A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record (original) (raw)
Abstract
Recent attempts to establish a molecular time-scale of eukaryote evolution failed to provide a congruent view on the timing of the origin and early diversification of eukaryotes. The major discrepancies in molecular time estimates are related to questions concerning the calibration of the tree. To limit these uncertainties, we used here as a source of calibration points the rich and continuous microfossil record of dinoflagellates, diatoms and coccolithophorids. We calibrated a small-subunit ribosomal RNA tree of eukaryotes with four maximum and 22 minimum time constraints. Using these multiple calibration points in a Bayesian relaxed molecular clock framework, we inferred that the early radiation of eukaryotes occurred near the Mesoproterozoic–Neoproterozoic boundary, about 1100 million years ago. Our results indicate that most Proterozoic fossils of possible eukaryotic origin cannot be confidently assigned to extant lineages and should therefore not be used as calibration points in molecular dating.
Keywords: molecular dating, origin of eukaryotes, protist phylogeny, relaxed molecular clock, small-subunit ribosomal RNA gene
1. Introduction
How ancient are eukaryotes, and when did the major eukaryotic groups diverge? These important evolutionary questions have recently been given increased attention. A common palaeontological interpretation of the fossil record suggests that eukaryotes originated about 2000 million years ago (Myr ago). This view is based on the presence of some putative eukaryotic fossils, including the ca 1850 Myr old spirally coiled ‘alga’ Grypania (Hoffman 1987) and large acritarchs from the 1800–1900 Myr old Chuanlinggou Formation (Zhang 1986). Some authors proposed an even older age for eukaryotes based on the presence of fossil biomarkers, such as steranes in the 2500–2800 Myr old shales from Australia (Brocks et al. 1999). However, neither Grypania nor such biomarkers can be unequivocally excluded from being of bacterial origin. A critical reassessment of these early fossils led Cavalier-Smith (2002_a_,b) to propose that eukaryotes originated only about 850 Myr ago, i.e. just before the Cryogenian glaciations.
Over the past few years, molecular data have been used to establish a time-scale of eukaryote evolution. These studies led to diametrically opposed conclusions and provoked a hot debate about the precision of molecular time estimates (Graur & Martin 2004; Hedges & Kumar 2004). On the one hand, it has been suggested that eukaryotes originated more than 2000 Myr ago, based on molecular clock analyses of genomic data (Hedges et al. 2001, 2004). On the other hand, the early diversification of eukaryotes was dated at 950–1259 Myr ago, according to an analysis of 129 proteins (Douzery et al. 2004).
There are two main sources of conflict between molecular and fossil dates: (i) biases due to the variations of the rates of molecular evolution and (ii) gaps in the fossil record and/or an inaccuracy of the calibration points (Benton & Ayala 2003). While new Proterozoic fossils are regularly described (e.g. Porter & Knoll 2000), a considerable effort has also been made to ameliorate molecular time estimates by using a relaxed molecular clock (RMC) approach (e.g. Aris-Brosou & Yang 2003). By contrast, relatively little has been done to improve the calibration step in molecular dating. Douzery et al. (2004) used multiple calibration points and discussed their utility. However, many studies used a single fossil event as the primary calibration point, sometimes adding secondary points inferred from molecular analyses (e.g. Hedges et al. 2004). The authors of these studies claimed that multiple calibrations lead to an underestimation of dates and are practically impossible when analysing large genomic databases with only few available taxa (Wang et al. 1999). The importance of calibration errors has been stressed by several authors (Graur & Martin 2004; Reisz & Müller 2004). Major sources of error associated with the imperfection of the fossil record for the calibration of molecular trees include: (i) the non-preservation of the earliest fossils of any lineage; (ii) uncertainties associated with the geological dating of fossils; and (iii) an incorrect taxonomic assignment of some fossils (Lee 1999).
In order to avoid some of these errors, we explored the potential of the well-documented, but largely ignored, continuous Phanerozoic microfossil record of protists as a source of calibration points. We selected 26 time constraints and used them to date a phylogeny of eukaryotes inferred from the small-subunit ribosomal RNA gene (SSU rRNA) in a Bayesian RMC framework. This approach allowed us to test whether or not the current interpretation of some key Proterozoic fossils as members of extant eukaryotic lineages is compatible with the Phanerozoic microfossil record.
