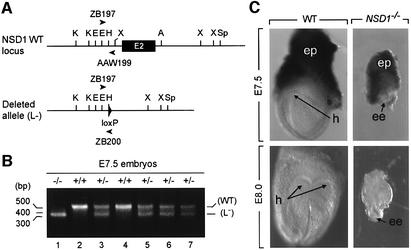
The two independent _NSD1_L3/+ ES cell lines, BT157 and BT259, were injected into C57BL/6 blastocysts to produce chimeric mice, and both contributed to the germline. Mice heterozygous for the targeted NSD1 gene (_NSD1_L3/+) were crossed with cytomegalovirus-Cre transgenic mice (_CMV-Cre_tg/0) expressing the Cre recombinase in the germline (Dupé et al., 1997). Tail DNA of the offspring was analyzed by genomic PCR to detect Cre-mediated excision (Figure 2A). Animals in which PCR assays detected an excised _L_– allele were crossed with wild-type C57BL/6 mice to produce Cre-negative _NSD1_L–/+ mice, hereafter referred to as _NSD1_–/+.
Fig. 2. Morphology of wild-type and _NSD1_–/– mutant embryos at E7.5 and E8.0. (A) PCR strategy for amplification of wild-type (WT) and excised (L–) NSD1 alleles. DNA samples were subjected to PCR amplification using a mixture of three primers (see Materials and methods). PCR amplification of the wild-type NSD1 allele by sense and antisense primers ZB197 and AAW199 produces a 550 bp DNA fragment (B, upper band), while PCR amplification of the excised allele by sense and antisense primers ZB197 and ZB200 produces a 350 bp DNA fragment (B, lower band). (B) Representative genotypic analysis of E7.5 embryos from an NSD1+/– intercross. (C) Dissected wild-type (left) and _NSD1_–/– mutant (right) littermates at E7.5 and E8.0. Abbreviations: ee, epiblast; ep, ectoplacental cone; h, head folds.
NSD1 is essential for early post-implantation development
Mice heterozygous for the NSD1 mutation were viable and fertile. Genotype analysis of progeny from heterozygote intercrosses revealed that 37% were wild type, 63% heterozygous and none homozygous (Table I), indicating that the NSD1 mutation is recessive embryonic lethal.
Table I. Genotype analysis of NSD1+/– intercross progeny.
| Stage | Genotype | Resorption | Total | ||
|---|---|---|---|---|---|
| +/+ | +/– | –/– | |||
| Newborn | 43 | 73 | 0 | – | 116 |
| E10.5 | 10 | 16 | 0 | 6 | 32 |
| E9.5 | 4 | 5 | 5a | – | 14 |
| E7.5 | 3 | 11 | 3a | – | 17 |
| E6.5 | 2 | 8 | 4a | – | 14 |
To determine the time of embryonic lethality, embryos from heterozygote intercrosses were collected at various stages of gestation and genotyped (Table I). No homozygous mutant embryos were recovered at, or after, E10.5 (Table I and data not shown), and the percentage of resorptions at this time was unusually high (19%; Table I). At E9.5, a set of degenerating embryos that consisted primarily of extraembryonic tissues was identified and genotyped as _NSD1_–/– mutants (Table I). At E8.0 and E7.5, all of the morphologically abnormal mutants recovered were also genotyped as mutants (Figure 2B and Table I); these homozygous mutant embryos were severely growth retarded when compared with their NSD1+/+ and NSD1+/– littermates (Figure 2C) and did not contain any structures that resemble those of control embryos (see Figure 2C legend). Mutant embryos generated from the two independently targeted ES cell clones showed identical phenotypes. These results indicate that the NSD1 gene is absolutely required for early post-implantation development in mice.
Disruption of the NSD1 gene leads to increased apoptosis and mesodermal defects
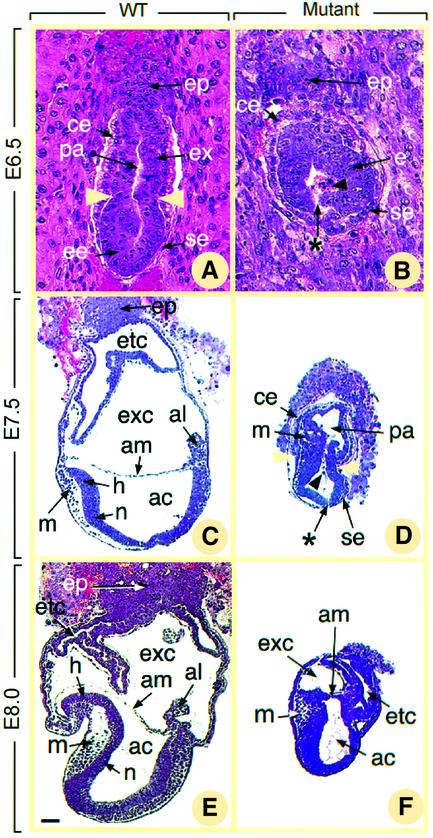
To analyze the NSD1 mutant phenotype further, we examined histological sections of embryos from heterozygote intercrosses, collected in utero from E6.5 to E8.0 (Figure 3). At the E6.5 egg cylinder stage, normal embryos displayed a well-organized structure, in which two layers of ectodermal and endodermal cells enclose the proamniotic cavity (pa, Figure 3A). A characteristic groove (yellow arrowheads in Figure 3A) separates the epiblast (ee) from the extraembryonic portion of the primitive ectoderm (ex, Figure 3A), the latter being capped by the ectoplacental cone (ep) which invades the maternal decidua. The visceral endodermal layer is composed of two cell subpopulations, the proximal cuboidal cells surrounding the extraembryonic ectoderm (ce) and the distal squamous cells surrounding the epiblast (se, Figure 3A). All abnormal presumptive _NSD1_–/– E6.5 embryos (n = 4), identified by their small size and absence of the typical egg cylinder shape, displayed the same histological phenotype (Figure 3B). The two cell types of the visceral endoderm and the ectoplacental cone were readily identified (ce, se and ep, Figure 3B). However, the ectoderm (e, Figure 3B) did not exhibit its characteristic groove and always showed an abnormal large gap (see asterisk in Figure 3B). Numerous dying cells marked by pyknotic nuclei were detected on each side of this gap and also within the proamniotic cavity (black arrowhead in Figure 3B, and data not shown).
