Gene Targeting of Cdc42 and Cdc42GAP Affirms the Critical Involvement of Cdc42 in Filopodia Induction, Directed Migration, and Proliferation in Primary Mouse Embryonic Fibroblasts (original) (raw)
Abstract
Recent studies in Cdc42 knockout mouse embryonic stem (ES) cells and ES-derived fibroblastoid cell lines raise concern on a body of literature derived by dominant mutant expression approach in a variety of cell lines implicating mammalian Cdc42 as a key regulator of filopodia induction, directional migration and cell cycle progression. To resolve the physiological function of mammalian Cdc42, we have characterized the Cdc42−/− and Cdc42GAP−/− primary mouse embryonic fibroblasts (MEFs) produced by gene targeting as the Cdc42 loss- or gain-of-activity cell model. The Cdc42−/− cells were defective in filopodia formation stimulated by bradykinin and in dorsal membrane ruffling stimulated by PDGF, whereas the Cdc42GAP−/− cells displayed spontaneous filopodia. The Cdc42 loss- or gain-of-activity cells were defective in adhesion to fibronectin, wound-healing, polarity establishment, and migration toward a serum gradient. These defects were associated with deficiencies of PAK1, GSK3β, myosin light chain, and FAK phosphorylation. Furthermore, Cdc42−/− cells were defective in G1/S-phase transition and survival, correlating with deficient NF-κB transcription and defective JNK, p70 S6K, and ERK1/2 activation. These results demonstrate a different requirement of Cdc42 activity in primary MEFs from ES or ES-derived clonal fibroblastoid cells and suggest that Cdc42 plays cell-type–specific signaling roles.
INTRODUCTION
The Rho family GTPase Cdc42 acts as an intracellular molecular switch cycling between the GTP-bound, active and the GDP-bound, inactive states (Van Aelst and D'Souza-Schorey, 1997; Bishop and Hall, 2000). The current biochemical model depicts that upon activation of a variety of cell surface receptors, Cdc42 can be activated to recognize specific downstream effectors to regulate multiple cell functions including actin cytoskeleton reorganization (Hall, 1998), directional migration, gene expression, and cell cycle S-phase progression (Kjoller and Hall, 1999; Ridley, 2001; Etienne-Manneville and Hall, 2002). Although genetic evidence for the related cell functions of Cdc42 is particularly strong in lower eukaryotes including Saccharomyces cerevisiae and Caenorhabditis elegans (Johnson and Pringle, 1990; Gotta et al., 2001; Kay and Hunter, 2001), genetic support for these roles of Cdc42 in mammalian cells is limited, in part because conventional gene targeting of Cdc42 leads to early embryonic lethality (Chen et al., 2000), and most published literature on mammalian Cdc42 cell function are derived by using dominant negative or constitutively active mutant expression approaches in clonal cell lines.
Although dominant mutant expression has been widely adopted in cell biological studies, it may have significant drawbacks (Feig, 1999). In principle, the dominant negative mutant of Cdc42 functions by sequestering the upstream Rho guanine nucleotide exchange factors and tends to be nonspecific in nature due to the recognized Rho GTPase substrate overlapping of multiple GEFs. Because of its noncatalytic nature, overexpression of the dominant negative mutant is typically needed to achieve effective blockage of the endogenous Cdc42 activity, further compounding the specificity issue with dosage dependency in the functional outcomes. On the other hand, constitutively active mutant of Cdc42 could also lack specificity for effector pathways since overexpression of the constitutively GTP bound Cdc42 could tie up individual effectors that are shared by other Rho GTPases such as Rac1 in a nonspatiotemporal manner. It is now known that multiple Cdc42 effectors exist in a given cell type, and their dynamic regulation by Cdc42 and possibly by other cellular factors is necessary for optimal Cdc42 signaling. Thus it is highly desirable to use genetic approaches with minimal perturbation of endogenous cellular environment to study Cdc42 function.
A number of recent studies have reported the use of Cdc42-specific gene targeting in mouse to define Cdc42 cell functions. Surprisingly, direct knockout of Cdc42 gene in embryonic stem (ES) cells does not appear to affect cell proliferative potential or alter the ERK1/2, p38, and JNK MAP kinase signaling that have previously been suggested to be regulated by Cdc42 (Chen et al., 2000). More intriguingly, a recent study using loxP/Cre-mediated deletion of Cdc42 from an ES-derived fibroblastoid cell line indicated that Cdc42 is dispensable for actin filopodia induction, directed migration, cell polarization, and mitosis (Czuchra et al., 2005). Combined with the rationale that the dominant mutant overexpression approaches may introduce cellular artifacts, these results cast doubts on our current understanding of mammalian Cdc42 cell function and raise the possibility that at least some of the previously assigned cellular role of Cdc42 might be attributed to experimental limitations.
To clarify the function of mammalian Cdc42, we have generated Cdc42 conditional knockout mice by a loxP/Cre recombination strategy to allow controlled deletion of Cdc42 in primary cells, as well as a Cdc42 gain-of-activity mouse model by gene targeting of a negative regulator of Cdc42, Cdc42GAP, that leads to specific upregulation of endogenous Cdc42 activity in most cell types (Wang et al., 2005, 2006). We report here the results obtained from the Cdc42-deficient and Cdc42GAP-deficient primary mouse embryonic fibroblasts (MEFs), which provide genetic evidence implicating Cdc42 in primary cell actin structural organization, migration, and proliferation. In addition, our examinations of Cdc42−/− and Cdc42GAP−/− MEF cells demonstrate that a tightly regulated Cdc42 activity is essential for proper signaling effects.
MATERIALS AND METHODS
Generation of Primary Cdc42−/− and Cdc42GAP−/− MEFs
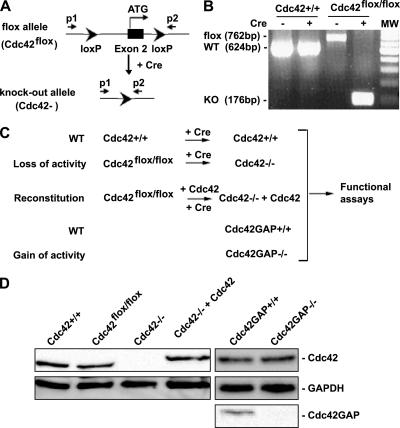
To produce Cdc42 conditional knockout mice, the guanine nucleotide binding sequence-encoding exon 2 of Cdc42 gene was flanked by a pair of loxP sequences (Figure 1A; the detailed strategy of derivation of the Cdc42flox/flox mice will be described elsewhere). Cdc42GAP gene targeted mice were generated previously (Wang et al., 2005). E12.5 old mouse embryos of the Cdc42+/+ and Cdc42flox/flox or Cdc42GAP+/+ and Cdc42GAP−/− genotypes were generated by cross-breeding Cdc42flox/+ or Cdc42GAP+/− heterozygous parents to provide MEFs. The MEFs were cultured in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) as described (Guo and Zheng, 2004). The genotypes of the cells and effective deletion and/or reconstitution of Cdc42 were confirmed by PCR-based protocols using relevant primers (Figure 1A; p1, p2) and by Western blotting using a Cdc42 mAb (BD Biosciences, San Jose, CA). The primer sequences are: p1: 5′-AGACAAAACAACAAGGTCCAGAAAC-3′; p2: 5′-CTGCCAACCATGACAACCTAAGTTC-3′. MEFs were used at low passage numbers (3–5) throughout the studies to avoid accumulative genetic abnormalities during passage. Primary MEFs derived from at least three independently bred mouse embryos were used in the cell functional assays described below.