2. Material and methods
An alignment of 240 SSU rRNA sequences from various eukaryotes, including an exhaustive sampling of all lineages for which a fossil record is known, was constructed manually with the Genetic Data Environment software (Larsen et al. 1993), following a secondary structure model (Wuyts et al. 2000). After determining possible calibration points from the literature through comparisons with available molecular data, the alignment was reduced to 83 eukaryotic sequences, due to computational limitations. A total of 1465 unambiguously aligned positions was used for phylogenetic analyses. An unrooted maximum-likelihood (ML) tree was inferred with the program PhyML (Guindon & Gascuel 2003), using the GTR+G+I model of evolution (Rodriguez et al. 1990). All parameters were estimated from the dataset. Additionally, a Bayesian analysis was conducted with MrBayes (Huelsenbeck & Ronquist 2001), with the same evolutionary model. Four simultaneous chains were run for 1 200 000 generations, and 12 000 trees were sampled, the first 2000 of which were discarded as the burn-in.
Divergence times were estimated under a Bayesian RMC framework with the multidistribute package (Kishino et al. 2001), and the program Baseml in the PAML package (Yang 1997) was used to estimate the parameters of the model. Two archaebacterial sequences were used to artificially constrain four possible positions for the root of the eukaryote tree. They were added to our dataset of 83 eukaryotic sequences for the dating analysis, and automatically pruned during the final step. Details of this procedure, and of the prior gamma distributions on the parameters of the relaxed clock model, can be found in the electronic supplementary material—Methods. All chains were started from random values, run for 1 000 000 generations and sampled every 100 generations, and the first 100 000 generations were discarded as the burn-in. The uncertainty of divergence time estimates was accounted for by using the 95% credibility intervals of the 10 000 samples.
3. Results and discussion
(a) Dating the eukaryote phylogeny
While the earliest known fossil of a given lineage merely provides a minimum date for the appearance of the lineage, true calibration points with both minimum and maximum time limits are rare and difficult to ascertain. In this study, we argue that organisms presenting a continuous fossil record represent the only unambiguous source of calibration points for molecular dating. Indeed, continuous records provide detailed information about the succession of different morphotypes in the different stratigraphic levels, so that the chance of underestimating the time of the first appearance of a given morphotype, because we simply missed it, is extremely reduced. Previous studies based on combined protein data generally used calibration points taken solely from the fossil records of plants, animals and fungi. However, because the fossils of multicellular organisms are rare and discrete, they are arguably best viewed as a source of minimum time estimates only.
By contrast, several lineages of protists have a rich and continuous microfossil record in the Phanerozoic, and are a potential source of accurate calibration points for molecular dating. Five groups of protists were considered for this study. After a careful comparison with available molecular data, two groups, Foraminifera and Radiolaria, were discarded because their SSU rRNA sequences were too divergent, while the remaining three groups, coccolithophorids, diatoms and dinoflagellates, allowed the selection of four maximum time constraints (MaxTCs) that could be confidently used as calibration points in our analysis (see the electronic supplementary material—Methods). In addition to these four MaxTCs, we also selected 22 minimum time constraints (MinTCs) among protists, plants, fungi and metazoans, based on the first appearances of some lineages in the fossil record. All Phanerozoic fossil events used as calibration points, and all Proterozoic fossils discussed in this study, are listed in the electronic supplementary material—table S1.
The four MaxTCs and the 22 MinTCs were used as prior time constraints in a Bayesian RMC dating of our SSU rRNA phylogeny of eukaryotes (figure 1). ML and Bayesian analyses yielded similar tree topologies (electronic supplementary material—figure S1), which are congruent with previously published eukaryote phylogenies (e.g. Baldauf et al. 2000; Nikolaev et al. 2004). The tree was rooted between unikonts (opisthokonts+Amoebozoa) and bikonts (Stechmann & Cavalier-Smith 2003). Three other possible rooting strategies were also considered; all of these led to similar time estimates, and the 95% confidence intervals of the dates at each node largely overlapped (electronic supplementary material—table S2). These observations support the hypothesis that whatever is the true position of the root of the eukaryotic tree, the early radiation of all extant eukaryotic supergroups probably occurred within a relatively short period of time (e.g. Philippe & Adoutte 1998).