Fig. 3. Histological sections of normal (A, C and E) and presumptive _NSD1_–/– (B, D and F) embryos at E6.5 (A and B), E7.5 (C and D) and E8.0 (E and F). In (A) and (B), deciduas were kept intact. In (C–F), embryos were dissected out of the decidua. Yellow arrowheads in (A) and (D) show the boundary between embryonic and extraembryonic regions. Asterisks in (B) and (D) indicate gaps in the mutant epiblast, and black arrowheads point to pyknotic cells in the proamniotic cavity. Abbreviations: ac, amniotic cavity; al, allantois; am, amnion; ce, cuboidal visceral endoderm; e, ectoderm; ee, epiblast; ep, ectoplacental cone; etc, ectoplacental cavity; ex, extraembryonic ectoderm; exc, exocoelom; h, head folds; m, mesoderm; n, neurectoderm; pa, proamniotic cavity; se, squamous visceral endoderm. Scale bar: 20 µm (A and B); 70 µm (C–F).
At E7.5 and E8.0, normal embryos have formed mesoderm (m) and neurectoderm (n, Figure 3C and E), as a consequence of gastrulation. Their anterior and posterior extremities are defined by the headfolds (h) and allantois (al) (Figure 3C and E), respectively; their proamniotic cavity has been partitioned by amniotic folds into amniotic (ac), exocolomic (exc) and ectoplacental (etc) cavities (Figure 3C and E). Abnormal presumptive _NSD1_–/– E7.5 embryos (n = 4) displayed a groove separating the embryonic and extraembryonic ectoderm (yellow arrowheads in Figure 3D). Few cells detaching from the ectoderm and resembling normal mesodermal cells (m, Figure 3D) were present between the ectoderm and the visceral endoderm. However, the headfolds and the allantois could not be identified, and the proamniotic cavity (pa, Figure 3D) remained undivided. At E8.0, abnormal presumptive _NSD1_–/– embryos (n = 4) had developed slightly further (Figure 3F) as they now displayed amniotic folds and three distinct cavities resembling those normally derived from the proamniotic cavity (ac, exc and etc, Figure 3F).
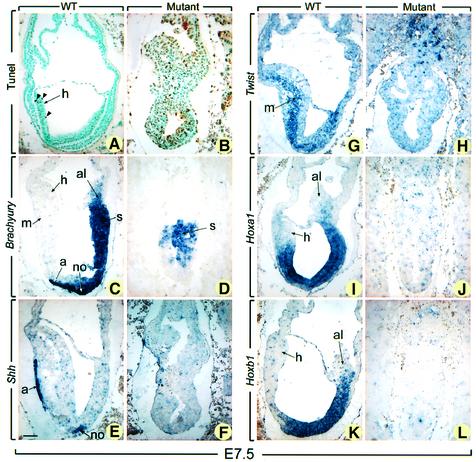
Ectodermal interruptions and the presence of many pyknotic nuclei in the ectoderm and the proamniotic or amniotic cavity were hallmarks of the presumptive _NSD1_–/– embryos at both E7.5 and E8.0 (asterisk and arrow in Figure 3D, and data not shown), suggesting that abnormal cell death occurs by apoptosis in the absence of NSD1. To confirm this hypothesis, TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick-end labeling) assays were performed to detect the fragmented DNA characteristic of apoptotic cells. At E7.5, numerous TUNEL-positive cells (7.25 ± 2.70%) were seen in the four presumptive mutant embryos, whereas <1% (0.57 ± 0.20%) of cells were positive in the littermate controls (compare Figure 4A and B). Altogether, these data indicate that _NSD1_–/– mutant embryos exhibit a retarded and abnormal gastrulation process, including a marked increase in apoptosis.
Fig. 4. TUNEL and in situ hybridization analyses of E7.5 normal and mutant embryos. Sections of E7.5 normal embryos (A, C, E, G, I and K) and their presumptive mutant littermates (B, D, F, H, J and L) were subjected to TUNEL reaction (A and B) and were hybridized with Brachyury (C and D), Shh (E and F), Twist (G and H), Hoxa1 (I and J), and Hoxb1 (K and L) antisense probes. In (A) and (B), brown-stained nuclei indicate end incorporation in DNA (arrowheads). Abbreviations: a, axial mesendoderm; al, allantois; h, head folds; m, mesoderm; no, node; s, primitive streak. Scale bar: 60 µm.