Figure 1.
Generation of Cdc42−/− and Cdc42GAP−/− primary MEFs by a loxP/Cre recombinase system or by a conventional gene targeting strategy respectively. (A) The loxP/Cre-mediated gene targeting strategy to generate Cdc42 gene–deleted (Cdc42−) allele. (B) Genotyping results of primary MEFs obtained from cross-breeding of Cdc42flox/+ heterozygous mice with or without retrovirus-mediated Cre transduction. The 762-base pair band represents the floxed (flox) allele, the 624-base pair band represents the WT (+) allele, and the 176-base pair band represents the Cdc42 KO (−) allele in the cells. (C) Depiction of the primary MEF cell models of Cdc42 loss or gain of activity and the relevant controls. The Cdc42−/− cells were generated by retrovirus-mediated Cre transduction into the Cdc42flox/flox primary MEFs, whereas the Cdc42GAP−/− cells were produced by direct extraction of MEFs from the Cdc42GAP−/− mouse embryos. The relevant control cells, including the genetically matched Cdc42+/+ or Cdc42GAP+/+ MEFs derived from the same litters of Cdc42flox/flox or Cdc42GAP−/− embryos and the (HA)3-tagged, WT Cdc42 reconstituted (Cdc42−/− + Cdc42) cells, were produced in parallel. All cells were used for the studies at comparable low passages (passages 3–5). (D) Expression of Cdc42 in various primary MEFs, including the exogenous (HA)3-Cdc42 reconstituted cells, was analyzed by anti-Cdc42 Western blotting. Cdc42GAP deletion in the Cdc42GAP−/− MEFs was confirmed by anti-Cdc42GAP immunoblotting.
Retroviral Transduction and Fluorescence-activated Cell Sorting Analysis
Cre-mediated deletion of the floxed Cdc42 alleles was achieved by transduction of the cells with Cre-expressing retrovirus or adenovirus to generate the Cdc42−/− (KO) as well as control Cdc42+/+ cells. Cdc42−/− cells reconstituted with wild-type (WT) Cdc42 (Cdc42−/− +Cdc42) or L61Rac1 (Cdc42−/− +L61Rac1) were generated by transduction of WT Cdc42- or L61Rac1- (HA-tagged) and Cre-expressing retroviruses. The Cre recombinase and HA-tagged WT Cdc42 or constitutively active L61Rac1 were cloned into a retrovirus vector of murine stem cell virus (MSCV)-internal ribosomal entry site (IRES) yellow fluorescent protein (MSCV-YFP) and the retrovirus vector MIEG3 containing internal ribosome entry site-enhanced green fluorescent protein (MIEG3-EGFP), respectively. Retroviral supernatant was prepared by using the Phoenix packaging cell system and subsequent infection were carried out in the presence of 8 μg/ml Polybrene (Sigma, St. Louis, MO) according to the described protocols (Guo and Zheng, 2004). Cells were harvested 48 h after infection and the EGFP and/or YFP-positive cells (typically 10–30%) were isolated by fluorescence-activated cell sorting (FACS). Deletion of the Cdc42 gene from Cdc42flox/flox cells was also achieved by using adenoviruses expressing the Cre recombinase (a generous gift from Dr. Jeff Molkentin, Cincinnati Children's Research Foundation, Cincinnati, OH).
Western Blotting and Immunofluorescence
MEFs were serum-starved for 16 h, stimulated with 10% FBS, and harvested at different time points. They were lysed in the RIPA buffer containing 10 mM Tris-HCl, pH 8.0, 0.14 M NaCl, 0.025% sodium azide, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 0.1 μM okadaic acid, 1 mM Na3VO4, 50 mM NaF, 50 mM sodium β-glycerophosphate, 1 mM PMSF, and protease inhibitors (Sigma). Protein extracts, 100 μg were resolved by electrophoresis and electrotransferred to PVDF membrane. The contents of GSK3β, PAK1, myosin light chain (MLC), cofilin, focal adhesion kinase (FAK), ERK1/2, JNK, p38, p70S6K, and/or their phosphorylated forms were determined by probing with respective antibodies obtained from Cell Signaling (Beverly, MA). For immunofluorescence, cells plated at 2 × 104 cells/slide were fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X-100. The cells were blocked with 2% BSA and stained with rhodamine-phalloidin (Molecular Probes, Eugene, OR) for actin, DAPI for nucleus and anti-vinculin (BD Biosciences) for focal complex. To assess the polarization of cells, we used a scoring assay based on the position of the microtubule organization center (MTOC) with respect to the wound direction using anti-α-tublin (Sigma) 16 h after wounding. Cells were viewed and photographed using a Leica DM IRB microscope (Deerfield, IL) equipped with an ORCA-ER C4742-95 camera (Hamamatsu, Bridgewater, NJ). Captured images were processed using OpenLab 3.1 software (Improvision, Lexington, MA).
Effector Domain Pulldown Assays
The relative levels of GTP-bound RhoA, Rac1, or Cdc42 were determined by an effector pulldown assay as described previously (Wang et al., 2003). Briefly, the log phase growing WT, knockout, or reconstituted cells were lysed in a buffer containing 1% Triton X-100 and 10 mM MgCl2, and the lysates were probed with glutathione-agarose immobilized GST-Rhotekin (for RhoA) or GST-PAK1 (for Cdc42 or Rac1) effector domain. Bound proteins were analyzed by immunoblotting with anti-RhoA, -Rac1, or -Cdc42 monoclonal antibodies (BD Biosciences).
Cell Proliferation, Cell Cycle, and Apoptosis Assays
Cell proliferation was determined by counting the number of cells in triplicate daily during the cell growth period. To determine the cell cycle profiles, MEFs were serum-starved for 48 h and then stimulated with 10% FBS. At different time points, the cells were subjected to PI/RNase staining followed by FACS analysis. Cell apoptosis was measured by the subG0/G1 percentage of the cell cycle analysis and alternatively, by AnnexinV-cy5 and 7AAD staining followed by FACS.
Cell Migration Assays (Transwell Assay)
Cell migration assays were performed using modified Boyden chambers (8.0-μm pore size, Becton Dickinson, Franklin Lakes, NJ). The lower chamber was filled with 600 μl of DMEM medium containing 10% FBS. Cells were harvested with trypsin/EDTA, resuspended to 2.5 × 105 cells/ml using serum-free DMEM medium and added to the upper chamber. The cells were allowed to migrate in 37°C at 5% CO2 for 16 h. Nonmigratory cells at the upper surface of the membrane were removed and the migrant cells attached to the lower surface were stained with 5% Giemsa solution and quantified. Each assay was performed three times in triplicates.
Wound Healing Motility Assay
MEF cells were plated at 1 × 106/well in six-well tissue culture plates to reach confluence in DMEM containing 10% FBS. Wounds were introduced to the confluent monolayer of cells with a yellow plastic pipette tip to create a cleared line. The medium was removed and replaced with IMDM with 0.5% FBS. The cells were incubated at 37°C, and cell movement into the wound area was photographed at different time points using a Leica DM IRB microscope.