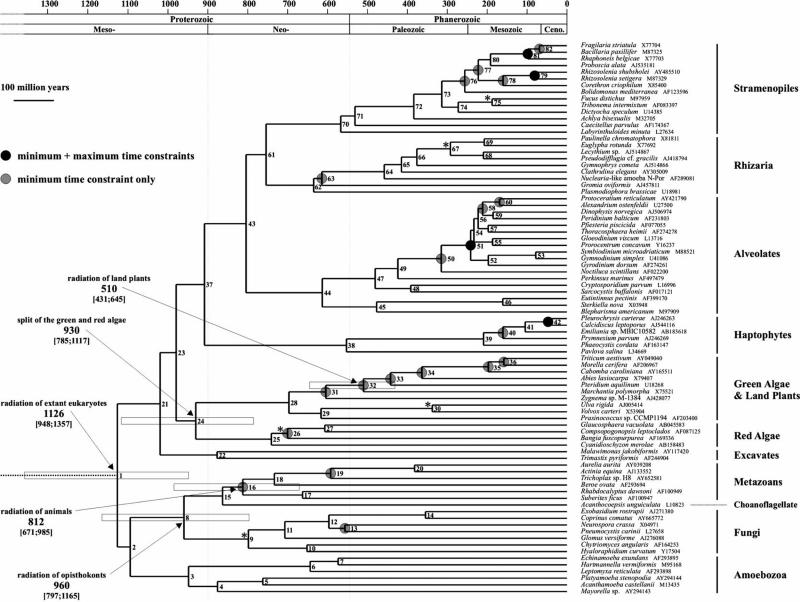
Figure 1.
A time-scale of eukaryote evolution, based on a Bayesian relaxed molecular clock applied to a dataset of 83 small-subunit ribosomal RNA gene sequences, calibrated using the Phanerozoic microfossil record. Branch lengths are proportional to the absolute ages of the subtending nodes, and the topology used was obtained by an ML analysis of the dataset using the GTR+G+I model of evolution (see §2). Species names, taxonomic position and GenBank accession numbers of the sequences are indicated on the right. White rectangles delimit 95% confidence intervals on ages of some key nodes. Confidence intervals of all other nodes can be found in the electronic supplementary material—table S3, according to their numbers. Circles indicate the 23 nodes under prior palaeontological calibration (light grey: lower bound only; dark grey: lower and upper bounds, except node 79 which had only an upper bound; see electronic supplementary material—table S1). Asterisks highlight the nodes to which additional minimum time constraints would be applied based on the existence of Proterozoic fossils putatively belonging to one of the lineages subtending the node. The two vertical dotted lines indicate the transition between Meso-/Neo-Proterozoic and the beginning of the Cambrian, respectively.
According to our time-scale, the basal radiation of extant eukaryotes (node 1 in figure 1) took place about 1126 Myr ago (range 948–1357 Myr ago). It was shortly followed by the radiations of amoebozoans, opisthokonts and bikonts, near the Mesoproterozoic–Neoproterozoic boundary. The basal radiations of animals, fungi, red algae and green algae occurred during the Neoproterozoic, leading to the Cambrian explosion of bilaterian animals and the dominance of green algae in Palaeozoic oceans. Interestingly, our results indicate that all other eukaryotic supergroups also radiated in the Neoproterozoic, including the chromalveolates, from which originate the ancestors of the three lineages (diatoms, dinoflagellates and coccolithophorids) that replaced the green algae as the dominant members of the eukaryotic phytoplankton at the end of the Palaeozoic (Falkowski et al. 2004).
The dating method used in this study largely depends on the prior gamma distributions on some of the parameters of the RMC model (e.g. Welch & Bromham 2005). In particular, the prior on the root age (a priori expected time between tips and root) has a direct influence on the posterior dates inferred during the analysis. To take this effect into account, we tested several plausible values for the a priori date at the root of the eukaryote radiation, and the posterior dates were found to converge to the values shown in figure 1. Moreover, our results are congruent with those of the recent study published by Douzery et al. (2004). In spite of recent criticisms (Blair & Hedges 2005), the maximum age limits used by Douzery et al. (2004) were apparently not too constraining, as the dates inferred in our time-scale are largely congruent with their estimations.