To characterize gastrulation defects further, presumptive _NSD1_–/– embryos were analyzed at E7.5 by in situ hybridization (ISH) using markers of mesodermal structures [Brachyury (T), Sonic hedgehog (Shh) and _Twist_] as well as positional markers of the anteroposterior body axis (Hoxa1 and Hoxb1). Expression of T is detectable in the early mesoderm as it ingresses through the primitive streak (s), in the node (no) and in the axial mesendoderm (a, Figure 4C) (Kispert and Herrmann, 1994). Shh is expressed in the node and the axial mesendoderm (Figure 4E; Echelard et al., 1993). Expression of Twist is observed in the paraxial and lateral mesoderm at a later stage than that of T (m, Figure 4G) (Stoetzel et al., 1995). Hoxa1 and Hoxb1 are expressed along the anteroposterior body axis with anterior boundaries that lie within the hindbrain (Figure 4I and K; Murphy and Hill, 1991). In the three presumptive _NSD1_–/– embryos analyzed, T was detected in an irregular cluster of embryonic cells (s, Figure 4D). No expression of the two other mesodermal markers (Shh and Twist) and of the two axial markers (Hoxa1 and Hoxb1) was observed in these embryos (Figure 4F, H, J and L). Thus, the primitive streak of mutant embryos has a markedly disorganized aspect, does not form a node and fails to produce mesendoderm and embryonic mesoderm. Moreover, the anteroposterior axis is not specified in these mutant embryos.
NSD1 expression during mouse development
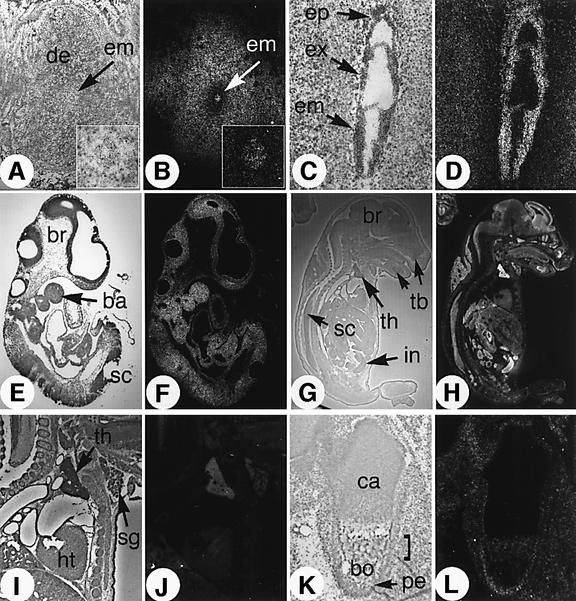
The expression pattern of NSD1 was examined by ISH at various developmental stages (Figure 5). At the early post-implantation E5.5 stage, NSD1 expression was detected in the developing embryo (em) as well as in the outer region of the uterine decidua (de, Figure 5A and B). In sections of gastrulation stage embryos (E7.5), we found NSD1 uniformly expressed throughout the embryo, in both embryonic and extraembryonic tissues (Figure 5C and D). This ubiquitous expression profile persisted until E14.5 (see E9.5 in Figure 5E and F, and data not shown). After this time, differential expression was seen, with the highest levels of NSD1 expression in proliferative cell populations. Enriched NSD1 levels were detected at E16.5 in the telencephalic region of the brain (br), spinal cord (sc), intestinal crypt cells (in), tooth buds (tb), thymus (th) and salivary glands (sg) (Figure 5G–J). NSD1 expression was also observed in the region of ossification of the developing bones (bo) and in the periosteum (pe), while it was absent in chondrocytes (ca, Figure 5K and L). Taken together, these results are consistent with the gastrulation defects displayed by the NSD1-deficient embryos and also suggest a critical role for NSD1 during post-gastrulation development (see Discussion).
Fig. 5. In situ hybridization analysis of NSD1 transcript distribution at various stages of mouse development. Brightfield and darkfield views of the histological sections are shown side by side (left and right panel, respectively), revealing the signal grain as white dots. (A and B) E5.5 embryo sectioned in utero. Insert panels show magnification of the embryo. (C and D) Ubiquitous expression of NSD1 in the ectoplacental cone (ep), extraembryonic (ex) and embryonic (em) germ layers of an E7.5 embryo. (E and F) Ubiquitous NSD1 expression in an E9.5 embryo. ba, branchial arches; br, brain; sc, spinal cord. (G and H) Enhanced expression of NSD1 in the brain (br), intestine (in), spinal cord (sc), thymus (th) and tooth buds (tb) of an E16.5 fetus. (I and J) Detail of the E16.5 heart (ht), thymus (th) and salivary gland area (sg). (K and L) Section through the ossification center of the femur. bo, bone tissue; ca, cartilage (chondrocytes); pe, periosteum.