Cell Adhesion Assays
Ninety-six–well plates (Linbro/Titertek, Flow Laboratories, Hamden, CT) were coated with fibronectin (Sigma) at 20 μg/ml concentrations for 2 h at 37°C. The wells were rinsed for two times with PBS and blocked with 100 μl 2% BSA for 30 min at room temperature, followed by three additional rinses with PBS. Cells (100 μl, 3 × 105 cells/ml) were added to each well in DMEM medium containing 10% FBS and were allowed to attach for 2 h. Nonadherent cells were removed by three washes with PBS, and the attached cells were harvested by trypsin-EDTA treatment. Nonspecific adhesion was taken into account by measurement of adherent cells on BSA-coated wells.
NF-κB and SRF Luciferase Reporter Assays
Cells were seeded at 60–70% confluency in 12-well dishes to grow overnight. NF-κB-, or SRF-luciferase and β-galactosidase reporter plasmid (internal control) were transfected using FuGENE 6. After a 4–5-h incubation with the transfection reagent, cells were allowed to recover overnight in DMEM containing 10% FBS. The cells were either serum-starved for another 24 h or cultured in the presence of serum for 24 h before harvesting according to manufacturer's instructions (Promega, Madison, WI). Luciferase levels were measured by using the luciferase reporter gene assay kit (Roche Applied Science, Indianapolis, IN) as described previously (Debidda et al., 2005) and normalized for transfection efficiency by β-galactosidase activity measured according to the manufacturer's instructions (BD Bioscience).
RESULTS
Generation of Cdc42−/− and Cdc42GAP−/− Primary MEFs
To clarify the role of Cdc42 in a primary cell setting, we used the loxP/Cre technology to generate Cdc42 gene deleted primary MEFs derived from the Cdc42flox/flox homozygous mice. The pair of loxP sites flanking exon 2 of cdc42 encoding the first 28 amino acids of the Cdc42 protein can be excised by Cre recombinase, resulting in a Cdc42 knockout allele (Cdc42−; Figure 1A). Effective removal of the Cdc42 exon 2 sequences from cell genomes is verified by a PCR-based protocol that shows the decrease in the predicted genomic DNA size sandwiched by the P1 and P2 PCR primer set from the floxed 762 base pairs to the knockout 176 base pairs (Figure 1B). After such a protocol, the Cdc42flox/flox and Cdc42+/+ MEFs derived from the matching embryos of the Cdc42flox/+ parents were treated by retrovirus expressing Cre recombinase/EGFP to produce the Cdc42+/+ and Cdc42−/− genotypes, and the Cdc42flox/flox cells were transduced with retroviruses expressing WT Cdc42/EGFP and Cre recombinase/YFP to produce the Cdc42 reconstituted Cdc42−/−/Cdc42 cells (Figure 1C). In parallel, the Cdc42GAP gene targeted MEFs (Cdc42GAP−/−) and the matching control cells (Cdc42GAP+/+) were prepared from Cdc42GAP+/− mouse breeding as described previously to provide a Cdc42 gain-of-activity cell model (Wang et al., 2005). Before functional assays, the protein levels of endogenous Cdc42 or exogenous (HA)3-Cdc42 in the low passage (passages 3–5) MEF cells isolated by FACS were analyzed by anti-Cdc42 Western blotting. As shown in Figure 1D, Cdc42+/+ and Cdc42flox/flox cells expressed levels of endogenous Cdc42 comparable to that of Cdc42GAP+/+ and Cdc42GAP−/− cells, whereas Cdc42−/− and Cdc42−/−+Cdc42 cells showed no detectable endogenous Cdc42 and Cdc42−/− +Cdc42 cells displayed a (HA)3-Cdc42 level comparable to that of the endogenous Cdc42 in Cdc42+/+ cells (Figure 1D). The Cdc42GAP−/− cells were defective in Cdc42GAP protein expression (Figure 1D). These Cdc42- or Cdc42GAP-gene–targeted primary MEFs at low passages (passages 3–5), together with their controls, constitute the cell systems we used for the present studies.
Cdc42 Cross-Talk with Rac1 and RhoA GTPases
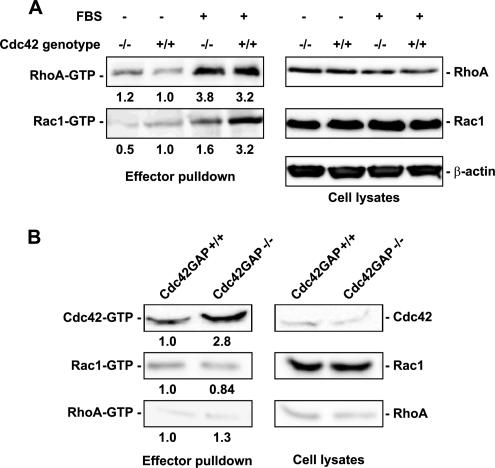
Cdc42 was thought to act upstream of Rac1 and RhoA in the course of growth factor–regulated actin reorganization in Swiss 3T3 fibroblasts (Nobes and Hall, 1995). To examine if Rac1 and RhoA activities were affected by Cdc42 loss or gain of activity in MEFs, we used the effector domain–binding assay to compare the relative Rac1-GTP and RhoA-GTP species in the Cdc42−/− and Cdc42GAP−/− cells. As shown in Figure 2A, loss of Cdc42 did not change RhoA expression level or activity but caused an approximately twofold reduction of the level of activated Rac1. In contrast, no activity or expression changes of Rac1 or RhoA were detected in the Cdc42GAP−/− cells in which Cdc42-GTP was significantly elevated compared with that of Cdc42GAP+/+ cells (Figure 2B). These results suggest that loss of Cdc42 is accompanied with a decrease in Rac1 activity without affecting the RhoA activation status, but a gain of Cdc42 activity is not sufficient to affect Rac1 or RhoA activity.
Figure 2.
Loss or gain of Cdc42 activity and effects on Rac1 and RhoA activities in Cdc42 (A) or Cdc42GAP (B) knockout cells. The expression and the relative level of GTP-bound forms of Cdc42, Rac1, and RhoA in various primary MEFs were probed by the effector-domain (GST-PAK1 for Cdc42-GTP and Rac1-GTP; GST-Rhotekin for RhoA-GTP) pulldown assays followed by anti-Cdc42, -Rac1, or -RhoA Western blotting. The lysate inputs are shown on the right-hand side and the corresponding quantifications are indicated below each pulldown blot. The results are representative of three independent measurements.