(b) Towards a reinterpretation of early eukaryotic fossils
In our molecular dating, we have consciously ignored all putative eukaryotic fossils from the Proterozoic to avoid problems related to their possible misidentification or erroneous assignment to extant taxa. However, to critically discuss the interpretation of these fossils, we have tested their compatibility with the dates inferred in our study. We focused on five fossils of particular interest: the arguable red alga Bangiomorpha (ca 1200 Myr old; e.g. Butterfield 2000), the putative xanthophyte Palaeovaucheria (ca 1000 Myr old; e.g. Woods et al. 1998), the possible cladophoracean green alga Proterocladus (ca 750 Myr old; Butterfield et al. 1994), the putative ‘higher’ fungus Tappania (ca 1400 Myr old; Butterfield 2005) and the vase-shaped microfossils (VSMs) attributed to extant lobose arcellinids or filose euglyphids (ca 750 Myr old; Porter & Knoll 2000; electronic supplementary material—table S1).
Our results suggest that a reinterpretation of the phylogenetic status of these five Proterozoic fossils is needed. First, the time-scale presented in figure 1 reveals that the four MaxTCs derived from the Phanerozoic fossil record are not compatible with the current interpretation of Bangiomorpha, Palaeovaucheria, Proterocladus and Tappania, or with an interpretation of VSMs as members of the euglyphid testate amoebae. Indeed, considering Bangiomorpha as a relative of extant Bangiales would imply a date of at least 1200 Myr ago for the separation between Bangia and other red algae (node 26 in figure 1), an event dated at 700 Myr ago (range 566–883 Myr ago) in our analyses. Similarly, considering Palaeovaucheria as a xanthophyte alga would imply a date of at least 1000 Myr ago for the separation between Xanthophyceae and Phaeophyceae (node 75 in figure 1), an event which is much younger according to our time-scale (187 Myr ago; range 119–275 Myr ago). Considering Proterocladus as a cladophoracean green alga would imply a date of at least 750 Myr ago for the separation between Chlorophyceae and Ulvophyceae (node 30 in figure 1), an event dated at only 337 Myr ago (205–513 Myr ago) in our analyses. Considering Tappania as a ‘higher’ fungus would imply a date of at least 1400 Myr ago for the divergence of Ascomycota+Basidiomycota from chytriids (node 9 in figure 1), an event which is 798 Myr old (634–1003 Myr old) according to our time-scale. Finally, considering the VSMs as euglyphid amoebae would imply a date of at least 750 Myr ago for the terminal radiation of the cercozoan filose amoebae (node 67 in figure 1), an event which is much younger according to our time-scale (292 Myr ago; range 195–416 Myr ago).
To test further the effect of these Proterozoic fossils on the eukaryote time-scale, the four MaxTCs used in our main analysis were removed and six new dating analyses were performed. First, we determined the date of the eukaryote radiation in the absence of any MaxTC. Then, the five possible MinTCs provided by the five Proterozoic fossils discussed above were added successively to five additional dating analyses (table 1). In the absence of any internal MaxTC (and apart from the direct influence of the prior on the root age; see above), the only upper limit in the dating process is the highest possible time between tips and root (the command ‘bigtime’), which was set at 4500 Myr ago. Consequently, all dates inferred without any MaxTC are significantly older than those presented in figure 1, and all confidence intervals are widened (line 2 in table 1). This clearly demonstrates the importance of using at least one internal MaxTC in dating analyses. Interestingly, in the presence of each of the additional MinTCs derived from the five Proterozoic fossils, all dates in the tree are displaced further back in time (see table 1), even in the case of the most conservative hypothesis (considering Bangiomorpha as a true red alga).
Table 1.