The SET domain of NSD1 methylates H3-K36 and H4-K20
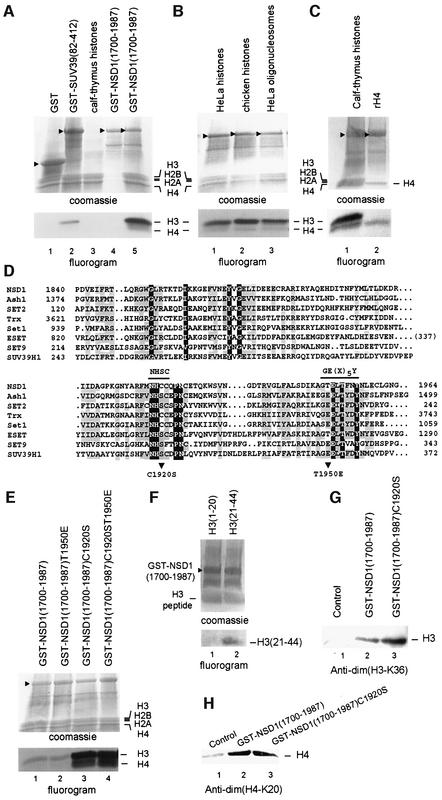
To investigate whether the SET domain of NSD1 exhibits HMTase activity, we performed an in vitro methylation assay using native histones as substrates. Recombinant GST–NSD1 (amino acids 1700–1987), GST–SUV39H1 (amino acids 82–412) and GST were incubated with a mixture of native calf thymus histones and _S_-adenosyl-[methyl-3H]l-methionine as a methyl donor. Reaction products were separated by SDS–PAGE, and methyl-3H-labeled proteins were visualized by fluorography (Figure 6A). As expected, GST–SUV39H1(82–412) showed HMTase activity towards H3 (Figure 6A, lane 2). GST–NSD1(1700–1987) also exhibited enzymatic activity and preferentially methylated H3 and, to a lesser extent, H4 (Figure 6A, lane 5), whereas, under similar conditions, no HMTase activity was observed with GST (Figure 6A, lane 1). GST–NSD1(1700–1987) was also tested for HMTase activity using purified HeLa nucleosomes as substrates. As shown in Figure 6B, GST–NSD1(1700–1987) methylated H3 and H4 when HeLa or chicken core histones (lanes 1 and 2) or HeLa oligonucleosomes (lane 3) were used as substrates. Similarly to native histones, recombinant H3 (Figure 6G) and H4 (Figure 6C, lane 2) were also methylated by GST–NSD1(1700–1987).
Fig. 6. The NSD1 SET domain has intrinsic HMTase activity with specificity for H3-K36 and H4-K20. (A) Coomassie blue-stained SDS–polyacrylamide gel and corresponding flurorogram of HMTase assays with the indicated GST fusion proteins and a mixture of purified calf thymus histones as substrates. (B) HMTase assays as in (A), except that reactions contained either a mixture of native core histones from HeLa cells (lane 1) or chicken (lane 2), or oligonucleosomes purified from HeLa cells (lane 3), and GST–NSD1(1700–1987) as enzyme. (C) HMTase assays as in (A), except that reactions contained either purified calf thymus histones (lane 1) or recombinant H4 (lane 2), and GST–NSD1(1700–1987) as enzyme. (D) Amino acid alignment of the NSD1 SET domain with other selected SET domains. The sequences were aligned using the CLUSTAL W program and manual adjustment. Invariant amino acids are highlighted in blue. Amino acids conserved in >60% of the proteins are highlighted in yellow. The two conserved amino acid stretches important for the methyltransferase catalytic activity of the SET domain are indicated. Mutations described in this study are indicated below the alignment. (E) The C1920S mutation generated a hyperactive SET domain mutant. Equal amounts of wild-type and mutant GST–NSD1 proteins (top panel) were compared for their HMTase activities (bottom panel) using equal amounts of purified calf thymus histones. (F) HMTase assays as in (A) using GST–NSD1(1700–1987) as enzyme and the indicated N-terminal peptides of H3. (G) Western blot analyses, using anti-dim(H3-K36), of HMTase assays containing recombinant H3 and the indicated GST–NSD1 proteins. (H) Western blot analyses, using anti-dim(H4-K20), of HMTase assays containing recombinant H4 and the indicated GST–NSD1 proteins.
Amino acid comparison of the SET domains from NSD1 and other well-known HMTases revealed extensive sequence similarity (Huang et al., 1998; and Figure 6D), except for the absence of two highly conserved residues within two short amino acid stretches, NHSC and GE(x)5Y, which previously have been shown to be critical for enzymatic activity (Rea et al., 2000; see also Marmorstein, 2003; and references therein). Instead of these conserved motifs, NSD1 contains NHCC and GT(x)5Y motifs (Figure 6D). We hypothesized that replacing the cysteine (C1920) and threonine (T1950) residues of NSD1 by serine (C1920S) and aspartate (T1950E), respectively, might increase the catalytic activity of NSD1 in vitro. To test this hypothesis, equivalent amounts of recombinant GST proteins, GST– NSD1(1700–1987)C1920S, GST–NSD1(1700–1987) T1950E and GST–NSD1(1700–1987)C1920ST1950E, were purified and tested for HMTase activity. As shown in Figure 6E, the single mutation T1950E did not interfere with the enzymatic activity of the NSD1 SET domain in vitro [compare GST–NSD1(1700–1987) with GST– NSD1(1700–1987)T1950E in Figure 6E, lanes 1 and 2]. On the other hand, the single mutation C1920S as well as the double mutation C1920ST14950E generated hyperactive mutants, which were ∼20-fold more efficient than wild type in methylating both H3 and H4 (Figure 6E, lanes 3 and 4). This result indicates that the C1920 residue in the NSD1 SET domain is a critical determinant of enzymatic activity.