Altered Morphology and Impaired Actin Responses in Cdc42 Loss or Gain of Activity MEFs
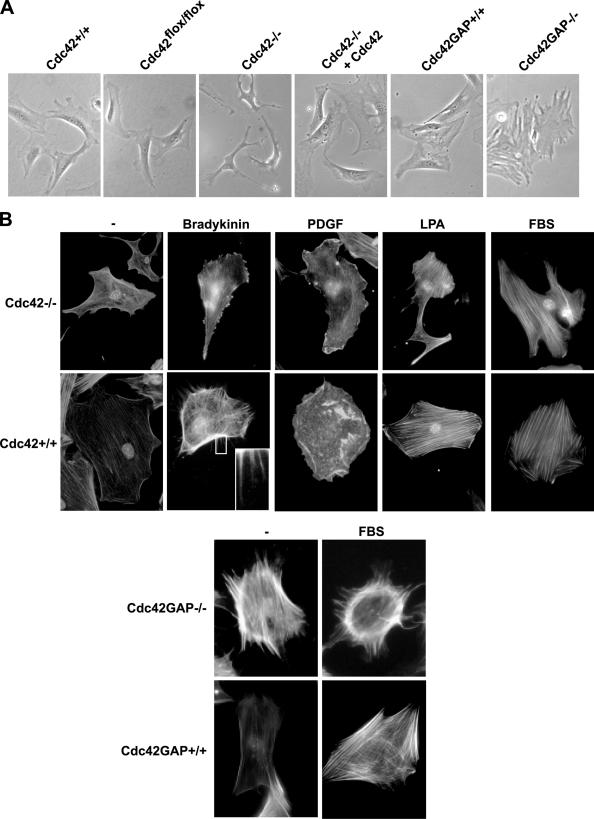
Although the Cdc42+/+ or parental Cdc42flox/flox MEFs showed a well-extended morphology, Cdc42−/− MEFs displayed a variety of irregular shapes including multiple extended protrusions, and contracted cell bodies under normal culture conditions. Cdc42GAP−/− cells, on the other hand, showed extended cell bodies with multiple “arms” (Figure 3A). Staining of the cells under serum-starved or ligand-stimulated conditions with fluorescent dye–conjugated phalloidin revealed that no filopodia was detectable in Cdc42−/− cells when the cells were challenged with bradykinin, whereas Cdc42GAP−/− cells displayed spontaneous filopodia (Figure 3B). Moreover, Cdc42−/− cells remained responsive to PDGF to form membrane ruffles at the cell periphery but had lost dorsal ruffles that were readily detected in WT MEFs upon PDGF stimulation (Figure 3B). Cdc42-deficient cells showed no apparent defects in actin stress fiber formation upon LPA stimulation compared with WT cells (Figure 3B). Reconstitution of constitutively active Rac1 mutant, L61Rac1, into the Cdc42−/− cell was not sufficient to reverse the morphological and actin organizational defects (Supplementary Figure S1). Thus, Cdc42 activity appears to be necessary and sufficient for filopodia induction and is involved in PDGF-mediated dorsal ruffling.
Figure 3.
Morphologies and actin cytoskeleton structures of Cdc42- or Cdc42GAP-deficient primary MEFs in response to stimuli. (A) The morphologies of Cdc42−/−, Cdc42GAP−/−, and control cells under normal tissue culture condition were examined by phase-contrast microscopy. (B) Various MEFs were grown on cover slides and stained with TRITC-phalloidin to reveal the F-actin structures. The Cdc42−/− and Cdc42+/+ cells were starved 16 h before stimulation with bradykinin (100 ng/ml), PDGF (10 ng/ml), LPA (20 ng/ml), or 10% FBS for 10 min. The Cdc42GAP−/− and matching Cdc42GAP+/+ cells were examined with or without 10% FBS stimulation.
Effects of Cdc42 Loss or Gain of Activity on Cell Adhesion and Migration
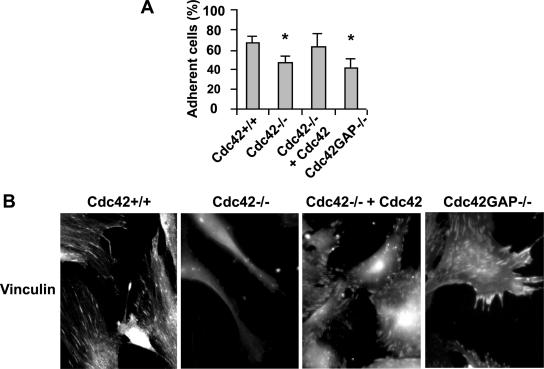
Cdc42-mediated actin reorganization may affect cell spreading, adhesion, and/or migration. We next examined the adhesion and migration properties of the Cdc42−/− and Cdc42GAP−/− MEFs. Comparisons of cell spreading at 0.5-, 1-, 3-, and 5-h time points after plating on the plastic surface of tissue culture dishes revealed distinct kinetics of WT, Cdc42−/−, and Cdc42GAP−/− cells (Supplementary Figure S2), in contrast with previously reported Cdc42−/− clonal fibroblastoid cells (Czuchra et al., 2005). The Cdc42−/− cells started to spread at 0.5 h and significantly higher percentage of the cells appeared well spread 1 h after plating (WT:Cdc42−/− = 17 ± 3.5%:37 ± 5.5%). At the 5-h time point, the attached Cdc42−/− cells showed a morphology characterized by retracted cell body and extension of branched and bead-like protrusions. Cdc42GAP−/− cells, on the other hand, showed branched and smaller feet-like protrusions at 5-h time point. Cdc42 reconstituted Cdc42−/− cells showed similar morphologies and spreading kinetics as WT cells, displaying well-spread edges and flat extensions (Supplementary Figure S2). Adhesion of both Cdc42- and Cdc42GAP-deficient cells to fibronectin-coated surface was significantly reduced (Figure 4A), and this is corroborated by further analysis of focal complexes with anti-vinculin immunofluorescence (Figure 4B). While the number of detectable adhesion complexes was significantly reduced in the Cdc42−/− cells, the size of the complexes appeared enlarged and the number reduced in the Cdc42GAP−/− cells (Figure 4B). These results suggest that both loss and gain of Cdc42 activity could impact on cell adhesion, but they may undergo different mechanisms.
Figure 4.
Effect of Cdc42 loss or gain of activity on adhesion to fibronectin and focal complex assembly. The percentage of cells adhered to the fibronectin matrix after a 2-h incubation (A), and the focal adhesion complex visualized by anti-vinculin immunostaining (TRITC; B) are shown. *p < 0.05 between the Cdc42−/− or Cdc42GAP−/− and respective control WT cells.