Dates calculated for the radiation of extant eukaryotes and the four nodes under prior maximum time constraints (MaxTCs) when using alternatively five new minimum time constraints (MinTCs) following the current taxonomic interpretation of some Proterozoic fossils.
| radiation of extant eukaryotes (node 1) | radiation of dinoflagellates (node 51) | radiation of the rhizosolenid diatoms (node 79) | radiation of the pennate diatoms (node 81) | radiation of extant coccolithophorids (node 41) | |
|---|---|---|---|---|---|
| calibration presented in figure 1 | 1126 (948–1357) | 241 (223–250) | 81 (61–90) | 98 (77–110) | 104 (54–176) |
| dates inferred in absence of any MaxTC, with only 22 MinTCs | 2331 (1432–3846) | 716 (380–1276) | 385 (179–740) | 407 (190–776) | 267 (105–552) |
| Bangiomorpha is a red alga (MinTC of 1200 Myr at node 26) | 2723 (1830–4182) | 841 (480–1421) | 451 (220–826) | 477 (238–875) | 314 (128–636) |
| Palaeovaucheria is a xanthophyte (MinTC of 1000 Myr at node 75) | 3868 (2929–4473) | 1223 (785–1740) | 480 (237–854) | 508 (256–906) | 336 (138–677) |
| Proterocladus is a cladophoracean (MinTC of 750 Myr at node 30) | 3233 (2094–4383) | 999 (570–1583) | 534 (262–923) | 563 (280–971) | 376 (155–736) |
| Tappania is a ‘higher’ fungus (MinTC of 1400 Myr at node 9) | 2909 (1994–4272) | 896 (513–1473) | 543 (278–936) | 577 (297–981) | 378 (157–738) |
| VSMs are filose testate amoebae (MinTC of 750 Myr at node 67) | 3270 (2216–4377) | 1017 (604–1577) | 709 (416–1101) | 755 (440–1155) | 455 (201–831) |
In our opinion, the best explanation of these observations is that the current interpretations of the Proterozoic fossils discussed here are erroneous. For some of them, this proposition does not sound surprising. For instance, the Mesozoic radiation of diatoms (about 222 Myr ago; node 77 in figure 1) is incompatible with a Proterozoic appearance of xanthophyte algae, as both of these lineages belong to the same radiation of autotrophic heterokont algae within the stramenopiles. This clearly excludes the possibility that Palaeovaucheria (ca 1000 Myr old) and even the younger Jacutianema (ca 750 Myr old; Butterfield 2004) belonged to Xanthophyceae.
Although considering the Proterozoic fossils presented above as members of extant lineages of eukaryotes makes poor sense in light of our data, it does not necessarily imply that all these fossils represent prokaryotes (mostly cyanobacteria) mistaken for eukaryotes, as proposed by Cavalier-Smith (2002_a_,b). Some of them might indeed be of bacterial origin, but given an initial radiation of extant eukaryotes 948–1357 Myr ago, it is also plausible that some (if not all) of the Proterozoic fossils, such as Bangiomorpha, Palaeovaucheria, Proterocladus and Tappania, correspond to extinct, basal lineages of eukaryotes that evolved morphological and/or ultrastructural features similar to those of extant lineages by convergence. We are thus not questioning the description of these fossils as possible eukaryotes, but rather their assignment to extant lineages.
The situation is different in the case of the VSMs. Contrary to the Proterozoic fossils we discussed above, VSMs could not be prokaryotes mistaken for eukaryotes, because their apertural structures indisputably indicate eukaryotic affinities (Porter & Knoll 2000). However, the exact affiliation of VSMs is controversial. Their morphology strongly suggests that they are the remnants of testate amoebae, but it is difficult to determine if they were related to extant lineages, such as the filose euglyphids or the lobose arcellinids. Our results indicate that they cannot have been closely related to euglyphids, which diverged in the Palaeozoic according to our time-scale. On the other hand, considering VSMs as relatives of extant arcellinids is compatible with our results. The few arcellinids sequenced to date belong to the main lineage of Amoebozoa (Smirnov et al. 2005), which diverged about 644 Myr ago (range 455–879 Myr ago; node 6 in figure 1). This indicates that VSMs might have belonged to the amoebozoan radiation, and might even have been related to extant arcellinids, which would substantially extend their fossil record (the next oldest fossil attributed to arcellinids is the 325 Myr old Prantlitina; Loeblich & Tappan 1964).