In an attempt to identify the target amino acid residue(s) of NSD1, HMTase assays were carried out with peptides consisting of amino acids 1–20 and 21–44 of H3 [see H3(1–20) and H3(21–44) in Figure 6F] using GST–NSD1(1700–1987) as enzyme. These assays revealed selective methylation of H3(21–44) (lane 2), whereas no signal was detected with H3(1–20) (lane 1). As Lys27 and/or Lys36 residue(s) might be methylated by NSD1, recombinant H3 was methylated by either GST–NSD1(1700–1987) or GST–NSD1(1700–1987)C1920S (Figure 6G) and analyzed by western blot analysis using site-specific histone methylation antibodies against dimethylated H3-K27 [anti-dim(H3-K27)] or dimethylated H3-K36 [anti-dim(H3-K36)]. Anti-dim(H3-K36) recognized GST–NSD1-methylated H3 (Figure 6G, lanes 2 and 3), but not unmethylated H3 (lane 1). In contrast, no methylation at Lys27 could be detected by anti-dim(H3-K27) (data not shown). These results indicate that the SET domain of NSD1 possesses H3-K36-specific HMTase activity in vitro. As H4-K20 is the only H4 lysine residue that is methylated in vivo, NSD1-methylated H4 was analyzed by immunoblotting using anti-dim(H4-K20) (Figure 6H). Recombinant H4 from methylation reactions including GST–NSD1(1700–1987) or GST–NSD1(1700– 1987)C1920S was strongly recognized by anti-dim(H4-K20) (Figure 6H, lanes 2 and 3) compared with control (minus GST fusions) reactions (lane 1), indicating that in addition to methylating H3-K36, the SET domain of NSD1 methylates H4-K20 in vitro.
Discussion
NSD1 is crucial for early post-implantation development
In the present study, we have examined the developmental consequences of a null mutation in the mouse NSD1 gene and found that NSD1 is required for early post-implantation development. In NSD1 mutant embryos, the primitive endoderm appears to develop normally, as indicated by the correct timing of differentiation of the squamous endodermal cell type. These embryos initiate gastrulation as they express Brachyury and form some extraembryonic mesoderm to generate amniotic folds (Tam and Behringer, 1997). However, mesodermal cells are scarce, the mesendoderm is absent and the anterioposterior axis is not specified. Additionally, and possibly as a primary consequence of the mutation, mutant embryos display enhanced apoptotic cell death in embryonic ectodermal cells as early as E6.5, the time at which generation of mesoderm from ectoderm normally begins. From these observations, we propose that NSD1 is not required for the initiation of mesoderm formation at the onset of gastrulation, but is critical for the survival of the embryonic ectoderm at this stage and hence the normal progression of gastrulation.
There are several SET protein-encoding genes, i.e. Ezh2, G9a, Suv39h1/h2 and Mll, that recently have been demonstrated to be embryonic lethal. Similarly to _NSD1_-deficient embryos, _Ezh2_–/– embryos fail to gastrulate properly (O’Carroll et al., 2001). In embryos lacking G9a, severe growth/developmental defects during gastrulation have also been reported and shown to be associated with apoptotic cell death (Tachibana et al., 2002). Recent inactivation of the Suv39h1- and Suv39h2-encoding genes demonstrated an essential role for these HMTases in pre- and postnatal development, with one-third of the double-mutant mice which survive to adulthood displaying growth retardation, infertility and chromosomal instabilities with an increased tumor risk (Peters et al., 2001). Mll (also known as All1, Hrx and Htrx) is another essential HMTase, and it controls embryo patterning via Hox gene expression (Yu et al., 1995). In this respect, we note that no expression of the Hoxa1 and Hoxb1 genes has been observed in E7.5 NSD1 mutant embryos. As both Hoxa1 and Hoxb1 contain retinoic acid response elements (RAREs) that are functional in vivo (Dupé et al., 1997; and references therein), it is tempting to speculate that NSD1 acting as a co-regulator for RARs (Huang et al., 1998) may play a direct role in mediating the Hoxa1 and Hoxb1 expression response to retinoic acid during early mouse development. However, although the RARE enhancer in close proximity to the Hoxa1 promoter has been shown to play a role in the control of Hoxa1 expression during gastrulation, deletion of this RARE did not abolish Hoxa1 expression (Dupé et al., 1997), which suggests a contribution of other transregulatory proteins and/or the presence of additional RAREs.
Of the various nuclear receptor cofactors identified to date, only a few have been reported to play a role in early embryogenesis, and many to exert in vivo function that can be compensated by functionally similar cofactors (Ciana et al., 2002; and references therein). Although it remains to be elucidated whether the observed defects in _NSD1_–/– embryos reflect a block(s) in nuclear receptor signaling or in the function of other transcription factors, our results clearly establish the absence of redundancy between NSD1 and any of the known nuclear receptor cofactors. They also provide genetic evidence that, at least during early embryogenesis, members of the NSD family including NSD1, NSD2/MMSET/WHSC1 and NSD3 (see references in Introduction), although structurally related, exert distinct, non-redundant functions.
Additional NSD1 function and human malignancy
NSD1 maps to human chromosome band 5q35 and was found fused to NUP98 in AML associated with the t(5;11)(q35;p15.5) translocation (Jaju et al., 2001). The NUP98 gene encodes a 98 kDa component of the nuclear pore complex that is thought to function as a docking protein through its conserved phenylalanine–glycine (FG) repeats (Radu et al., 1995). These repeats have been shown to bind transcription factors and to be retained in various oncogenic fusion transcripts, including the NUP98–NSD1 transcript in which they are fused to the conserved C-terminal region of NSD1 containing the five PHD fingers, the PWWP and SET domains, and the C5HCH motif (see Figure 1B), whereas the reciprocal NSD1–NUP98 transcript includes the N-terminal NIDs of NSD1 fused to the C-terminal RNA-binding domain of NUP98 (Jaju et al., 2001). Although functional analyses of these chimeric transcripts are required to elucidate their specific role in leukemogenesis, their identification provides strong evidence for a connection between NSD1 and cancer.