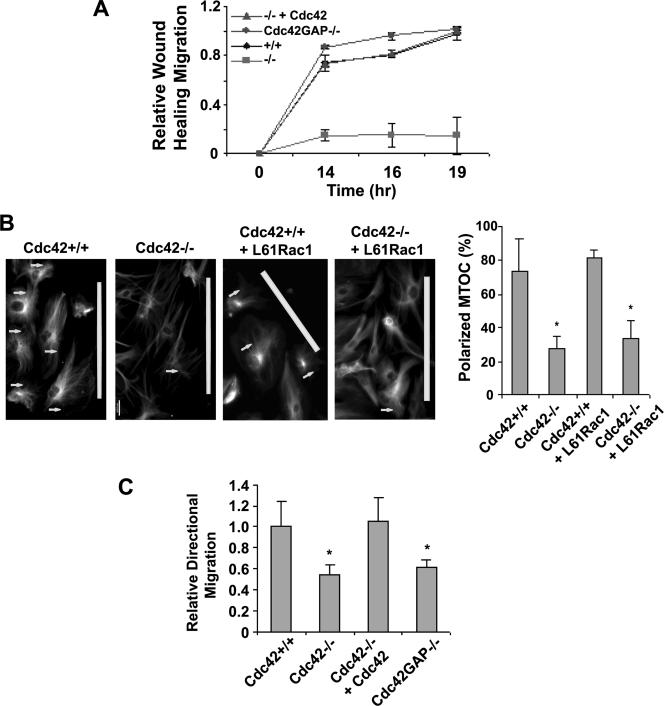
To assess the effect of Cdc42 loss or gain of activity on cell migration, we carried out wound healing assays to determine if Cdc42 activity regulates the motility of cells after a wound is introduced and transwell migration assays to examine if Cdc42 is required for directional migration toward a serum gradient. Figure 5A shows that within 14 h of wound introduction, Cdc42+/+ cells had migrated into the cleared section of cell confluent monolayer, whereas few Cdc42−/− cells were able to move into the wound opening (Figure 5A and Figure S3). Cdc42 reconstituted Cdc42−/− cells and Cdc42GAP−/− cells showed similar ability to migrate toward the wound opening as WT cells. During the course of the migration, WT as well as the Cdc42−/−+Cdc42 cells displayed lamellipodia and actin microspikes at the leading edge, but no such structures were detected in the Cdc42−/− cells (unpublished data). Immunostaining of α-tubulin of the leading edge migrating cells revealed that unlike the well-ordered orientation of the MTOC in WT cells (i.e., toward the wound induction site), the orientation of MTOC in Cdc42−/− cells was mostly disordered, and this effect could not be rescued by L61Rac1 mutant expression (Figure 5B). In serum-induced transwell assays, the number of Cdc42−/− or Cdc42GAP−/− cells migrated across the transwell pores after a 16-h incubation was approximately half of that of the WT or Cdc42 reconstituted Cdc42−/− cells (Figure 5C). These results indicate that Cdc42 activity is required for cell movement, polarity, and directed migration, but constitutively elevated Cdc42 activity results in a loss of directionality. Because the L61Rac1 mutant failed to rescue the adhesion or migration defect of the Cdc42−/− cells (Supplementary Figure S1), we conclude that the reduced Rac1 activity in Cdc42−/− cells does not contribute to these cellular phenotypes.
Figure 5.
Effect of loss or gain of Cdc42 activity on cell migration. (A) Wound-healing assays were carried out on a monolayer of confluent cells to determine cell movement toward the wound gap. The average migration distances to the center of the wound were quantified at the indicated times after the wound was introduced. (B) Immunostaining of α-tubulin revealed the MTOC orientation (arrows) toward the wound induction site (marked with white bars) 16 h after the wounds were introduced. The number of cells containing a wound-directed MTOC orientation was quantified under a microscope. (C) Transwell cell migration toward 10% FBS was measured to determine cell directional movement. The number of cells migrated across the membrane toward the serum gradient into the lower chamber of the Transwells was quantified 16 h after plating. The results are representative of three independent measurements. *p < 0.05 between the Cdc42−/−, Cdc42GAP−/−, or Cdc42−/−+L61Rac1 and the respective control cells.
Defective Signaling Related to Actin Structure and Migration in the Absence of Cdc42
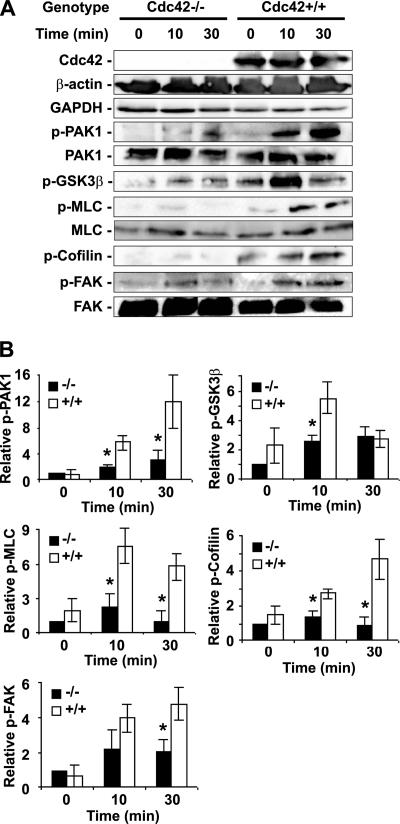
The signaling components involved in regulating cell actin organization and migration include PAK1, GSK3β, MLC, cofilin, and FAK. We examined possible activity changes of these molecules in response to serum stimulation affected by Cdc42 deletion by phosphorylation-specific Western blotting of various MEF lysates. Cdc42-deficient cells showed little change in phospho-PAK1 content after serum stimulation, whereas Cdc42+/+ cells displayed significant increase in phospho-PAK1 at the 10- or 30-min time point (Figure 6). Phospho-MLC and -cofilin showed a similar pattern of lack of response to serum in the Cdc42−/− cells. GSK3β, a suggested target of the Par6/PKCζ complex (Etienne-Manneville and Hall, 2001, 2003; Tzima et al., 2003), also displayed significantly dampened phosphorylation response at the 10-min time point (Figure 6). These signaling molecules also showed a lower level of phosphorylation at basal state before serum stimulation. Interestingly, phospho-FAK level in the Cdc42−/− cells was detectably altered 30 min after serum stimulation, suggesting that FAK regulation by Cdc42 may be involved in cell adhesion maintenance. Taken together, these data indicate that loss of Cdc42 activity leads to multiple downstream signaling defects induced by serum that could contribute to the observed actin and migration phenotypes.
Figure 6.
Cdc42 activity regulates signaling responses to serum stimulation relevant to cell actin organization and migration. (A) Western blotting of p-GSK3β, p-PAK1, p-MLC, p-Cofilin, p-FAK, and relevant controls in the Cdc42−/− and Cdc42+/+ MEFs after 0, 10, or 30 min of 10% FBS stimulation after a 16-h serum withdrawal. (B) The relative Western blot levels were quantified by using a densitometer (mean ± SD from three independent experiments). All data were normalized to those of Cdc42−/− cells at time 0. *p < 0.05 between the Cdc42−/− and Cdc42+/+ cells.