4. Conclusions
By using the continuous microfossil record to calibrate the eukaryote phylogeny, we estimated that the radiation of early eukaryotes occurred near the Mesoproterozoic–Neoproterozoic border, about 1100 Myr ago. This result is congruent with the increasing number of microfossils appearing in the Neoproterozoic (Knoll 1994). Several of these fossils may represent early eukaryotes, but as shown by our study their assignment to recent lineages is highly uncertain. Therefore, we believe they should not be used as calibration points in molecular dating. Recently, several studies challenged isotopic and microfossil evidence for an early origin of life on Earth and the presence of true fossils in rocks older than 2000 Myr (e.g. Brasier et al. 2002; van Zuilen et al. 2002). In fact, the first undisputable traces of life on Earth might be the bacterial fossils of the 1900 Myr old Gunflint Formation of Ontario (Moorbath 2005). In agreement with this idea, our results favour the hypothesis that the history of extant eukaryotes did probably not span more than one-quarter of the Earth's history.
Acknowledgments
We are indebted to Benoît Stadelmann and Jérôme Flakowski for helpful discussion, and thank Sam Bowser and two anonymous referees for useful comments on the manuscript. This work was supported by Swiss National Science Foundation grant 3100A0-100415.
Footnotes
†
Present address: Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK.
Supplementary Material
Methods and supplementary figure and tables.
Additional details on the phylogenetic and dating analyses and on our choice of four maximum time constraints, and description of all fossil events used or discussed in this study.
References
- Aris-Brosou S, Yang Z. Bayesian models of episodic evolution support a late Precambrian explosive diversification of the Metazoa. Mol. Biol. Evol. 2003;20:1947–1954. doi: 10.1093/molbev/msg226. 10.1093/molbev/msg226 [DOI] [PubMed] [Google Scholar]
- Baldauf S.L, Roger A.J, Wenk-Siefert I, Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. 10.1126/science.290.5493.972 [DOI] [PubMed] [Google Scholar]
- Benton M.J, Ayala F.J. Dating the tree of life. Science. 2003;300:1698–1700. doi: 10.1126/science.1077795. 10.1126/science.1077795 [DOI] [PubMed] [Google Scholar]
- Blair J.E, Hedges S.B. Molecular clocks do not support the Cambrian explosion. Mol. Biol. Evol. 2005;22:387–390. doi: 10.1093/molbev/msi039. 10.1093/molbev/msi039 [DOI] [PubMed] [Google Scholar]
- Brasier M.D, Green O.R, Jephcoat A.P, Kleppe A.K, Van Kranendonk M.J, Lindsay J.F, Steele A, Grassineau N.V. Questioning the evidence for Earth's oldest fossils. Nature. 2002;416:76–81. doi: 10.1038/416076a. 10.1038/416076a [DOI] [PubMed] [Google Scholar]
- Brocks J.J, Logan G.A, Buick R, Summons R.E. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. 10.1126/science.285.5430.1033 [DOI] [PubMed] [Google Scholar]
- Butterfield N.J. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. [Google Scholar]
- Butterfield N.J. A vaucheriacean alga from the middle Neoproterozoic of Spitsbergen: implications for the evolution of Proterozoic eukaryotes and the Cambrian explosion. Paleobiology. 2004;30:231–252. [Google Scholar]
- Butterfield N.J. Probable Proterozoic fungi. Paleobiology. 2005;31:165–182. [Google Scholar]
- Butterfield N.J, Knoll A.H, Swett K. Paleobiology of the Neoproterozoic Svanbergfjellet Formation, Spitsbergen. Fossil Strata. 1994;34:1–84. [Google Scholar]
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002a;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002b;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Douzery E.J.P, Snell E.A, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl Acad. Aci. USA. 2004;101:15 386–15 391. doi: 10.1073/pnas.0403984101. 10.1073/pnas.0403984101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski P.G, Katz M.E, Knoll A.H, Quigg A, Raven J.A, Schofield O, Taylor F.J.R. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. 10.1126/science.1095964 [DOI] [PubMed] [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. 10.1016/j.tig.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hedges S.B, Kumar S. Precision of molecular time estimates. Trends Genet. 2004;20:242–247. doi: 10.1016/j.tig.2004.03.004. 10.1016/j.tig.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Hedges S.B, Chen H, Kumar S, Wang D.Y, Thompson A.S, Watanabe H. A genomic timescale for the origin of eukaryotes. BMC Evol. Biol. 2001;1:4. doi: 10.1186/1471-2148-1-4. 