Further supporting this link is the recent identification of a nonsense and three frameshift mutations in the NSD1 gene that cause SOTOS syndrome (Kurotaki et al., 2002), a disorder that is characterized by the overgrowth of neural tissues, heart defects, advanced bone age, developmental delays and increased risk of cancers. Although the heterozygous NSD1+/– mice displayed a normal growth rate, indicating that the SOTOS phenotype in mice may be more subtle than in man and could be only observed after careful analysis of the growth pattern from birth onwards (M.Mark, O.Wendling, R.Losson and P.Chambon, in progress), our ISH data on murine embryonic sections revealed an expression pattern providing strong support for the involvement of NSD1 in the development of the SOTOS phenotype. Notably, NSD1 expression was observed in the brain as well as in the region of ossification of the developing bones and in the periosteum. Thus, disruption of NSD1 function in these tissues may result in an overgrowth as seen in SOTOS patients. To validate this hypothesis and to gain further insights into the post-gastrulation functions of NSD1, tissue-specific NSD1 ablations using mice homozygous for the floxed _NSD1_L2 allele described in this study will be required.
The SET domain of NSD1 is a novel HMTase domain
Our data indicate that the NSD1 SET domain has an intrinsic HMTase activity capable of methylating recombinant, native free and nucleosomal histones H3 and H4 with specificity for H3-K36 and H4-K20. Each of these two lysine residues previously has been shown to be targeted by specific SET domain-containing HMTases (see Introduction). However, none of these HMTases has been implicated in combining both modifications. Thus, although the precise substrate specificity of NSD1 in vivo remains to be determined, our findings raise the interesting possibility that NSD1 may represent a novel HMTase leading to a unique divalent methylation pattern. Until now, Set2 was the only known SET protein with HMTase activity specific to H3-K36 (Strahl et al., 2002). Interestingly, this HMTase isolated from S.cerevisiae belongs to the same subclass of SET domain-containing proteins as NSD1 (Schneider et al., 2002), and recently has been shown to be recruited to chromatin through selective interaction with the phosphorylated C-terminal domain (CTD) of RNA polymerase II (Xiao et al., 2003), thus suggesting a role for Set2 and H3-K36 in transcription elongation. Although an interaction between NSD1 and the RNA polymerase II CTD has not yet been investigated, the fact that, outside the SET domain, there is no obvious homology between NSD1 and Set2 may indicate that both proteins are targeted to chromatin through distinct molecular mechanisms. Moreover, the ability of NSD1 to bind directly to nuclear receptors is rather in favor of a model in which NSD1 functions in transcription initiation. Among the other known HMTases containing a SET domain, three have been reported to methylate H4-K20: the human SET8 protein and its Drosophila counterpart (Fang et al., 2002), the human PR-Set7 protein (Nishioka et al., 2002b) and Drosophila Ash1 (Beisel et al., 2002). Whereas SET8 and PR-Set7 are both H4-K20-selective HMTases, Ash1 has a broader substrate specificity, methylating H3-K4 and H3-K9 in addition to H4-K20. Comparison of the SET domains of these HMTases has enabled the classification of Ash1 in the same subfamily as Set2 and NSD1 (Huang et al., 1998; Schneider et al., 2002).
When directly tethered to a promoter region through fusion to a heterologous DNA-binding domain, Set2 exerts a transcriptional repression function dependent on its HMTase activity (Strahl et al., 2002), indicating a role for H3-K36 methylation in downregulating gene expression. Recent studies have also associated H4-K20 methylation with transcriptionally inactive chromatin (Nishioka et al., 2002b). Thus, based on these findings and our present data, it is tempting to speculate an essential function for NSD1 in the transcriptional silencing of developmentally regulated genes by methylation of H3-K36 and H4-K20. Note, however, that combining these two modifications could also specify a novel epigenetic code leading to a functional effect different from that generated by each modification, such as gene activation versus gene silencing. For instance, it has been shown recently that methylation of H3-K9 is not a determinant of gene silencing in the context of a nucleosome also methylated at H3-K4 and H4-K20 (Beisel et al., 2002).
Materials and methods
Construction of the targeting vector
Mouse NSD1 genomic clones were obtained by screening a 129/Sv ES cell-derived DNA library with a mouse NSD1 full-length cDNA probe. To create the 5′ homologous arm, a 4.9 kb NSD1 genomic fragment containing the 3′ 2.5 kb of intron 1 and the 5′ 2425 bp of exon 2 was first sucloned into pBluescriptS SK**+** to generate pNSD1-12. A floxed neomycin selection cassette was then introduced in the 5′ to 3′ orientation into the intron 1 _Hin_dIII site of pNSD1-12, yielding pNSD1-17. The 3′ homologous arm was constructed from a 4.5 kb NSD1 genomic fragment containing the 3′ 984 bp of exon 2 and the 5′ 3.5 kb of intron 2. First, this fragment was subcloned into pBluescript SK**+, yielding pNSD1-2. Then, a double-stranded oligonucleotide containing a loxP site (underlined), a _Hin_dIII site immediately upstream of the loxP site, and _Afl_II overhangs was produced by annealing two synthetic oligonucleotides XZ182 (5′-ACTGCTTAAGAAGCTTATAACTTCGTATAATGTATGCTATACG AAGTTATCTTAAGCGAT-3′) and XZ 183 (5′-ATCGCTTAAGATAA CTTCGTATAGCATACAT TATACGAAGTTATAAGCTTCTTAAG CAGT-3′), and was cloned into the intron 2 _Afl_II site of pNSD1-2, yielding pNSD1-32. The _loxP_-containing _Sma_I–_Xba_I fragment of pNSD1-32 was inserted into the _Sma_I and _Xba_I site of pBluescript SK+**, yielding pNSD1-8. The final targeting vector pNSD1-3 [also called pNSD1(LNL:L) in Figure 1A] was obtained by inserting a _Kpn_I–_Sma_I fragment of pNSD1-17 (which contains the floxed Neo cassette) into the _Kpn_I and _Sma_I sites of pNSD1-8.