Effect of Cdc42 Deficiency on Cell Proliferation
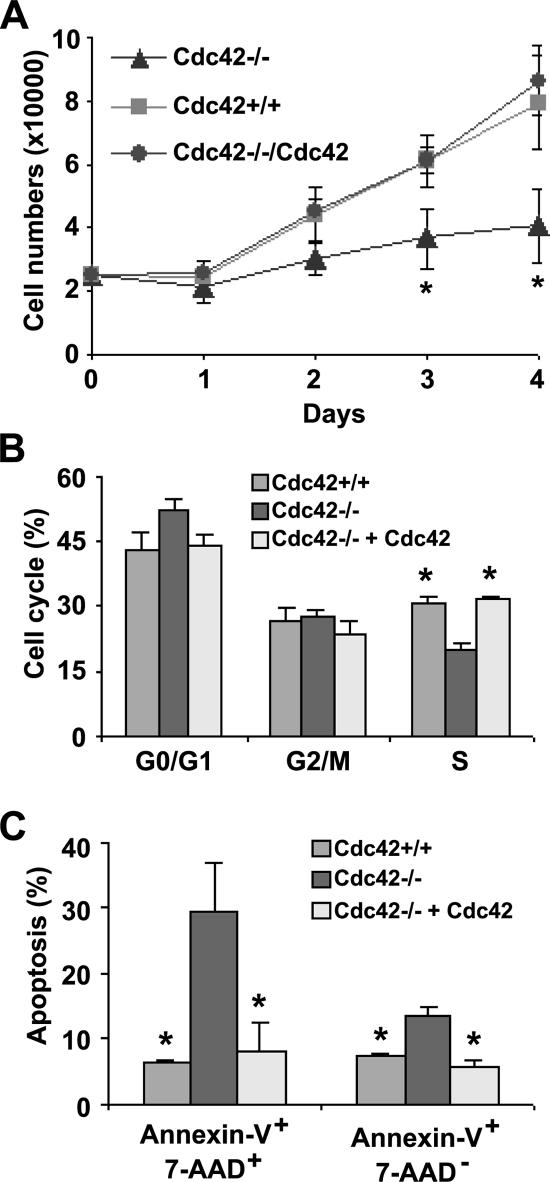
Constitutively active or fast cycling Cdc42 mutant overexpression can lead to uncontrolled cell growth in NIH 3T3 cells (Lin et al., 1997, 1999), prompting us to examine the effect of Cdc42 loss or gain of activity on primary MEF proliferation. Growth of Cdc42-deficient cells showed drastic defects compared with Cdc42+/+ or Cdc42−/−+Cdc42 cells under normal cell culture conditions (Figure 7A). To a lesser extent, Cdc42GAP−/− MEFs were also found to be defective in growth (Wang et al., 2005). The slowed growth of Cdc42−/− cells was associated with cell cycle S-phase transition defect as revealed by PI-staining and FACS analysis (Figure 7B). In addition, Cdc42−/− cells showed a significant increase in Annexin-V+ population after serum starvation for 48 h (Figure 7C). Interestingly, Cdc42GAP-deficient MEFs showed only a survival defect without detectable cell cycle alteration (Wang et al., 2005). These results indicate that Cdc42 loss or gain of activity is disadvantageous to primary MEF growth and that Cdc42 is required for normal G1/S-phase transition and cell survival.
Figure 7.
Cdc42 activity is critical for primary MEF proliferation, cell cycle progression, and cell survival. (A) Cell proliferation potential of the respective MEFs under regular serum conditions was determined. MEF cells were plated in 12-well plates in triplicates in the presence of 10% FBS, and at the varying time points the cell numbers were quantified by Trypan blue exclusion analysis. (B) Various MEFs were labeled with PI/RNase, and the cell populations at various cell cycle phases were analyzed by FACS after 48-h serum starvation followed by a 24-h 10% FBS incubation. (C) The MEFs were labeled with Annexin-V/7AAD and subjected to FACS analysis to determine the percent of cell populations undergo apoptosis after 16-h serum starvation followed by 6-h stimulation by 10% FBS/DMEM. *p < 0.05 between the Cdc42−/− and Cdc42+/+ cells.
Effects of Cdc42 Loss or Gain of Activity on NF-κB and SRF Transcription
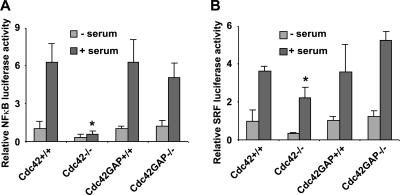
It has been shown that Cdc42, among other Rho GTPases, is a critical regulator of gene transcription of several cell growth regulatory molecules including NF-κB and SRF in clonal cell lines (Hill et al., 1995; Perona et al., 1997; Montaner et al., 1998, 1999). Activation of the NF-κB reporter gene under serum stimulation was abolished in the Cdc42−/− cells but appeared normal in the Cdc42GAP−/− cells (Figure 8A), suggesting that Cdc42 activity is necessary but not sufficient for serum-induced transcriptional activation of NF-κB. However, the SRF reporter activities in the Cdc42−/− and Cdc42GAP−/− cells were comparable with that of WT controls (Figure 8B), indicating that Cdc42 is neither required nor sufficient for SRF activation. These data suggest a distinct role of Cdc42 activity in the transcription regulation of NF-κB and SRF in primary MEFs from previously suggested functions (Hill et al., 1995; Perona et al., 1997; Montaner et al., 1998, 1999).
Figure 8.
Effects of Cdc42 loss or gain of activity on NF-κB and SRF transcription activities. NF-κB (A) or SRF (B) transcriptional reporter plasmids were cotransfected into the respective MEF cells with a β-gal expressing reporter. After a 4–5-h incubation, cells were allowed to recover overnight in normal culture media. After 24 h with or without starvation, the luciferase activities were assayed of the cells. The luciferase reporter activities were normalized to those of the cotransfected β-gal activities and are expressed as fold of changes compared with those of WT control cells. *p < 0.05 between the Cdc42−/− and Cdc42+/+ cells under similar treatment.
Defective MAPK and p70S6K Signaling Activities in the Cdc42 Loss or Gain of Activity MEFs
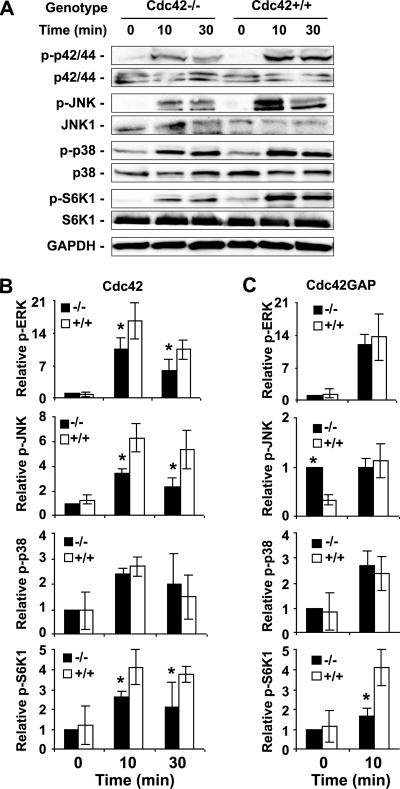
Multiple signaling pathways downstream of Cdc42 may mediate its effect on cell proliferation and transcription. To begin to examine the signaling pathways that are regulated by Cdc42 activity in the MEFs, we probed the phosphorylation states of a few previously implicated Cdc42 effector kinases by Western blotting. We found that ERK1/2 response to serum was dampened in the Cdc42−/− MEF but was unaffected in the Cdc42GAP−/− cells (Figure 9, A and B), suggesting that Cdc42 activity is necessary but not sufficient for ERK activation. Cdc42 deletion led to defective JNK activation after serum stimulation, whereas in Cdc42GAP−/− cells the basal phospho-JNK was significantly elevated but the response to serum challenge was not affected (Figure 9), consistent with the notion that Cdc42 activity is necessary and sufficient for JNK activation. However, there was no detectable alteration in phsopho-p38 status in either gain or loss of Cdc42 activity cells compared with WT controls (Figure 9), indicating that the p38 MAPK pathway is Cdc42-independent in primary MEFs. In addition to MAP kinase modules, p70S6K has been implicated to be an effector for Cdc42 which could mediate Cdc42-regulated cell cycle G1 progression (Chou et al., 2003). Both the Cdc42-deficent and Cdc42GAP-deficient MEFs showed similar basal p70S6K phosphorylation patterns as the WT cells but yielded reduced response to serum stimulation (Figure 9). Taken together, these results strongly indicate that Cdc42 is involved in the regulation of ERK1/2, JNK, and p70S6K activities, but a balanced Cdc42 activity is important for proper signaling responses.
Figure 9.