10.1186/1471-2148-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B, Blair J.E, Venturi M.L, Shoe J.L. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. 10.1186/1471-2148-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P.H. Early Proterozoic foredeeps, foredeep magmatism, and superior-type iron-formations of the Canadian Shield. In: Kröner A, editor. Proterozoic lithospheric evolution. American Geophysical Union; Washington, DC: 1987. pp. 85–98. [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Kishino H, Thorne J.L, Bruno W.J. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol. Biol. Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- Knoll A.H. Proterozoic and early Cambrian protists: evidence for accelerating evolutionary tempo. Proc. Natl Acad. Sci. USA. 1994;91:6743–6750. doi: 10.1073/pnas.91.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N, Olsen G.J, Maidak B.L, McCaughey M.J, Overbeek R, Macke T.J, Marsh T.L, Woese C.R. The ribosomal database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S. Molecular clock calibrations and metazoan divergence dates. J. Mol. Evol. 1999;49:385–391. doi: 10.1007/pl00006562. 10.1007/PL00006562 [DOI] [PubMed] [Google Scholar]
- Loeblich A.R, Tappan H. Sarcodina—chiefly ‘Thecamoebians’ and Foraminiferida. In: Moore R.C, editor. Treatise on invertebrate paleontology, Part C, Protista 2. University of Kansas Press; Lawrence, KS: 1964. pp. 1–900. [Google Scholar]
- Moorbath S. Palaeobiology: dating earliest life. Nature. 2005;434:155. doi: 10.1038/434155a. 10.1038/434155a [DOI] [PubMed] [Google Scholar]
- Nikolaev S.I, Berney C, Fahrni J.F, Bolivar I, Polet S, Mylnikov A.P, Aleshin V.V, Petrov N.B, Pawlowski J. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc. Natl Acad. Sci. USA. 2004;101:8066–8071. doi: 10.1073/pnas.0308602101. 10.1073/pnas.0308602101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Adoutte A. The molecular phylogeny of Protozoa: solid facts and uncertainties. In: Combs G.H, Vickerman K, Sleigh M.A, Warren A, editors. Evolutionary relationships among Protozoa. Chapman & Hall; London, UK: 1998. pp. 25–56. [Google Scholar]
- Porter S.M, Knoll A.H. Testate amoebae in the Neoproterozoic era: evidence from vase-shaped microfossils in the Chuar Group, Grand Canyon. Paleobiology. 2000;26:360–385. [Google Scholar]
- Reisz R.R, Müller J. Molecular timescales and the fossil record: a paleontological perspective. Trends Genet. 2004;20:237–241. doi: 10.1016/j.tig.2004.03.007. 10.1016/j.tig.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Oliver J.L, Marin A, Medina J.R. The general stochastic model of nucleotide substitution. J. Theor. Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Smirnov A, Nassonova E, Berney C, Fahrni J, Bolivar I, Pawlowski J. Molecular phylogeny and classification of the lobose amoebae. Protist. 2005;156:129–142. doi: 10.1016/j.protis.2005.06.002. 10.1016/j.protis.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr. Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. 10.1016/S0960-9822(03)00602-X [DOI] [PubMed] [Google Scholar]
- van Zuilen M.A, Lepland A, Arrhenius G. Reassessing the evidence for the earliest traces of life. Nature. 2002;418:627–630. doi: 10.1038/nature00934. 10.1038/nature00934 [DOI] [PubMed] [Google Scholar]
- Wang D.Y, Kumar S, Hedges S.B. Divergence time estimates for the early history of animal phyla and the origin of plants, animals, and fungi. Proc. R. Soc. B. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. 10.1098/rspb.1999.0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.J, Bromham L. Molecular dating when rates vary. Trends Ecol. Evol. 2005;20:320–327. doi: 10.1016/j.tree.2005.02.007. 10.1016/j.tree.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Woods K.N, Knoll A.H, German T. Xanthophyte algae from the Mesoproterozoic/Neoproterozoic transition: confirmation and evolutionary implications. Geol. Soc. Am. Abstr. Program. 1998;30:232. [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. 10.1093/nar/28.23.4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Clastic facies microfossils from the Chuanlinggou Formation (1800 Ma) near Jixian, North China. J. Micropaleontol. 1986;5:9–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods and supplementary figure and tables.
Additional details on the phylogenetic and dating analyses and on our choice of four maximum time constraints, and description of all fossil events used or discussed in this study.