Generation of mutant mice
The targeted ES cells containing an L3 allele were injected into C57BL/6 blastocysts to produce male chimeric offspring. These were backcrossed with C57BL/6 females, and the germline transmission of the targeted allele was determined by PCR analysis using a sense primer located in intron 1 upstream of the 5′ _Hin_dIII site (primer ZB197, 5′-GTCTGC ATTAAGTAATTGTGCCCTGAAG-3′) and an antisense primer located downstream of the 5′ loxP site in the Neo cassette (primer ZB198, 5′-TGTGTGCGAGGCCAGAGGCCACTTGTGTAG-3′). These primers generated a 350 bp DNA fragment from the targeted allele. Mice heterozygous for the targeted NSD1 gene (_NSD1_L3/+) were crossed with _CMV-Cre_tg/0 transgenic mice (Dupé et al., 1997). Tail DNA of the offspring was analyzed by genomic PCR using either the sense primer ZB197 and an antisense primer located in intron 1 downstream of the 3′ loxP site of the Neo cassette (AAW199, 5′-ACTGACTCCTCTTCTG GAGATCCTGAGTTC-3′) to amplify the wild-type allele (550 bp long), or the sense primer ZB197 and an antisense primer located in the 3′ loxP site (primer ZB200, 5′-ACTGTGGCATAGCATACTTAGCACATCTC-3′) to amplify the excised _L_– allele (350 bp long).
Histological analysis
Mouse embryos were fixed for 14 h in Bouin’s fluid, dehydrated, and embedded in paraffin. Sections were stained with hematoxylin and eosin according to standard procedures.
In situ hybridization
ISH was performed on sections pre-fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C for 12 h and embedded in paraffin using either digoxigenin-labeled riboprobes or an 35S-labeled riboprobe for NSD1 (region + 3456 to + 5070 of the mouse NSD1 cDNA encoding amino acids 1152–1690).
TUNEL assay
TUNEL assay was performed on paraffin sections using the ApopTag Plus Peroxidase In situ Apoptosis Detection kit (Quantum Appligene, France) according to the manufacturer’s protocol. TUNEL-positive cells were quantified by counting 400–1000 nuclei per embryo (excluding the ectoplacental cone) in eight normal and four presumptive _NSD1_–/– embryos.
Expression and purification of GST fusion proteins
Recombinant GST and GST fusion proteins were expressed from the pGEX-2T vector in Escherichia coli BL21 strain and purified on glutathione–Sepharose beads (Pharmacia) as previously described (Rea et al., 2000).
In vitro histone methyltransferase assays
HMTase assays were performed as previously reported (Strahl et al., 2002). Briefly, 50 µl of reaction containing substrates (20 µg of a mixture of native calf thymus histones, 2 µg of a mixture of purified HeLa or chicken core histones, 2 µg of HeLa oligonucleosomes, 5 µg of recombinant histones or 10 µg of histone peptides), enzymes (∼20 µg of GST-bound proteins) and 500 nCi of _S_-adenosyl-[methyl-3H]l-methionine (25 µCi/ml) (Amersham) in methyltransferase activity buffer [MAB: 50 mM Tris–HCl pH 9.0, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM β-mercaptoethanol] was incubated for 30 min at 30°C. Reactions were stopped by boiling in SDS loading buffer, and reaction products were separated by 4–20% SDS–PAGE and visualized by Gel code (Pierce) staining and fluorography. For the western blot analyses, the reaction products were loaded onto a 4–20% SDS–polyacrylamide gel, electrophoresed, and transferred onto a PVDF membrane. Anti-dim(K36-H3) (Upstate), anti-dim(K20-H4) (Upstate) and anti-dim(K27-H3) (a gift from Y.Zhang, University of California, San Francisco, CA) were used at 1:1000, 1:1000 or 1:1500 dilutions, respectively, and signals were detected by enhanced chemiluminescence. Native calf thymus histones were purchased from Boehringer Mannheim. Native oligonucleosomes were purified from HeLa cells as previously described (Steger et al., 1997). Recombinant histones and histone tail peptides were obtained from Upstate Biotechnology, USA.
Acknowledgments
Acknowledgements
We are grateful to D.Metzger for materials gifts, technical advice and helpful discussions, X.Belin and M.Cerviño for technical assistance, the staff of the ES cell culture and animal facility, and B.G.Herrmann, T.Jenuwein, R.Krumlauf, A.P.McMahon, F.Perrin-Smitt, Y.Zhang for reagents. This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, l’Hôpital Universitaire de Strasbourg (HUS), the Association pour la Recherche sur le Cancer, the Collège de France, the Fondation pour la Recherche Médicale (FRM) and Bristol-Myers-Squibb.