Altered MAPK and p70S6K signaling activities in Cdc42−/− and Cdc42GAP−/− MEFs in response to serum stimulation. (A) Western blotting of p-ERK1/2, p-p38, p-JNK1/2 and p-S6K, and relevant controls as performed using MEF cell lysates after stimulation of the cells with 10% FBS for the indicated times after a 16-h serum withdrawal. (B and C) The relative Western blot levels were quantified by using a densitometer (mean ± SD from three independent experiments). *p < 0.05 between the knockout and WT cells.
DISCUSSION
Although the cellular functions of mammalian Cdc42 has been well investigated by the dominate negative or constitutively active mutant expression approach in a variety of clonal cell lines, it has become clear that experimental limitations of the conventional methods could lead to erroneous interpretations. Recent studies of Cdc42 function using genetic knockout approach in ES and ES-derived fibroblastoid clones (Chen et al., 2000; Czuchra et al., 2005) raise concerns about the physiological significance of some of previous findings. Our present work demonstrating that Cdc42 is critically involved in actin filopodia formation, cell motility, directional migration, and cell growth and is required for the serum regulation of PAK1, GSK3β, MLC, ERK1/2, JNK, and NF-κB pathways in primary MEFs help affirm that Cdc42 indeed can play these roles in primary cells. The comparative studies of Cdc42 loss-of-activity and gain-of-activity primary MEFs further suggest that a balanced Cdc42 activity is important for proper signaling output and cell regulation.
In Swiss 3T3 fibroblast cells, it was proposed that a hierarchy of signaling cascade from Cdc42 to Rac1 to RhoA may be at work in mediating cell actin reorganization (Nobes and Hall, 1995). Such cross-talk among the Rho GTPases is likely cell type and clonal dependent. In primary MEFs, Cdc42-deficiency leads to reduced Rac1 activity but normal RhoA activity under both serum-free and serum-stimulation conditions, whereas Cdc42GAP knockout cells display constitutively elevated Cdc42 activity without detectable alteration of Rac1 or RhoA activity (Figure 2), suggesting that Cdc42 activity is necessary but not sufficient for Rac1 activation. Because most phenotypes of Cdc42−/− cells cannot be rescued by expression of an active Rac1 mutant, Cdc42 appears to regulate cell behaviors including actin organization, adhesion, and directed migration in a Rac1 activity–independent manner. However, it remains possible that certain phenotypes we observed in the Cdc42−/− cells are associated with other related Rho GTPase activities that are regulated by Cdc42.
Cdc42 regulation of actin polymerization and filopodia formation has been shown in a wide variety of mammalian cell types as well as in yeast, flies, and worms. Studies carried out in Cdc42-deficient ES cells provided supporting evidence of its essential role in actin polymerization and actin microfilament formation. However, recent studies in clonal fibroblastoid cells generated by differentiation and immortalization of ES cells showed normal formation of filopodia and lamellipodia in the absence of Cdc42, raising the possibility that Cdc42-related Rho GTPases such as Wrch2 or TC10 could be playing a redundant role. In our primary cell setting, MEFs did not form filopodia after bradykinin stimulation or at the leading edge after wound damage upon Cdc42 deletion, whereas spontaneous filopodia were abundant in Cdc42GAP−/− cells (Figure 3), suggesting that Cdc42 activity is necessary and sufficient for filopodia induction. One unexpected observation of the Cdc42−/− cells is that no dorsal lamellipodia was formed in the absence of Cdc42 upon PDGF stimulation when peripheral lamellipodia was evident. Takenawa's group has shown that WAVE1 and WAVE2 play differential roles in dorsal and peripheral lamellipodia formation, with WAVE1 being essential for dorsal ruffling and WAVE2 key for peripheral ruffle formation (Suetsugu et al., 2003). In this context, our data suggest that Cdc42 contributes to the regulation of WAVE1 or a related pathway in dorsal lamellipodia induction.
Cdc42 was found to regulate adhesion complex formation and cell migration (Nobes and Hall, 1999). In macrophages inhibition of Cdc42 blocks chemotaxis toward a CSF gradient without affecting cell mobility (Ridley, 2001). In Drosophila Cdc42 loss of function does not affect the migration of peripheral glial cells (Sepp and Auld, 2003). In our studies, Cdc42 deletion in primary MEFs causes abnormal cell spreading, reduced adhesion to fibronectin, defective mobility in wound healing, and decreased chemotaxis toward a serum gradient (Figures 4 and 5). Some of these effects may be related to impaired formation of filopodia and/or defective polarity of the cells. In the Cdc42 gain-of-activity Cdc42GAP−/− MEFs, adhesion and directional migration were inhibited but motility in wound healing was not (Figure 5), consistent with the notion that Cdc42 activity is involved in directional movement. We further found that the serum-regulated signaling components including PAK1, GSK3β, MLC, cofilin, and FAK were affected by Cdc42 deletion (Figure 6), supporting previous findings that the Cdc42-PAK1-MLC/Cdc42-PAK1-cofilin pathways are important for actin reorganization, the Cdc42-GSK3β signaling axis is important for cell polarized migration, and the Cdc42 may regulate FAK activity in modulating adhesion (Etienne-Manneville and Hall, 2002; Ridley, 2001).
In Cdc42-deficient ES or ES-derived fibroblastoid cells, cell growth proceeds normally without mitotic defects (Chen et al., 2000; Czuchra et al., 2005). In primary MEFs, however, Cdc42 activity is critical for cell proliferation, as loss or gain of activity of Cdc42 affects cell cycle progression and/or survival (Figure 7; Wang et al., 2005). The cell growth defects of the Cdc42−/− cells correlate with defects in ERK1/2, JNK, and/or p70S6K activity, and with a defect in transcriptional activation of NF-κB (Figures 8 and 9). Although our results are mostly consistent with previous reports in the literature where Cdc42 is known to be essential in eukaryotic cell growth and can regulate apoptosis and G1/S-phase transition under dominant mutant overexpression conditions, they provide the first genetic evidence that Cdc42 plays an important cell growth regulatory role in a mammalian primary cell setting.
Direct comparison of the proliferation properties of the Cdc42 loss- and gain-of-activity cell models leads to a few unexpected observations of Cdc42 signaling and function. First, both loss and gain of Cdc42 activities in MEFs cause cell growth inhibition. Loss of Cdc42 affects both the cell cycle G1/S-phase transition and cell survival, whereas gain of Cdc42 activity increases spontaneous apoptosis only (Wang et al., 2005). Second, loss of Cdc42 dampens serum-induced PAK1 and JNK activities, whereas gain of Cdc42 activity promotes PAK1 and JNK activation. The PAK1-JNK axis appears to be the only pathway examined that show a “linear” tendency of regulation by Cdc42 activity in the two knockout cell models. Other related pathways such as ERK1/2, p38, or p70S6K show either no effect or disparate effect by Cdc42 or Cdc42GAP deletion (e.g., both Cdc42 and Cdc42GAP knockout cells show a decrease in p70S6K response, whereas Cdc42−/− cells are deficient in ERK1/2 response but Cdc42GAP−/− cells appear normal in ERK1/2 activity). Third, although Cdc42 activity is necessary but not sufficient for serum-induced NF-κB transcription, it is neither necessary nor sufficient for SRF transcription activation. It remains to be seen if most of these altered signaling pathways are involved in the growth phenotypes of the Cdc42 and Cdc42GAP knockout cells, because the elevated JNK activity in Cdc42GAP−/− MEFs is responsible for increased cell apoptosis. Overall, our parallel examination of Cdc42−/− and Cdc42GAP−/− MEF cells suggests that a balanced, or tightly regulated, Cdc42 activity is essential for proper signaling effects.
Given the multiple effector pathways implicated in Cdc42 signaling to the actin cytoskeleton and nucleus, including the PAK1 regulated MLC and MEK1 and the Par3/Par6/aPKC complex-mediated GSK3β, it was somewhat surprising that Cdc42-deficient ES cells exhibited normal phosphorylation patterns of mitogen- and stress-activated protein kinases and other related signaling kinases, including GSK3β, ERK1/2, JNK, p38, or AKT, and that Cdc42−/− fibroblastoid cells also appeared normal in these signaling responses. In this context, our primary Cdc42 loss- or gain-of-activity MEFs display mostly similar pattern of signaling alterations as expected in the literature. One explanation of the discrepancies between our observations and the ES or ES-derived fibroblastoid studies may be the cell-type differences. Because Cdc42 conventional knockout mice die at the gastrulation stage, it is possible that Cdc42 is dispensable during early embryonic development and ES cell growth. Recent gene targeting studies implicating Cdc42 in keratinocyte stem cell differentiation (Wu et al., 2006) and in hematopoietic stem cell quiescence maintenance (Yang et al., 2005) further highlight a cell lineage–specific role of Cdc42. Another explanation could come from differences in primary and clonal cell genetic backgrounds. Cdc42−/−;p53−/− MEFs are spontaneously immortalized and can grow similarly like WT MEFs (our unpublished observation), suggesting that Cdc42 is not essential for cell proliferation once immortalized, as is the case in the ES-derived fibroblastoid cell lines. These explanations may also apply to the differences between our results and previous observations in actin organization and cell migration (Czuchra et al., 2005). Because Cdc42 is ubiquitously expressed and is capable of mediating signal transduction in multiple pathways, a major challenge in future studies is to define its cell-type– and stimulus-specific signaling mechanism and function in diverse cell settings under physiological conditions.
Supplementary Material
[Supplemental Material]
ACKNOWLEDGMENTS
We thank James F. Johnson and Clara Blair for expert technical assistance. This work was supported by National Institutes of Health Grants CA105117 and HL085362.
Abbreviations used:
BSA
bovine serum albumin
EGFP
enhanced green fluorescence protein
ES
embryonic stem
FACS
fluorescence-activated cell sorting
FBS
fetal bovine serum
GAP
GTPase-activating protein
MEF
mouse embryonic fibroblast
MTOC
microtubule organization center.
Footnotes
REFERENCES
- Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- Chen F., et al. Cdc42 is required for PIP(2)-induced actin polymerization and early development but not for cell viability. Curr. Biol. 2000;10:758–765. doi: 10.1016/s0960-9822(00)00571-6. [DOI] [PubMed] [Google Scholar]
- Chou M. M., Masuda-Robens J. M., Gupta M. L. Cdc42 promotes G1 progression through p70 S6 kinase-mediated induction of cyclin E expression. J. Biol. Chem. 2003;278:35241–35247. doi: 10.1074/jbc.M305246200. [DOI] [PubMed] [Google Scholar]
- Czuchra A., Wu X., Meyer H., van Hengel J., Schroeder T., Geffers R., Rottner K., Brakebusch C. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol. Biol. Cell. 2005;16:4473–4484. doi: 10.1091/mbc.E05-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debidda M., Wang L., Zang H., Poli V., Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J. Biol. Chem. 2005;280:17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Feig L. A. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M. C., Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- Guo F., Zheng Y. Rho family GTPases cooperate with p53 deletion to promote primary mouse embryonic fibroblast cell invasion. Oncogene. 2004;23:5577–5585. doi: 10.1038/sj.onc.1207752. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Wynne J., Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Johnson D. I., Pringle J. R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay A. J., Hunter C. P. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 2001;11:474–481. doi: 10.1016/s0960-9822(01)00141-5. [DOI] [PubMed] [Google Scholar]
- Kjoller L., Hall A. Signaling to Rho GTPases. Exp. Cell Res. 1999;253:166–179. doi: 10.1006/excr.1999.4674. [DOI] [PubMed] [Google Scholar]
- Lin R., Bagrodia S., Cerione R., Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- Lin R., Cerione R. A., Manor D. Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J. Biol. Chem. 1999;274:23633–23641. doi: 10.1074/jbc.274.33.23633. [DOI] [PubMed] [Google Scholar]
- Montaner S., Perona R., Saniger L., Lacal J. C. Multiple signalling pathways lead to the activation of the nuclear factor kappaB by the Rho family of GTPases. J. Biol. Chem. 1998;273:12779–12785. doi: 10.1074/jbc.273.21.12779. [DOI] [PubMed] [Google Scholar]
- Montaner S., Perona R., Saniger L., Lacal J. C. Activation of serum response factor by RhoA is mediated by the nuclear factor-kappaB and C/EBP transcription factors. J. Biol. Chem. 1999;274:8506–8515. doi: 10.1074/jbc.274.13.8506. [DOI] [PubMed] [Google Scholar]
- Nobes C. D., Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Nobes C. D., Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona R., Montaner S., Saniger L., Sanchez-Perez I., Bravo R., Lacal J. C. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- Ridley A. J. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–477. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- Sepp K. J., Auld V. J. RhoA and Rac1 GTPases mediate the dynamic rearrangement of actin in peripheral glia. Development. 2003;130:1825–1835. doi: 10.1242/dev.00413. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Yamazaki D., Kurisu S., Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Tzima E., Kiosses W. B., del Pozo M. A., Schwartz M. A. Localized cdc42 activation, detected using a novel assay, mediates microtubule organizing center positioning in endothelial cells in response to fluid shear stress. J. Biol. Chem. 2003;278:31020–31023. doi: 10.1074/jbc.M301179200. [DOI] [PubMed] [Google Scholar]
- Van Aelst L., D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Wang L., Yang L., Burns K., Kuan C. Y., Zheng Y. Cdc42GAP regulates c-Jun N-terminal kinase (JNK)-mediated apoptosis and cell number during mammalian perinatal growth. Proc. Natl. Acad. Sci. USA. 2005;102:13484–13489. doi: 10.1073/pnas.0504420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang L., Filippi M. D., Williams D. A., Zheng Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood. 2006;107:98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang L., Luo Y., Zheng Y. A novel strategy for specifically down-regulating individual Rho GTPase activity in tumor cells. J. Biol. Chem. 2003;278:44617–44625. doi: 10.1074/jbc.M308929200. [DOI] [PubMed] [Google Scholar]
- Wu X., Quondamatteo F., Lefever T., Czuchra A., Meyer H., Chrostek A., Paus R., Langbein L., Brakebusch C. Cdc42 controls progenitor cell differentiation and b-catenin turnover in skin. Genes Dev. 2006;20:571–585. doi: 10.1101/gad.361406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang L., Cancelas J. A., Williams D. A., Zheng Y. A critical role of the Rho family GTPase Cdc42 in hematopoietic stem cell mobilization, homing, engraftment and differentiation. Blood. 2005;106:269a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]