References
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Angrand P.O., Apiou,F., Stewart,A.F., Dutrillaux,B., Losson,R. and Chambon,P. (2001) NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics, 74, 79–88. [DOI] [PubMed] [Google Scholar]
- Beisel C., Imhof,A., Greene,J., Kremmer,E. and Sauer,F. (2002) Histone methylation by the Drosophila epigenetic regulator Ash1. Nature, 419, 857–862. [DOI] [PubMed] [Google Scholar]
- Chesi M., Nardini,E., Lim,R.S., Smith,K.D., Kuehl,W.M. and Bergsagel,P.L. (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood, 92, 3025–3034. [PubMed] [Google Scholar]
- Ciana P., Vegeto,E., Beato,M., Chambon,P., Gustafsson,J.A., Parker,M., Wahli,W. and Maggi,A. (2002) Looking at nuclear receptors from the heights of Erice. Workshop on nuclear receptor structure and function. EMBO Rep., 3, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupé V., Davenne,M., Brocard,J., Dolle,P., Mark,M., Dierich,A., Chambon,P. and Rijli,F.M. (1997) In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE). Development, 124, 399–410. [DOI] [PubMed] [Google Scholar]
- Echelard Y., Epstein,D.J., St-Jacques,B., Shen,L., Mohler,J., McMahon,J.A. and McMahon,A.P. (1993) Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell, 75, 1417–1430. [DOI] [PubMed] [Google Scholar]
- Fang J. et al. (2002) Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol., 12, 1086–1099. [DOI] [PubMed] [Google Scholar]
- Huang N., vom Baur,E., Garnier,J.-M., Lerouge,T., Vonesch,J.-L., Lutz,Y., Chambon,P. and Losson,R. (1998) Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J., 17, 3398–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaju R.J. et al. (2001) A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood, 98, 1264–1267. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol., 11, 266–273. [DOI] [PubMed] [Google Scholar]
- Kispert A. and Herrmann,B.G. (1994) Immunohistochemical analysis of the Brachyury protein in wild-type and mutant mouse embryos. Dev. Biol., 161, 179–193. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2002) Histone methylation in transcriptional control. Curr. Opin. Genet. Dev., 12, 198–209. [DOI] [PubMed] [Google Scholar]
- Kurotaki N. et al. (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nature Genet., 30, 365–366. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. (2003) Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem. Sci., 28, 59–62. [DOI] [PubMed] [Google Scholar]
- Murphy P. and Hill,R.E. (1991) Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development, 111, 61–74. [DOI] [PubMed] [Google Scholar]
- Nishioka K., Chuikov,S., Sarma,K., Erdjument-Bromage,H., Allis,C.D., Tempst,P. and Reinberg,D. (2002a) set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin modification. Genes Dev., 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K. et al. (2002b) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell, 9, 1201–1213. [DOI] [PubMed] [Google Scholar]
- O’Carroll D. et al. (2000) Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol., 20, 9423–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D., Erhardt,S., Pagani,M., Barton,S.C., Surani,M.A. and Jenuwein,T. (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol., 21, 4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H.F.M. et al. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, 323–337. [DOI] [PubMed] [Google Scholar]
- Radu A., Moore,M.S. and Blobel,G. (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell, 81, 215–222. [DOI] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Robyr D, Wolffe,A.P. and Wahli,W. (2000) Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol. Endocrinol., 14, 329–347. [DOI] [PubMed] [Google Scholar]
- Rosati R., La Starza,R., Veronese,A., Aventin,A., Schwienbacher,C., Vallespi,T., Negrini,M., Martelli,M.F. and Mecucci,C. (2002) NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood, 15, 3857–3860. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M.G. and Glass,C.K. (2001) Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem., 276, 36865–36868. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider,R., Bannister,A.J., Sheriff,J., Bernstein,B.E., Emra,N.C.T., Schreiber,S.L., Mellor,J. and Kouzarides,T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature, 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Schneider R., Bannister,A.J. and Kouzarides,T. (2002) Unsafe SETs: histone lysine methyltransferase and cancer. Trends Biochem. Sci., 27, 396–402. [DOI] [PubMed] [Google Scholar]
- Stec I., Wright,T.J., van Ommen,G.J., de Boer,P.A., van Haeringen,A., Moorman,A.F., Altherr,M.R. and den Dunnen,J.T. (1998) WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet., 7, 1071–1082. [DOI] [PubMed] [Google Scholar]
- Stec I., Nagl,S.B., van Ommen,G.J. and den Dunnen,J.T. (2000) The PWWP domain: a potential protein–protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett., 473, 1–5. [DOI] [PubMed] [Google Scholar]
- Steger D.J., Owen-Hughes,T., John,S. and Workmann,J.L. (1997) Analysis of transcription factor-mediated remodeling of nucleosomal arrays in a purified system. Methods, 12, 276–285. [DOI] [PubMed] [Google Scholar]
- Stoetzel C., Weber,B., Bourgeois,P., Bolcato-Bellemin,A.L. and Perrin-Schmitt,F. (1995) Dorso-ventral and rostro-caudal sequential expression of M-twist in the postimplantation murine embryo. Mech. Dev., 51, 251–263. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. et al. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol., 22, 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M. et al. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev., 16, 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P. and Behringer,R.R. (1997) Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev., 68, 3–25. [DOI] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann,A., Krauss,V., Dorn,R., Korge,G. and Reuter,G. (1994) The protein encoded by the Drosophila position-effect-variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J., 13, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cao,R., Xia,L., Erdjument-Bromage,H., Borchers,C., Tempst,P. and Zhang,Y. (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell, 8, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Xiao T., Hall,H., Kizer,K.O., Shibata,Y., Hall,M.C., Borchers,C.H. and Strahl,B.D. (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev., 17, 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xia,L., Wu,D.Y., Wang,H., Chansky,H.A., Schubach,W.H., Hickstein,D.D. and Zhang,Y. (2002) Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene, 21, 148–152. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hess,J.L., Horning,S.E., Brown,G.A. and Korsmeyer,S.J. (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature, 378, 505–508. [DOI] [PubMed] [Google Scholar]